Abstract
Arenaviruses comprise a diverse family of rodent-borne viruses that are responsible for recurring and emerging outbreaks of viral hemorrhagic fevers worldwide. The Junín virus, a member of the New World arenaviruses, is endemic to the pampas grasslands of Argentina and is the etiologic agent of Argentine hemorrhagic fever. In this study, we have analyzed the assembly and function of the Junín virus envelope glycoproteins. The mature envelope glycoprotein complex is proteolytically processed from the GP-C precursor polypeptide and consists of three noncovalently associated subunits, G1, G2, and a stable 58-amino-acid signal peptide. This tripartite organization is found both on virions of the attenuated Candid 1 strain and in cells expressing the pathogenic MC2 strain GP-C gene. Replacement of the Junín virus GP-C signal peptide with that of human CD4 has little effect on glycoprotein assembly while abolishing the ability of the G1-G2 complex to mediate pH-dependent cell-cell fusion. In addition, we demonstrate that the Junín virus GP-C signal peptide subunit is myristoylated at its N-terminal glycine. Alanine substitution for the modified glycine residue in the GP-C signal peptide does not affect formation of the tripartite envelope glycoprotein complex but markedly reduces its membrane fusion activity. In contrast to the classical view that signal peptides act primarily in targeting nascent polypeptides to the endoplasmic reticulum, we suggest that the signal peptide of the arenavirus GP-C may serve additional functions in envelope glycoprotein structure and trafficking.
Arenaviruses are the etiological agents of acute hemorrhagic fevers with high mortality in humans. Old World arenaviruses, such as Lassa virus and lymphocytic choriomeningitis virus (LCMV), and New World arenaviruses, such as Junín and Machupo viruses, are endemic to rodent populations and are transmitted to humans through casual or aerosol contact (11, 19). Recurring and emerging outbreaks throughout the world represent an ongoing public health problem. The potential for use of these agents in biological warfare has elevated this family of viruses to national concern. Here, we study the Junín virus, a member of the New World Tacaribe complex of arenaviruses that is responsible for recurring outbreaks of Argentine hemorrhagic fever (AHF) in the pampas grasslands of Argentina.
The arenaviruses are enveloped, bisegmented RNA viruses (11, 19). The genome consists of two single-stranded RNA molecules, designated L (∼7.2 kb) and S (∼3.4 kb). The S segment encodes the major structural components of the virion: the internal nucleocapsid protein (N) and the external envelope glycoprotein precursor (GP-C). The L segment encodes the RNA-dependent RNA polymerase (L) and a zinc-binding matrix protein (Z). The genomic RNAs are used to produce genomic-complementary and genomic-sense RNAs that serve as ambisense templates for mRNA synthesis (51). Arenaviral particles assemble at the plasma membrane and bud to produce infectious virions. Specific pathways and determinants of virion assembly and budding remain poorly understood.
The viral envelope glycoprotein is an essential component in the assembly and release of infectious virions and is responsible for virus entry into target cells. The arenavirus GP-C envelope glycoprotein precursor contains a signal peptide (SP) sequence that targets the nascent polypeptide to the rough endoplasmic reticulum (ER) where high-mannose glycan cores are added at potential glycosylation sites (10, 27, 31). During transit to the cell surface, the GP-C precursor is proteolytically cleaved by the cellular subtilase SKI-1/S1P (4, 45) to yield the mature glycoproteins G1 and G2. The G1-G2 complex extends 5 to 10 nm on the virion particle and is anchored to the viral membrane by the C-terminal transmembrane domain of G2 (10) (Fig. 1).
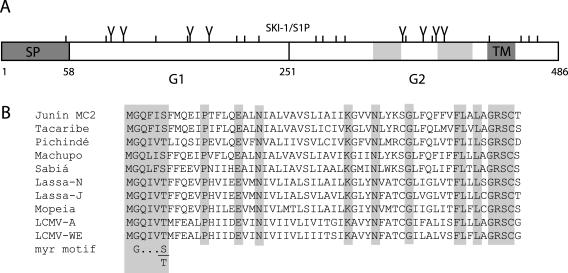
FIG. 1.
Schematic representation of the Junín virus GP-C glycoprotein and SP. (A) The Junín virus MC2 GP-C glycoprotein is depicted. Amino acids are numbered from the initiating methionine, and cysteine residues (|) and potential glycosylation sites (Y) are marked. The SP cleavage site (after amino acid 58) and the SKI-1/S1P cleavage site (after amino acid 251) and the resulting SP, G1 and G2 subunits are indicated. Within G2, the C-terminal transmembrane domain (TM) is shown, as are heptad-repeat regions (light gray shading) that may be involved in membrane fusion. The N termini of the arenavirus G1 and G2 glycoproteins have been previously determined experimentally (10, 12, 27, 45). (B) A comparison of the signal peptides of New World (Junín MC2 [accession no. D10072], Tacaribe [accession no. M20304], Pichindé [accession no. M16735], Machupo [accession no. AY129248], and Sabiá [accession no. U41071]) and Old World (Lassa-Nigeria [Lassa-N, accession no. X52400], Lassa-Josiah [Lassa-J, accession no. M15076], Mopeia [accession no. M33879], LCMV-Armstrong [LCMV-A, accession no. M20869] and LCMV-WE [accession no. M22138]) arenaviruses is shown. Identically conserved regions are highlighted, as is the conserved myristoylation (myr) motif G-X3-S/T (59).
Arenavirus entry into host cells is initiated by G1 binding to cell surface receptors followed by endocytosis of the virion into smooth vesicles (7). Although α-dystroglycan serves as a binding receptor for the Old World arenaviruses (13), the receptor utilized by the major group of New World arenaviruses, including Junín virus, is unknown (64). In the arenaviruses, membrane fusion is pH dependent and is activated upon acidification of the maturing endosome (7, 15, 21, 22). The ectodomain of G2 contains two heptad-repeat sequences characteristic of coiled coils found in other viral membrane fusion proteins including human immunodeficiency virus type 1 gp41, influenza HA2, and Ebola virus Gp2 (33). The pH-dependent activation and reorganization of the Junín virus envelope glycoprotein complex likely involves the formation of a helical hairpin structure in G2. Formation of the highly stable helical bundle is thought to be directly coupled to initiation of membrane fusion and entry of the infectious virion core (see references 24, 39, and 69).
The envelope glycoprotein complex thus provides a viable target for antiviral intervention. Passive administration of neutralizing antibodies has been shown to be effective in the treatment of Junín virus infection (28), but their role in Lassa fever and LCMV infection is less clear (29). Similarly, the ability of antiviral peptides to inhibit membrane fusion by preventing the formation of the helix bundle structure (42, 70) suggests that a similar therapeutic strategy may extend to the arenaviruses.
In this report, we examine the biosynthesis and assembly of the Junín virus envelope glycoprotein complex. We show that the 58-amino-acid residue SP of GP-C is myristoylated and stably associated with the mature G1-G2 complex. Substitution by the CD4 SP sequence or mutation of the myristoylation motif dramatically reduces the ability of the envelope glycoprotein complex to mediate pH-dependent membrane fusion. The existence of an essential SP subunit within the envelope glycoprotein complex points to a unique feature in the morphogenesis and life cycle of arenaviruses.
MATERIALS AND METHODS
Cells, viruses, and monoclonal antibodies.
The African green monkey kidney cell line Vero 76 (CRL-1587) was obtained from the American Type Culture Collection (Manassas, Va.) and grown in Dulbecco's modified Eagle medium (DMEM; Gibco) containing high glucose, sodium pyruvate, glutamine, penicillin-streptomycin, and 10% fetal bovine serum (FBS). The attenuated vaccine strain of Junín virus, Candid 1 (1, 3, 49), was kindly provided by R. Shope and R. Tesh, World Health Organization Reference Center for Arboviruses (University of Texas Medical Branch) and was propagated in Vero cells under biosafety level 2 conditions (16). Mouse monoclonal antibodies (MAbs) directed against the Junín virus envelope glycoprotein were prepared against intact, gamma-irradiated Junín virus particles (61) and kindly provided by A. Sanchez and T. Ksiazek (Special Pathogens Branch, Centers for Disease Control and Prevention). The six MAbs (GB03-BE08, GD01-AG02, QC03-BF11, EC05-AA04, LD05-BF09, and QD04-AF03) recognize the G1 subunit (unpublished data). Plasma from convalescent-phase AHF patients was kindly provided by Delia Enría (Instituto Nacional de Enfermedades Virales Humanas, Pergamino, Argentina). Recombinant vaccinia viruses expressing the bacteriophage T7 polymerase (vTF7-3) (32) and the β-galactosidase gene under the control of a T7 promoter (vCB21R-lacZ) (54) were obtained from T. Fuerst and B. Moss and C. Broder, P. Kennedy, and E. Berger, respectively, through the National Institutes of Health (NIH) AIDS Research and Reference Reagent Program, which also provided the human CD4 cDNA (48).
Junín virus envelope glycoprotein constructs.
The GP-C coding region from the pathogenic Junín virus strain MC2 (36) was introduced into the mammalian expression vector pcDNA 3.1+ (by using an AvrII restriction site 72 bp upstream of the GP-C initiation codon and a SmaI site 26 bp downstream of the termination codon) to yield the plasmid pcDNA-JGPC. The DNA sequence of the MC2 GP-C gene was determined, and the corrected sequence is appended to accession no. D10072. It should be noted that the amino acid sequence of the MC2 GP-C gene differs from that of the attenuated Candid 1 strain by only two conservative substitutions in G2 (I427 versus F in the transmembrane domain and S446 versus T in the cytoplasmic domain of MC2) and that the basis for attenuation in the Candid 1 strain is unknown (1, 35). An adventitious polyadenylation motif (AATAAA) in the MC2 GP-C open reading frame was mutated (GATCAA) without changing the encoded amino acids to obviate potential problems in expression. A 15-amino-acid residue S-peptide (Spep) affinity tag (KETAAAKFERQHMDS) (43) was introduced at the C-terminal cytoplasmic end of GP-C (pcDNA-JGPC/Spep) to facilitate biochemical analysis (34). A mutation to generate an SKI-1/S1P cleavage-defective GP-C gene (RRSLK to AASLK) (4, 44) (cd JGPC/Spep) was introduced by QuikChange mutagenesis (Stratagene). The SP of human CD4 (48) was adapted by PCR and used to replace the Junín virus GP-C SP sequence in the CD4sp-JGPC construct. DNA sequencing was used to confirm all manipulations, and three independent clones of each were originally evaluated to assure a consistent phenotype.
Expression and analysis of Junín virus envelope glycoproteins.
The attenuated Junín virus, Candid 1, was grown in Vero cells and quantitated by focus formation after 4 days of growth under agarose overlay. Foci of infected cells were identified by indirect immunostaining of cold methanol-acetone (1:1)-fixed monolayers by using AHF plasma or mouse MAbs directed to G1 (61), horseradish peroxidase-conjugated secondary antibodies, and diaminobenzidine substrate. The Junín virus GP-C envelope glycoprotein was transiently expressed in Vero cells by using either FuGene-6 (Roche Biochemicals) or Metafectene (Biontex, Munich, Germany) reagent for transfection as described by the respective manufacturers. Optimal expression of the Junín virus GP-C gene was achieved by using the T7 promoter of the pcDNA vector and a recombinant vaccinia virus expressing the T7 polymerase (vTF7-3) (32). In these studies, Vero cells in 10-cm-diameter culture dishes were infected with vTF7-3 for 30 min at a multiplicity of infection of 2 in DMEM containing 2% FBS and 10 μM cytosine arabinoside (araC) to limit vaccinia virus DNA replication and cytopathic effect (41). Cultures were then transfected with plasmid DNA and continued in medium containing 10% FBS and araC. Expression was detected by immunostaining after 24 h as described above. For metabolic labeling, cultures were grown for 6 h following transfection and subsequently labeled for 18 h with 125 μCi (each) of [35S]methionine and [35S]cysteine (Amersham Biosciences) in cysteine- and methionine-free DMEM. Cultures infected with Candid 1 virus were labeled for 18 h starting 5 days after infection with a multiplicity of approximately 0.5 focus-forming units per cell. In some studies, cultures were labeled with 250 μCi of [3H]myristic acid (Amersham Biosciences) in DMEM containing 5 mM sodium pyruvate and delipidated FBS (Cocalico Biologicals, Inc.).
Metabolically labeled cultures were washed in physiologic buffered saline (PBS), and cells were subsequently lysed with cold buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% Triton X-100, and protease inhibitors (1 μg [each] of aprotinin, leupeptin, and pepstatin/ml). A soluble fraction was prepared by centrifugation at 15,000 × g for 30 min at 4°C. Cell culture supernatants were filtered (0.2-μm pore size) and chilled prior to the addition of Triton X-100 to 1%. Immunoprecipitation with convalescent-phase AHF plasma or G1-specific MAbs was performed at 4°C for 1 h in lysis buffer, and immune complexes were isolated with protein A-Sepharose (Sigma). Spep-tagged glycoproteins were precipitated at 4°C for 12 h with S-protein agarose beads (SAG; Novagen). For all precipitations, cell and viral lysates were precleared with protein A-Sepharose or bovine serum albumin-conjugated agarose beads (Sigma), respectively. In some studies, the isolated glycoproteins were subsequently deglycosylated with peptide N-glycosidase F (PNGase F; New England Biolabs) as described by the manufacturer (73).
Isolated proteins were analyzed by Laemmli sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), generally following the reduction of disulfide linkages by boiling in 100 mM dithiothreitol. In some studies, commercial NuPAGE Bis-Tris gels were used (Invitrogen). 35S-labeled proteins were visualized by phosphorimaging with a Fuji FLA-3000G instrument. Tritium-labeled proteins were detected by fluorography following treatment with Amplify (Amersham Biosciences). Spep-tagged proteins were detected by Western blot analysis with S-protein-conjugated horseradish peroxidase (Novagen).
Analysis of cell-cell fusion.
Cell-cell fusion mediated by the Junín virus envelope glycoprotein was assessed in Vero cells by the formation of multinucleated syncytia and by the β-galactosidase fusion reporter gene assay (54). In the syncytium-formation assay, vTF7-3-infected cell cultures expressing the Junín virus envelope glycoproteins were grown on poly-d-lysine-coated plates for 24 h in the presence of araC as described above. Cells were pulsed for 1 h with neutral pH medium or with medium adjusted to pH 5.0 [DMEM containing 10 mM HEPES, 10 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES), and 10 μM araC]. Previous studies had identified pH 5.0 as optimal for cell-cell fusion of Junín virus-infected cells (14). Cultures were then continued for 2 h in neutral growth medium containing araC and subsequently fixed with cold methanol-acetone (1:1). Junín virus envelope glycoprotein-expressing cells and syncytia were detected by immunostaining as described above. In the β-galactosidase fusion reporter gene assay, Vero cells infected with a recombinant vaccinia virus bearing the β-galactosidase gene under the control of a T7 promoter (vCB21R-lacZ) served as the target for JGPC-mediated cell-cell fusion (54). In this assay, cultures were infected with vCB21R-lacZ for 90 min at a multiplicity of infection of 2 in DMEM containing 2% FBS. Cells were then washed and used to seed 96-well poly-d-lysine-coated microculture dishes (15,000 cells/well) in DMEM containing 10% FBS and 100 μg of rifampin (Sigma)/ml to minimize vaccinia virus assembly and cytopathic effect (41). After overnight incubation, parallel cultures of vTF7-3-infected and JGPC-expressing cells were harvested in PBS containing 0.1 mM EDTA, resuspended in growth medium containing 2% FBS, 10 μM araC, and 100 μg of rifampin/ml, and 15,000 cells were added to each microculture containing the vCB21R-lacZ-infected target cells. After 4 h, the cocultures were pulsed for 1 h with neutral or pH 5.0 medium containing araC and rifampin and were subsequently refed with neutral medium (containing araC and rifampin), and culture continued for 5 h. At this time, cultures were washed and fixed for 5 min at 4°C in PBS containing 2% formaldehyde and 0.2% glutaraldehyde (54). Staining for β-galactosidase activity was in PBS containing 5 mM (each) potassium ferricyanide and potassium ferrocyanide, 2 mM MgCl2, and 1 mg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal; Sigma)/ml. In other experiments, cocultures were solubilized with lysing solution (Tropix) and the expressed β-galactosidase was quantitated by using the chemiluminescent substrate Galacto-Lite plus (Tropix) as recommended by the manufacturer. Chemiluminescence was determined by using a Tropix TR717 microplate luminometer.
Sequence analysis.
Sequence comparisons among arenavirus GP-C proteins used the NIH BLAST server at http://www.ncbi.nlm.nih.gov/BLAST. Predictive analyses of protein sequences utilized the PredictProtein suite of programs (60). SP sequences were analyzed by using the SignalP algorithms (versions 1.1 and 2.0) (52, 53) available at http://www.cbs.dtu.dk/services/SignalP/. Myristoylation motifs were identified by using ProSite at http://www.expasy.ch/prosite/ (40) and were further evaluated by using the MYRbase server at http://mendel.imp.univie.ac.at/myristate (50).
RESULTS
pH-dependent membrane fusion by the Junín virus envelope glycoprotein.
Entry of the arenavirus into the target cell is mediated by conformational changes in the envelope glycoprotein complex induced upon acidification of the endosome (7, 15, 21, 22). Membrane fusion can also be induced by the envelope glycoprotein on the surface of infected cells by exposing the cultures to acidic pH (14). To examine the fusogenic properties of the Junín virus GP-C, Vero cell cultures expressing the molecularly cloned GP-C gene of the pathogenic MC2 isolate of Junín virus (JGPC) were pulsed with medium adjusted to pH 5.0 (14). As shown in Fig. 2A, cell-cell fusion of JGPC-expressing cells to form multinucleated syncytia was only observed upon low pH treatment. The ability to generate syncytia was unaffected by the addition of the C-terminal affinity tag in JGPC/Spep. In contrast, mutation of the SKI-1/S1P recognition site to prevent G1-G2 cleavage (4, 44) abolished pH-dependent cell-cell fusion (cd JGPC/Spep).
FIG. 2.
pH-dependent fusion mediated by Junín virus envelope glycoproteins. Vero cells expressing either the unmodified Junín virus GP-C glycoprotein (JGPC), the Spep-tagged JGPC/Spep, or the SKI-1/S1P cleavage defective mutant cd JGPC/Spep were tested for their ability to mediate cell-cell fusion. In panel A, cultures of expressing cells were pulsed with medium at pH 5.0 or with neutral medium for 1 h and then incubated for 2 h prior to fixation with cold methanol-acetone (1:1). Cells infected with the T7 polymerase-expressing vaccinia virus vTF7-3 but not transfected with envelope glycoprotein plasmid (vaccinia only) served as negative controls. Cultures were stained with anti-G1 MAb AF03, a secondary antibody conjugated to horseradish peroxidase and diaminobenzidine substrate. Multinucleated syncytia were visualized microscopically. In panel B, parallel assays were performed by using the β-galactosidase fusion reporter gene assay (54). Here, envelope glycoprotein-expressing cells were cocultured with Vero cells infected with a recombinant vaccinia virus bearing the β-galactosidase gene under the control of the T7 promoter (vCB21R-lacZ). Cocultures were then pulsed with medium at pH 5.0 or neutral pH and continued for 5 h. Following fixation with formaldehyde-glutaraldehyde, cell-cell fusion and β-galactosidase expression were detected with X-Gal substrate. Cocultures of cells infected with the respective vaccinia viruses but without an envelope glycoprotein plasmid (vaccinia only) served as negative controls.
Fusion activity was also assessed by using the β-galactosidase reporter gene assay (54). In this format, Vero cells expressing JGPC under the control of the T7 RNA polymerase produced by vTF7-3 were cocultured with cells infected by the recombinant vaccinia virus vCB21R-lacZ containing the LacZ gene under the control of the T7 promoter (54). Cell-cell fusion enables expression of β-galactosidase, which is visualized by X-Gal staining (Fig. 2B) or quantitated by using a chemiluminescent β-galactosidase substrate (see Fig. 5). The results obtained with this reporter assay were concordant with those deduced from syncytium formation. Taken together, these assays demonstrate that the wild-type and Spep-tagged Junín virus GP-C genes expressed in Vero cells are competent to promote pH-dependent cell-cell fusion. Elimination of the SKI-1/S1P proteolytic cleavage site abolishes fusogenicity.
FIG. 5.
Quantitative analysis of cell-cell fusion by mutant Junín virus envelope glycoproteins. Vero cells infected with vTF7-3 and expressing the Junín virus envelope glycoproteins and cells infected with vCB21R-lacZ were cocultured as described for the β-galactosidase fusion reporter gene assay (see legend to Fig. 2B). β-Galactosidase expression was quantitated by chemiluminescence with Galacto-Lite Plus (Tropix) substrate and is presented in relative light units. Cultures pulsed with medium at pH 5.0 are indicated by dark grey bars, and cultures treated at neutral pH are indicated by light grey bars. Cocultures of cells infected with the respective vaccinia viruses but without the GP-C expression plasmid are shown with the label vLacZ+vT7. Standard deviations among replicate cocultures are shown. The extent of pH-dependent cell-cell fusion is determined after subtracting the relative light unit value of the culture treated at neutral pH.
Expression of Junín virus envelope glycoprotein.
To determine the basis for membrane fusion by the Junín virus envelope glycoproteins, we examined the envelope glycoprotein complex in cells infected with the attenuated vaccine strain of Junín virus, Candid 1 (1, 3, 49), and in cells expressing the JGPC plasmid constructs. Cells were metabolically labeled with [35S]methionine and [35S]cysteine, and the expression of the GP-C glycoproteins was examined in nonionic detergent lysates of cells and in the cell culture supernatant.
The immunoprecipitation of Candid 1-infected cells with MAb AF03 directed to G1 resulted in the isolation of a band of 35 kDa and a more heterogeneous species of 30 to 35 kDa (Fig. 3A, lane 1), consistent with one or both of the viral glycoproteins G1 and G2. In addition, a 60-kDa glycoprotein representing the uncleaved G1-G2 precursor (GP-C) was also detected. In the culture supernatant of Candid 1-infected cells (Fig. 3B, lane 1), virion particles contained a predominant smear of the 30- to 35-kDa glycoprotein(s). The unprocessed GP-C precursor was largely excluded from packaging in the virion particle (see also reference 44).
FIG. 3.
Expression and biogenesis of the Junín virus GP-C. Vero cells infected with Candid 1 virus or expressing recombinant forms of the GP-C were metabolically labeled with [35S]methionine and [35S]cysteine, and the envelope glycoproteins were immunoprecipitated with the anti-G1 MAb AF03. Panels A and B represent envelope glycoproteins from cell lysates and cell culture supernatants, respectively. Panels C and D represent the same samples, respectively, treated with PNGase F to generate the deglycosylated polypeptides. Recombinant forms of the envelope glycoprotein gene are unmodified Junín virus GP-C (JGPC), the Spep-tagged JGPC/Spep, and the SKI-1/S1P cleavage defective mutant cd JGPC/Spep. Mock-transfected cells are also represented. Lanes are numbered, and specific comparisons among samples are discussed in detail in the text. The 14C-labeled-protein markers (Amersham Biosciences) are indicated in kilodaltons and shown by dots. The positions of the GP-C precursors and G2, G1, and SSP subunits are labeled. SSP bands in PNGase F-treated samples are distorted by nonionic detergent. In panel D (left), the source of the diffuse bands (below the G1 polypeptide and at ≈16 kDa) is unclear.
Further definition of the viral glycoproteins was obtained by deglycosylation with PNGase F. In this analysis (Fig. 3C and D, lanes 1), the expected 25- and 28-kDa polypeptides of G1 and G2, respectively, were readily distinguished and both G1 and G2 glycoproteins were present in infected cells and on virion particles. The finding that G2 could be precipitated by using MAbs directed to G1 indicated that the subunits remain associated upon solubilization of the virion in nonionic detergent. SDS-PAGE analysis of the glycoprotein complex under nonreducing conditions demonstrated that the association between heterologous G1 and G2 subunits and among homologous subunits is through noncovalent interactions (data not shown).
Examination of the immunoprecipitated glycoproteins also reveals a small polypeptide of approximately 5 kDa in the G1-G2 complex isolated from infected cells and virion particles (Fig. 3A and B, lanes 1). A similarly sized polypeptide has been characterized as a stable form of the GP-C SP in cells expressing the Lassa virus GP-C (27) and in LCMV virions (31). Nonetheless, the inclusion of the SP within the Candid 1 virion and, importantly, in association with the G1-G2 complex was unexpected. As judged by SDS-PAGE under nonreducing conditions, the stable SP (SSP) is noncovalently associated with the G1-G2 complex (see below).
The pattern of GP-C expression in JGPC- and JGPC/Spep-transfected cells was fundamentally similar to that in Candid 1-infected cells, except for the appearance of a 65-kDa form of GP-C precursor in the transfected cells (Fig. 3A and C, lanes 3 and 4). The 60-kDa precursor found in virus-infected cells was also present. We, as others (27, 31), suggest that the larger precursor represents the entire GP-C glycoprotein, with the 58-amino-acid SP uncleaved. The significant levels of the two precursor glycoproteins in transfected cells, relative to that in Candid 1-infected cells, is likely due to the saturation of the cell's proteolytic pathways upon overexpression (5). Despite the abundance of the 65-kDa full-length GP-C precursor, we found no evidence of a 35- to 40-kDa precursor containing only the SP and G1, suggesting that the SP and SKI-1/S1P cleavages are ordered during biosynthesis and that cleavage of the former is essential for subsequent maturation by SKI-1/S1P (see also reference 27). By contrast, the SP cleavage event was unaffected by the defect at the SKI-1/S1P cleavage site in cd JGPC-Spep (Fig. 3A and C, lanes 5). The identification of the 28-kDa deglycosylated polypeptide as G2 was further validated by the higher molecular size of the Spep-tagged G2 polypeptide from JGPC/Spep (Fig. 3C, lanes 3 versus 4) and by Western blot analysis with S-protein-conjugated horseradish peroxidase (data not shown).
Importantly, the mature glycoproteins G1 and G2 were generated in JGPC- and JGPC/Spep-expressing cells, as was the 5-kDa SSP. Moreover, the three subunits were all coprecipitated by using either the anti-G1 MAb (Fig. 3A and C) or SAG (data not shown). Coprecipitation was observed upon lysis in our standard 1% Triton X-100 nonionic detergent as well as in mixtures with 60 mM β-octyl glucoside (data not shown); the latter is a nonionic detergent known to efficiently extract glycoproteins from Triton-resistant membranes (8). Thus, the association among subunits is not dependent upon colocalization in detergent-resistant membrane fragments but appears to be mediated by protein-protein interactions.
The growth medium of cultures expressing either JGPC or JGPC/Spep contained G1 but not G2 (Fig. 3B and D, lanes 3 and 4). This suggests that the arenavirus envelope glycoproteins alone are not sufficient to mediate budding and the release of membrane-bound particles. The G1 found in the medium is likely released by shedding from the noncovalently associated G1-G2 complex on the cell surface. As expected, neither G1 nor G2 was found upon expression of the cd JGPC/Spep plasmid nor was the 5-kDa polypeptide released to the medium (Fig. 3B, lanes 3 and 4). This suggests that the SP remains associated with the cell and virion membranes, presumably via the hydrophobic region, as a signal anchor.
Taken together, this analysis suggests that the Junín virus envelope glycoproteins assemble as a tripartite complex of the SSP, G1, and G2 subunits. Proteolytic cleavage of SSP from the GP-C precursor may in fact be mediated by the cellular signal peptidase. The SignalP algorithm (version 1.1) (52), trained to recognize signal sequences and signal peptidase cleavage sites, is able to identify the authentic cleavage site (SCT-EE), albeit among several potential cleavage sites (data not shown). A recent SignalP algorithm utilizing a hidden Markov model (version 2) (53) is, however, unable to determine a nominal signal peptidase cleavage site in the Junín virus GP-C sequence (data not shown). Although typical SP n and h (hydrophobic) regions (52) are detected in SSP with modest scores by this latter algorithm (0.35 versus 1.0 in the well-characterized CD4 signal sequence), a canonical c region and cleavage motif was not found (cleavage probability, 0.025). Thus, the SSP region likely serves to target the nascent polypeptide to the ER membrane, but the biogenesis and function of this unusual signal sequence is unclear.
Role of SSP in envelope glycoprotein assembly and function.
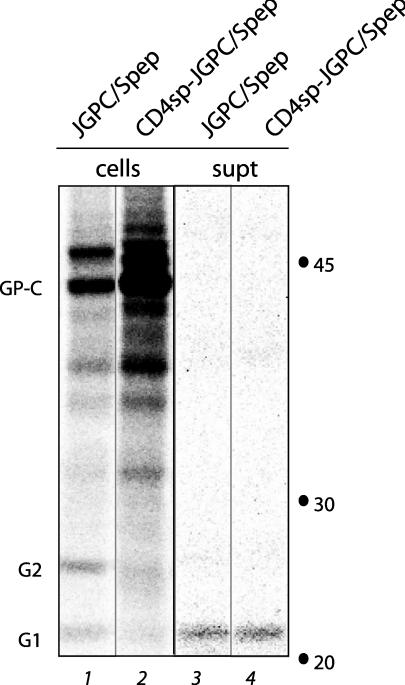
To determine whether the SSP functions solely as a signal sequence, we replaced SSP in JGPC/Spep with the 25-amino-acid SP of human CD4. Upon expression in Vero cells, the chimeric CD4sp-JGPC/Spep precursor glycoprotein was synthesized and proteolytically processed to yield G1 and G2 similarly to the native JGPC/Spep glycoprotein. SDS-PAGE analysis of the expressed glycoproteins is illustrated in Fig. 4 following deglycosylation with PNGase F. The efficiency of proteolytic maturation appeared somewhat altered in the CD4sp chimera relative to the native JGPC/Spep glycoprotein (Fig. 4, lanes 1 and 2), but the G1-G2 complex remained associated in the chimera and could be precipitated by either anti-G1 MAbs (Fig. 4, lanes 1 and 2) or SAG (data not shown). As judged by the extent of G1 shedding (Fig. 4, lanes 3 and 4), the transport and integrity of the G1-G2 complex was unaffected by substitution of the CD4 SP.
FIG. 4.
Analysis of expression of GP-C bearing CD4 SP. GP-C constructs were expressed in Vero cells and metabolically labeled with [35S]methionine and [35S]cysteine. In the CD4sp-JGPC/Spep construct, the GP-C SP has been replaced with that of CD4. The glycoproteins were immunoprecipitated from cell lysates (cells, left panel) or cell culture supernatants (supt, right panel) with the anti-G1 MAb BF11. All isolated glycoproteins were subsequently deglycosylated with PNGase F to better resolve the G1 and G2 polypeptides. Markers and labels are as described in the legend to Fig. 3.
These data suggest that the SSP subunit of the Junín virus GP-C is not required for the biogenesis of the G1-G2 complex. Therefore, we were surprised to find that the G1-G2 complex produced in conjunction with the CD4 SP was entirely unable to mediate membrane fusion (CD4sp-JGPC/Spep) (Fig. 5). This finding suggests that SSP plays an integral role in the function of the envelope glycoprotein complex.
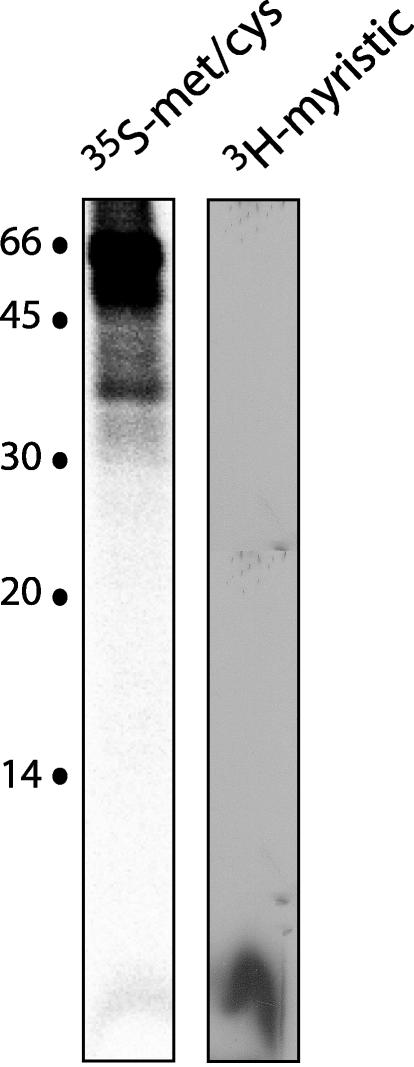
SSP subunit is myristoylated.
The potential involvement of the SSP in envelope glycoprotein function was further suggested by the presence of a canonical myristoylation motif (G-X3-S/T) (59) at the amino terminus. This motif is conserved among all of the arenavirus SSP sequences (Fig. 1) and suggested to us that myristoylation may be an important component in the biosynthesis and assembly of the arenavirus envelope glycoprotein complex. Fatty acid addition is a widely recognized form of protein modification that is often associated with regulatory and membrane trafficking roles (reviewed in reference 59). To determine whether the Junín virus SSP subunit was myristoylated, JGPC/Spep-expressing cells were metabolically labeled with [3H]myristic acid. Upon immunoprecipitation with a G1-specific MAb, the 5-kDa SSP subunit was clearly detected by fluorography (Fig. 6). Neither G1 nor G2, or either of the GP-C precursors, was labeled with the [3H]myristic acid. Thus, the conserved myristoylation motif in SSP is utilized in the Junín virus envelope glycoprotein. Furthermore, the 5-kDa myristoylated SSP was detected regardless of whether the immunoprecipitated complex had been reduced prior to SDS-PAGE (data not shown), indicating that SSP is not disulfide linked to G1 or G2.
FIG. 6.
Myristoylation of the Junín virus SSP. Vero cells expressing JGPC/Spep were metabolically labeled with either [35S]methionine and [35S]cysteine or [3H]myristic acid as described in Materials and Methods. Cell lysates were immunoprecipitated with anti-G1 MAb BF11 and analyzed by SDS-PAGE. In this experiment, protein samples were not reduced prior to electrophoresis.
Role of myristoylation in envelope glycoprotein biogenesis and function.
To investigate the specific role of myristoylation of the SSP subunit, we replaced the critical glycine of the G-X3-S/T motif with alanine. This relatively conservative substitution at the N-terminal glycine (following the initiating methionine) has been shown in a number of studies to specifically prevent myristoylation and affect membrane association. The G2A mutation was thus introduced into JGPC/Spep to assess the effects on envelope glycoprotein biogenesis and function. As shown in Fig. 7A, we could detect no differences in the biosynthesis, proteolytic maturation, assembly, or shedding of the G2A JGPC/Spep glycoproteins relative to the wild type. Importantly, the G2A SSP subunit was coprecipitated with G1 and G2 by using the anti-G1 MAb BF11 (Fig. 7A, lanes 3 and 4). Metabolic labeling of the G2A mutant SSP with [3H]myristic acid confirmed the lack of myristoylation and also the location and identity of the 3H-fatty acid in the wild-type SSP (data not shown). Despite the lack of myristoylation, the G2A SSP was not detected in the cell culture medium, suggesting that the SP sequence was sufficient to maintain membrane anchorage. Therefore, myristoylation per se did not appear to grossly affect the assembly or transport of the SSP-G1-G2 envelope glycoprotein complex.
FIG. 7.
Analysis of expression of the nonmyristoylated G2A mutant of GP-C. Wild-type JGPC/Spep and the G2A myristoylation site mutant G2A-JGPC/Spep were expressed in Vero cells and metabolically labeled with [35S]methionine and [35S]cysteine. (A) Glycoproteins from cell culture supernatants (supt) or cell lysates (cells) were analyzed following immunoprecipitation with the anti-G1 MAb BF11 by PAGE with a 10% Bis-Tris NuPAGE gel and morpholineethanesulfonic acid (MES) running buffer. A sample from a mock-transfected culture is shown to identify nonspecific background bands. (B) The immunoprecipitated glycoproteins from cell lysates were deglycosylated with PNGase F prior to electrophoresis. Markers and labels are as described in the legend to Fig. 3.
By contrast, the G2A mutant of JGPC was markedly defective in mediating cell-cell fusion. In the quantitative β-galactosidase reporter gene assay (Fig. 5), cell-cell fusion by the G2A mutant was reduced to 25% of the wild-type level. Whereas this finding is consistent with that obtained with the CD4 SP construct CD4sp-JGPC/Spep, the G2A mutant specifically demonstrates that myristoylation of the SSP subunit is necessary for membrane fusion. Apparently, the nonmyristoylated SSP is able to associate with the G1-G2 complex but in a manner that is unable to support efficient envelope glycoprotein complex-mediated fusion.
DISCUSSION
This study demonstrates that the 58-amino-acid SP of the Junín virus GP-C envelope glycoprotein precursor is myristoylated and integrally involved in the structure and function of the envelope glycoprotein. We show that the mature envelope glycoprotein complex comprises three subunits derived from GP-C by two ordered proteolytic cleavage events. The initial cleavage results in the generation of the N-terminal 58-amino-acid polypeptide containing a hydrophobic core region that acts as a signal sequence (see also references 25and 31). In contrast to conventional SPs, however, the GP-C SP is stable and is found associated with G1 and G2 on the mature Junín virion. Moreover, this polypeptide is specifically acylated at an N-terminal myristoylation motif that is conserved in all arenaviral GP-C glycoproteins. The protease responsible for the SSP cleavage event is uncertain, but some predictive algorithms suggest that the cellular signal peptidase may be involved. The second cleavage event is mediated by the cellular SKI-1/S1P subtilase (4, 45). This cleavage generates the two conventional envelope glycoprotein subunits: the receptor-binding G1 moiety and the transmembrane G2 protein. Our results indicate that this latter cleavage is independent of the SSP per se but does require a functional SP such as that provided by CD4. These ordered cleavage events likely occur during progress of the nascent envelope glycoprotein complex along the ER and Golgi membranes.
The cleaved SSP, G1, and G2 remain noncovalently associated on the mature virion. This tripartite organization of the envelope glycoprotein complex is distinct from that in many other viruses. Although the incorporation of small virion membrane polypeptides within viral envelopes is not unprecedented (for examples, see references 47 and 72), the topology of the SSP subunit and the organization of the Junín virus envelope glycoprotein complex are largely unknown (see references 26, 31). In the native complex, the hydrophobic core of the SSP subunit may act in part as a signal anchor to mediate membrane attachment. This function may be enhanced by myristoylation of the SSP N terminus. It is also possible that SSP interacts with transmembrane region of G2.
Although neither the SSP subunit nor the myristoylation event is essential for assembly and transport of the G1-G2 complex, the myristoylated SSP subunit is required for the mature envelope glycoprotein complex to mediate pH-dependent membrane fusion. Interestingly, a functional SSP of the cd JGPC complex can act in trans to rescue fusion activity of the CD4sp-JGPC complex (unpublished data; see also reference 25). Furthermore, a mutant envelope glycoprotein containing a G2A mutation that prevents myristoylation is markedly defective in promoting cell-cell fusion. Thus, the myristoylated SSP subunit appears to not be required for membrane association per se but rather for the ultimate fusion activity of the mature complex. This finding is unexpected because protein acylation often acts to modulate membrane association (reference 59 and references therein). For instance, myristoylation of the retrovirus matrix protein is essential for the stable association of the Gag precursor with cellular membranes and, thus, for assembly of the virion core (9, 37, 38, 56). The role of acylation is more complex for transmembrane viral envelope glycoproteins. Mutations that prevent palmitoylation of the envelope glycoproteins of orthomyxoviruses (74) and retroviruses (23, 30, 55, 63) interfere with virion assembly, although the nonacylated forms of these glycoproteins appear to retain their intrinsic ability to mediate membrane fusion (65, 71).
The precise role of the myristoylated SSP in the Junín virus envelope glycoprotein complex is not known. It is possible that the myristic acid moiety of the SSP serves an integral structural role, as it does in picornavirus assembly (2, 18). The SSP polypeptide may itself be important in maintaining envelope glycoprotein structure to enable proper assembly and/or function of the complex. One may imagine a role in stabilizing the prefusogenic form of the envelope glycoprotein. Because virus-mediated membrane fusion is a cooperative process involving multiple envelope glycoprotein oligomers in the stabilization of the fusion pore (6, 46), higher-order interactions among SSP subunits may also be involved.
The myristoylated SSP may also be involved in modulating membrane fusion through effects on membrane trafficking of the envelope glycoprotein complex. In particular, cellular membranes are not homogeneous but rather contain specialized regions called rafts that are enriched in cholesterol and sphingolipids and form ordered microdomains that function as platforms for protein-protein interactions and for macromolecular assembly (see references 62 and 67 and references therein). Recent studies highlight the critical role of rafts in the assembly, budding, and infectivity of diverse viruses (see references 17, 58, and 68 and references therein), and it is possible that myristoylation of the SSP may be important in directing the Junín virus envelope glycoprotein complex to membrane rafts. In the case of the influenza virus hemagglutinin, the increased local concentration of envelope glycoprotein within rafts has been shown to facilitate the highly cooperative process of membrane fusion (68). The defect in membrane fusion by the nonmyristoylated Junín virus envelope glycoprotein may likewise derive from a failure to concentrate in membrane rafts. Further studies are needed to determine whether or not the Junín virus envelope glycoproteins are associated with membrane rafts. Because raft-associated glycoproteins typically distribute between raft and nonraft membranes, it is not inconsistent for us to find the Junín virus envelope glycoproteins in the detergent-soluble (i.e., nonraft) cell fraction examined in the present work.
The importance of myristoylation determined in our genetic studies is consistent with the reported sensitivity of the Junín virus to analogs of myristic acid (20). In the previous study, the expression of the Junín virus envelope glycoproteins was not affected by the analogs, although the basis for the defect in infectivity—in virion assembly, budding, or entry—was not addressed. It should be noted, however, that the arenavirus Z (or matrix) (57, 66) protein contains an N-terminal myristoylation motif that may or may not be utilized, and therefore, the precise target of myristic acid analogues is unclear. It is possible that the arenaviruses utilize myristoylation to colocalize the envelope glycoproteins and the virion matrix protein for virion assembly. Further analysis of arenavirus assembly and envelope glycoprotein function may point to specific targets for antiviral intervention. The development of safe and effective vaccines and therapeutics to combat arenaviral hemorrhagic fever will have broad implications for public health and biodefense.
Acknowledgments
We thank Robert Shope and Robert Tesh (WHO Reference Center for Arboviruses, University of Texas Medical Branch) for providing the Candid 1 virus and Tom Ksiazek and Tony Sanchez (Special Pathogens Branch, CDC) for Junín virus GP-C MAbs. We are grateful to Meg Trahey (The University of Montana) for insightful suggestions during the development of this project and in manuscript preparation. The recombinant vaccinia viruses vTF7-3 and vCB21R-lacZ and the human CD4 cDNA were obtained through the NIH AIDS Research and Reference Reagent Program, and we thank the following contributors: Tom Fuerst, Bernard Moss, Christopher Broder, Paul Kennedy, Edward Berger, and Richard Axel. Human plasma from convalescent-phase AHF patients was kindly provided by Delia Enría (Instituto Nacional de Enfermedades Virales Humanas). We also thank Ed Berger (NIH) for advice on the fusion reporter assay, Christina Jambor (University of Alabama, Birmingham) for advice on [3H]myristic acid labeling, Sudhakar Agnihothram and Kimberly Hardwick for technical assistance, and Kathryn Follis for editorial review of the manuscript. We are grateful to James Meegan (NIH) for encouragement in this project.
V.R. holds a research career award from Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), Argentina.
REFERENCES
- 1.Albarino, C. G., P. D. Ghiringhelli, D. M. Posik, M. E. Lozano, A. M. Ambrosio, A. Sanchez, and V. Romanowski. 1997. Molecular characterization of attenuated Junin virus strains. J. Gen. Virol. 78:1605-1610. [DOI] [PubMed] [Google Scholar]
- 2.Ansardi, D. C., D. C. Porter, and C. D. Morrow. 1992. Myristoylation of poliovirus capsid precursor P1 is required for assembly of subviral particles. J. Virol. 66:4556-4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrera Oro, J. G., and K. T. McKee, Jr. 1991. Toward a vaccine against Argentine hemorrhagic fever. Bull. Pan. Am. Health Organ. 25:118-126. [PubMed] [Google Scholar]
- 4.Beyer, W. R., D. Popplau, W. Garten, D. von Laer, and O. Lenz. 2003. Endoproteolytic processing of the lymphocytic choriomeningitis virus glycoprotein by the subtilase SKI-1/S1P. J. Virol. 77:2866-2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Binley, J. M., R. W. Sanders, B. Clas, N. Schuelke, A. Master, Y. Guo, F. Kajumo, P. J. Maddon, W. C. Olson, and J. P. Moore. 2000. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J. Virol. 74:627-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blumenthal, R., D. P. Sarkar, S. Durell, D. E. Howard, and S. J. Morris. 1996. Dilation of the influenza hemagglutinin fusion pore revealed by the kinetics of individual cell-cell fusion events. J. Cell Biol. 135:63-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borrow, P., and M. B. A. Oldstone. 1994. Mechanism of lymphocytic choriomeningitis virus entry into cells. Virology 198:1-9. [DOI] [PubMed] [Google Scholar]
- 8.Brown, D. A., and J. K. Rose. 1992. Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell 68:533-544. [DOI] [PubMed] [Google Scholar]
- 9.Bryant, M., and L. Ratner. 1990. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc. Natl. Acad. Sci. USA 87:523-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buchmeier, M. J. 2002. Arenaviruses: protein structure and function. Curr. Top. Microbiol. Immunol. 262:159-173. [DOI] [PubMed] [Google Scholar]
- 11.Buchmeier, M. J., M. D. Bowen, and C. J. Peters. 2001. Arenaviruses and their replication, p. 1635-1668. In D. M. Knipe and P. M. Howley (ed.), Fields Virology, vol. 2. Lippincott, Williams, & Wilkins, Philadelphia, Pa.
- 12.Burns, J. W., and M. J. Buchmeier. 1991. Protein-protein interactions in lymphocytic choriomeningitis virus. Virology 183:620-629. [DOI] [PubMed] [Google Scholar]
- 13.Cao, W., M. D. Henry, P. Borrow, H. Yamada, J. H. Elder, E. V. Ravkov, S. T. Nichol, R. W. Compans, K. P. Campbell, and M. B. A. Oldstone. 1998. Identification of alpha-dystroglycan as a receptor for lymphocytic choriomeningitis virus and Lassa fever virus. Science 282:2079-2081. [DOI] [PubMed] [Google Scholar]
- 14.Castilla, V., and S. E. Mersich. 1996. Low-pH-induced fusion of Vero cells infected with Junin virus. Arch. Virol. 141:1307-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Castilla, V., S. E. Mersich, N. A. Candurra, and E. B. Damonte. 1994. The entry of Junin virus into Vero cells. Arch. Virol. 136:363-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Centers for Disease Control and Prevention. 1999. Biosafety in microbiological and biomedical laboratories, 4th ed. Centers for Disease Control and Prevention, Atlanta, Ga.
- 17.Chazal, N., and D. Gerlier. 2003. Virus entry, assembly, budding, and membrane rafts. Microbiol. Mol. Biol. Rev. 67:226-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chow, M., J. F. Newman, D. Filman, J. M. Hogle, D. J. Rowlands, and F. Brown. 1987. Myristoylation of picornavirus capsid protein VP4 and its structural significance. Nature 327:482-486. [DOI] [PubMed] [Google Scholar]
- 19.Clegg, J. C. S., M. D. Bowen, M. J. Buchmeier, J.-P. Gonzalez, I. S. Lukashevich, C. J. Peters, R. Rico-Hesse, and V. Romanowski. 2000. Arenaviridae, p. 633-640. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy: classification and nomenclature of viruses. Seventh report of the international Committee on Taxonomy of Viruses. Academic Press, San Diego, Calif..
- 20.Cordo, S. M., N. A. Candurra, and E. B. Damonte. 1999. Myristic acid analogs are inhibitors of Junin virus replication. Microbes Infect. 1:609-614. [DOI] [PubMed] [Google Scholar]
- 21.Di Simone, C., and M. J. Buchmeier. 1995. Kinetics and pH dependence of acid-induced structural changes in the lymphocytic choriomeningitis virus glycoprotein complex. Virology 209:3-9. [DOI] [PubMed] [Google Scholar]
- 22.Di Simone, C., M. A. Zandonatti, and M. J. Buchmeier. 1994. Acidic pH triggers LCMV membrane fusion activity and conformational change in the glycoprotein spike. Virology 198:455-465. [DOI] [PubMed] [Google Scholar]
- 23.Dubay, J. W., S. J. Roberts, B. H. Hahn, and E. Hunter. 1992. Truncation of the human immunodeficiency virus type 1 transmembrane glycoprotein cytoplasmic domain blocks virus infectivity. J. Virol. 66:6616-6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eckert, D. M., and P. S. Kim. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777-810. [DOI] [PubMed] [Google Scholar]
- 25.Eichler, R., O. Lenz, T. Strecker, M. Eickmann, H. D. Klenk, and W. Garten. 2003. Identification of Lassa virus glycoprotein signal peptide as a trans-acting maturation factor. EMBO Rep. 4:1084-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eichler, R., O. Lenz, T. Strecker, M. Eickmann, H. D. Klenk, and W. Garten. 2004. Lassa virus glycoprotein signal peptide displays a novel topology with an extended ER-luminal region. J. Biol. Chem. 279:12293-12299. (First published 6 January 2004; 10.1074/jbc.M312975200.) [DOI] [PubMed] [Google Scholar]
- 27.Eichler, R., O. Lenz, T. Strecker, and W. Garten. 2003. Signal peptide of Lassa virus glycoprotein GP-C exhibits an unusual length. FEBS Lett. 538:203-206. [DOI] [PubMed] [Google Scholar]
- 28.Enria, D. A., A. M. Briggiler, N. J. Fernandez, S. C. Levis, and J. I. Maiztegui. 1984. Importance of dose of neutralising antibodies in treatment of Argentine haemorrhagic fever with immune plasma. Lancet ii:255-256. [DOI] [PubMed] [Google Scholar]
- 29.Fisher-Hoch, S. P., and J. B. McCormick. 2001. Towards a human Lassa fever vaccine. Rev. Med. Virol. 11:331-341. [DOI] [PubMed] [Google Scholar]
- 30.Freed, E. O., and M. A. Martin. 1996. Domains of the human immunodeficiency virus type 1 matrix and gp41 cytoplasmic tail required for envelope incorporation into virions. J. Virol. 70:341-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Froeschke, M., M. Basler, M. Groettrup, and B. Dobberstein. 2003. Long-lived signal peptide of lymphocytic choriomeningitis virus glycoprotein pGP-C. J. Biol. Chem. 278:41914-41920. [DOI] [PubMed] [Google Scholar]
- 32.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gallaher, W. R., C. DiSimone, and M. J. Buchmeier. 2001. The viral transmembrane superfamily: possible divergence of Arenavirus and Filovirus glycoproteins from a common RNA virus ancestor. BMC Microbiol. 1:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gallina, A., T. M. Hanley, R. Mandel, M. Trahey, C. C. Broder, G. A. Viglianti, and H. J. Ryser. 2002. Inhibitors of protein-disulfide isomerase prevent cleavage of disulfide bonds in receptor-bound glycoprotein 120 and prevent HIV-1 entry. J. Biol. Chem. 277:50579-50588. [DOI] [PubMed] [Google Scholar]
- 35.Ghiringhelli, P. D., C. G. Albarino, M. Piboul, and V. Romanowski. 1997. The glycoprotein precursor gene of the attenuated Junin virus vaccine strain (Candid #1). Am. J. Trop. Med. Hyg. 56:216-225. [DOI] [PubMed] [Google Scholar]
- 36.Ghiringhelli, P. D., R. V. Rivera-Pomar, M. E. Lozano, O. Grau, and V. Romanowski. 1991. Molecular organization of Junin virus S RNA: complete nucleotide sequence, relationship with other members of the Arenaviridae and unusual secondary structures. J. Gen. Virol. 72:2129-2141. [DOI] [PubMed] [Google Scholar]
- 37.Gottlinger, H. G., J. G. Sodroski, and W. A. Haseltine. 1989. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 86:5781-5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hermida-Matsumoto, L., and M. D. Resh. 1999. Human immunodeficiency virus type 1 protease triggers a myristoyl switch that modulates membrane binding of Pr55(gag) and p17MA. J. Virol. 73:1902-1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hernandez, L. D., L. R. Hoffman, T. G. Wolfsberg, and J. M. White. 1996. Virus-cell and cell-cell fusion. Annu. Rev. Cell Dev. Biol. 12:627-661. [DOI] [PubMed] [Google Scholar]
- 40.Hofmann, K., P. Bucher, L. Falquet, and A. Bairoch. 1999. The PROSITE database, its status in 1999. Nucleic Acids Res. 27:215-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hruby, D. E., D. L. Lynn, and J. R. Kates. 1980. Identification of a virus-specified protein in the nucleus of vaccinia virus-infected cells. J. Gen. Virol. 47:293-299. [DOI] [PubMed] [Google Scholar]
- 42.Kilby, J. M., S. Hopkins, T. M. Venetta, B. DiMassimo, G. A. Cloud, J. Y. Lee, L. Alldredge, E. Hunter, D. Lambert, D. Bolognesi, T. Matthews, M. R. Johnson, M. A. Nowak, G. M. Shaw, and M. S. Saag. 1998. Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat. Med. 4:1302-1307. [DOI] [PubMed] [Google Scholar]
- 43.Kim, J.-S., and R. T. Raines. 1993. Ribonuclease S-peptide as a carrier in fusion proteins. Protein Sci. 2:348-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kunz, S., K. H. Edelmann, J.-C. de la Torre, R. Gorney, and M. B. A. Oldstone. 2003. Mechanisms for lymphocytic choriomeningitis virus glycoprotein cleavage, transport, and incorporation into virions. Virology 314:168-178. [DOI] [PubMed] [Google Scholar]
- 45.Lenz, O., J. ter Meulen, H.-D. Klenk, N. G. Seidah, and W. Garten. 2001. The Lassa virus glycoprotein precursor GP-C is proteolytically processed by subtilase SKI-1/S1P. Proc. Natl. Acad. Sci. USA 98:12701-12705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin, X., C. A. Derdeyn, R. Blumenthal, J. West, and E. Hunter. 2003. Progressive truncations C terminal to the membrane-spanning domain of simian immunodeficiency virus Env reduce fusogenicity and increase concentration dependence of Env for fusion. J. Virol. 77:7067-7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lindemann, D., T. Pietschmann, M. Picard-Maureau, A. Berg, M. Heinkelein, J. Thurow, P. Knaus, H. Zentgraf, and A. Rethwilm. 2001. A particle-associated glycoprotein signal peptide essential for virus maturation and infectivity. J. Virol. 75:5762-5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maddon, P. J., A. G. Dalgleish, J. S. McDougal, P. R. Clapham, R. A. Weiss, and R. Axel. 1986. The T4 gene encodes the AIDS virus receptor and is expressed in the immune system and the brain. Cell 47:333-348. [DOI] [PubMed] [Google Scholar]
- 49.Maiztegui, J. I., K. T. McKee, Jr., J. G. Barrera Oro, L. H. Harrison, P. H. Gibbs, M. R. Feuillade, D. A. Enria, A. M. Briggiler, S. C. Levis, A. M. Ambrosio, N. A. Halsey, and C. J. Peters. 1998. Protective efficacy of a live attenuated vaccine against Argentine hemorrhagic fever. AHF Study Group. J. Infect. Dis. 177:277-283. [DOI] [PubMed] [Google Scholar]
- 50.Maurer-Stroh, S., B. Eisenhaber, and F. Eisenhaber. 2002. N-terminal N-myristoylation of proteins: prediction of substrate proteins from amino acid sequence. J. Mol. Biol. 317:541-557. [DOI] [PubMed] [Google Scholar]
- 51.Meyer, B. J., J. C. de la Torre, and P. J. Southern. 2002. Arenaviruses: genomic RNAs, transcription, and replication. Curr. Top. Microbiol. Immunol. 262:139-157. [DOI] [PubMed] [Google Scholar]
- 52.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 53.Nielsen, H., and A. Krogh. 1998. Prediction of signal peptides and signal anchors by hidden Markov model, p. 122-130. In Proceedings of the Sixth International Conference on Intelligent Systems for Molecular Biology. AAAI Press, Menlo Park, Calif. [PubMed]
- 54.Nussbaum, O., C. C. Broder, and E. A. Berger. 1994. Fusogenic mechanisms of enveloped-virus glycoproteins analyzed by a novel recombinant vaccinia virus-based assay quantitating cell fusion-dependent reporter gene activation. J. Virol. 68:5411-5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ochsenbauer-Jambor, C., D. C. Miller, C. R. Roberts, S. S. Rhee, and E. Hunter. 2001. Palmitoylation of the Rous sarcoma virus transmembrane glycoprotein is required for protein stability and virus infectivity. J. Virol. 75:11544-11554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pal, R., M. S. Reitz, Jr., E. Tschachler, R. C. Gallo, M. G. Sarngadharan, and F. D. Veronese. 1990. Myristoylation of gag proteins of HIV-1 plays an important role in virus assembly. AIDS Res. Hum. Retrovir. 6:721-730. [DOI] [PubMed] [Google Scholar]
- 57.Perez, M., R. C. Craven, and J. C. de la Torre. 2003. The small RING finger protein Z drives arenavirus budding: implications for antiviral strategies. Proc. Natl. Acad. Sci. USA 100:12978-12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rawat, S. S., M. Viard, S. A. Gallo, A. Rein, R. Blumenthal, and A. Puri. 2003. Modulation of entry of enveloped viruses by cholesterol and sphingolipids. Mol. Membr. Biol. 20:243-254. [DOI] [PubMed] [Google Scholar]
- 59.Resh, M. D. 1999. Fatty acylation of proteins: new insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim. Biophys. Acta 1451:1-16. [DOI] [PubMed] [Google Scholar]
- 60.Rost, B. 1996. PHD: predicting 1D protein structure by profile based neural networks. Methods Enzymol. 266:525-539. [DOI] [PubMed] [Google Scholar]
- 61.Sanchez, A., D. Y. Pifat, R. H. Kenyon, C. J. Peters, J. B. McCormick, and M. P. Kiley. 1989. Junin virus monoclonal antibodies: characterization and cross-reactivity with other arenaviruses. J. Gen. Virol. 70:1125-1132. [DOI] [PubMed] [Google Scholar]
- 62.Simons, K., and E. Ikonen. 1997. Functional rafts in cell membranes. Nature 387:569-572. [DOI] [PubMed] [Google Scholar]
- 63.Spies, C. P., G. D. Ritter, Jr., M. J. Mulligan, and R. W. Compans. 1994. Truncation of the cytoplasmic domain of the simian immunodeficiency virus envelope glycoprotein alters the conformation of the external domain. J. Virol. 68:585-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Spiropoulou, C. F., S. Kunz, P. E. Rollin, K. P. Campbell, and M. B. A. Oldstone. 2002. New World arenavirus clade C, but not clade A and B viruses, utilizes alpha-dystroglycan as its major receptor. J. Virol. 76:5140-5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Steinhauer, D. A., S. A. Wharton, D. C. Wiley, and J. J. Skehel. 1991. Deacylation of the hemagglutinin of influenza A/Aichi/2/68 has no effect on membrane fusion properties. Virology 184:445-448. [DOI] [PubMed] [Google Scholar]
- 66.Strecker, T., R. Eichler, J. Meulen, W. Weissenhorn, H. D. Klenk, W. Garten, and O. Lenz. 2003. Lassa virus Z protein is a matrix protein and sufficient for the release of virus-like particle. J. Virol. 77:10700-10705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Suomalainen, M. 2002. Lipid rafts and assembly of enveloped viruses. Traffic 3:705-709. [DOI] [PubMed] [Google Scholar]
- 68.Takeda, M., G. P. Leser, C. J. Russell, and R. A. Lamb. 2003. Influenza virus hemagglutinin concentrates in lipid raft microdomains for efficient viral fusion. Proc. Natl. Acad. Sci. USA 100:14610-14617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weissenhorn, W., A. Dessen, L. J. Calder, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1999. Structural basis for membrane fusion by enveloped viruses. Mol. Membr. Biol. 16:3-9. [DOI] [PubMed] [Google Scholar]
- 70.Wild, C., T. Greenwell, D. Shugars, L. Rimsky-Clarke, and T. Matthews. 1995. The inhibitory activity of an HIV type 1 peptide correlates with its ability to interact with a leucine zipper structure. AIDS Res. Hum. Retrovir. 11:323-325. [DOI] [PubMed] [Google Scholar]
- 71.Yang, C., C. P. Spies, and R. W. Compans. 1995. The human and simian immunodeficiency virus envelope glycoprotein transmembrane subunits are palmitoylated. Proc. Natl. Acad. Sci. USA 92:9871-9875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yao, J. S., E. G. Strauss, and J. H. Strauss. 1996. Interactions between PE2, E1, and 6K required for assembly of alphaviruses studied with chimeric viruses. J. Virol. 70:7910-7920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.York, J., and J. H. Nunberg. 2004. Role of hydrophobic residues in the central ectodomain of gp41 in maintaining the association between human immunodeficiency virus type 1 envelope glycoprotein subunits gp120 and gp41. J. Virol. 78:4921-4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang, J., A. Pekosz, and R. A. Lamb. 2000. Influenza virus assembly and lipid raft microdomains: a role for the cytoplasmic tails of the spike glycoproteins. J. Virol. 74:4634-4644. [DOI] [PMC free article] [PubMed] [Google Scholar]