Abstract
Vertebrates mount a strong innate immune response against viruses, largely by activating the interferon system. Double-stranded RNA (dsRNA), a common intermediate formed during the life cycle of many viruses, is a potent trigger of this response. In contrast, no general inducible antiviral defense mechanism has been reported in any invertebrate. Here we show that dsRNA induces antiviral protection in the marine crustacean Litopenaeus vannamei. When treated with dsRNA, shrimp showed increased resistance to infection by two unrelated viruses, white spot syndrome virus and Taura syndrome virus. Induction of this antiviral state is independent of the sequence of the dsRNA used and therefore distinct from the sequence-specific dsRNA-mediated genetic interference phenomenon. This demonstrates for the first time that an invertebrate immune system, like its vertebrate counterparts, can recognize dsRNA as a virus-associated molecular pattern, resulting in the activation of an innate antiviral response.
In vertebrates, receptors of the innate immune system recognize pathogen-associated molecular patterns in order to activate early defense mechanisms. Innate immune recognition plays an important role not only in clearing the majority of invading pathogens but also in priming and directing the adaptive immune response. The most prominent of the innate immune receptors belong to the Toll-like receptor (TLR) family. TLRs share structural features with the mammalian interleukin-1 receptor and function as activators of intracellular signaling in response to infection. In mammals the TLRs are involved in recognition of diverse microbial products including lipopolysaccharide (LPS) (23, 37), lipoteichoic acids (39), unmethylated CpG-rich DNA (6), bacterial flagellin (20), and double-stranded RNA (dsRNA) (4). Upon their activation, TLRs engage a variety of intracellular adaptor molecules, leading to signal transduction events that regulate the expression of genes of the immune system (reviewed in reference 3). This general model for cell-based immunity (i.e., microbial recognition, signal transduction, and transcriptional activation) is also used by invertebrates. In Drosophila melanogaster, recognition of bacterial peptidoglycan triggers immune responses through activation of the Toll and immune deficiency (Imd) pathways (29; reviewed in reference 21). These two distinct signal transduction pathways are involved in the fly's responses to fungi and gram-positive bacteria and to gram-negative bacteria, respectively.
Although the invertebrate immune system has been well studied in the context of antibacterial and antifungal responses, there is no information at the cellular or molecular level regarding the invertebrate immune response directed against viruses. In mammals, antiviral and antibacterial innate responses include partially overlapping, but distinct pathways. The most prominent innate antiviral response in vertebrates is the interferon (IFN) system. IFNs comprise a family of cytokines expressed in response to viral infection and other insults and regulate a myriad of cellular and systemic responses directed to control viral propagation (see reference 30 for a review). Upon induction of cells by circulating IFNs, signal transduction through the Janus kinase/signal transducer and activator of transcription (JAK/STAT) system results in the induction of hundreds of genes (14, 15). IFN-inducible genes include those encoding RNA-dependent protein kinase (PKR), the Mx protein, oligoadenylate synthetase, and IFNs themselves. This self-amplifying system can be triggered not only by IFN, but also directly by viral components. A potent inducer of the IFN response is dsRNA, a molecule that often occurs during viral infection as a result of viral genomic replication and viral RNAs with extensive secondary structure (reviewed in reference 25). In mammals dsRNA is recognized by TLR3, which activates myeloid differentiation factor 88 (Myd88)-dependent and independent signal transduction cascades, leading to the expression of IFN-β (4, 36). dsRNA also induces antiviral responses intracellularly, by directly activating PKR, which leads to inhibition of cellular and viral protein synthesis via phosphorylation of eukaryotic translation initiation factor 2α (eIF2α) (34). It has been generally accepted that these dsRNA-induced immune responses are absent from invertebrates, a conclusion supported by the lack of genes homologous to IFNs or to the major effectors of the IFN response (e.g., PKR) in several fully sequenced invertebrate genomes (1, 10, 13, 22).
The complexity of the biological properties of dsRNA in vivo became more apparent with the discovery of dsRNA-mediated postranscriptional gene silencing (PTGS or RNA interference [RNAi]). This phenomenon has been described in both plants and animals, including nematodes (16), insects (35), and mammals (42). Typically, dsRNA is processed into short duplexes 21 to 25 bp long, known as short interfering RNAs by the RNase III Dicer (7). These short interfering RNAs are utilized to recognize homologous mRNAs and trigger their degradation by RNase activities associated with the RNA-induced silencing complex (18, 19). It has been proposed that RNAi represents an ancient antiviral mechanism used to inhibit the expression of viral gene products in infected cells because viral dsRNAs often occur in the course of infection. For instance, viral strategies to evade the RNAi pathway exist in both plant and animal viruses (31, 32, 40).
The sequence-specific effects of dsRNA that result in endogenous RNA degradation are widely conserved and probably present in most invertebrates. In contrast, the sequence-independent induction of antiviral immunity by dsRNA has long been thought to be exclusive to vertebrates. Here we report the induction of an antiviral state by nonspecific dsRNA in the marine shrimp Litopenaeus vannamei. Our findings show for the first time that invertebrates can display inducible antiviral immunity in response to a bona fide virus-associated molecular structure. Moreover, this opens the possibility that innate antiviral immunity in invertebrates shares some of the molecular features of vertebrate antiviral responses.
MATERIALS AND METHODS
Animals and experimental viral infection.
The bioassay system, experimental animals, and White spot syndrome virus (WSSV) and Taura syndrome virus (TSV) inocula used in these studies have been described elsewhere (38). Briefly, shrimp (L. vannamei) were stocked individually in 260-ml tissue culture flasks and acclimated for a 2- to 3-day period, with a 100% daily water exchange (artificial seawater; Marine Environment), and fed approximately half a pellet of commercial feed every day. When indicated (see Fig. 2b), shrimp were kept in 10-gallon tanks (10 to 15 shrimp/tank) connected to a recirculation system with constant water flow and air supply. After acclimation, shrimp were treated by intramuscular injection of dsRNA and infected by intramuscular injection of 0.45-μm-pore-size-filtered extracts of infected shrimp. In the case of TSV, the final dilution was 10−8 or 10−5, and in the case of WSSV the final dilution was 4 × 10−8 to 6 × 10−8 (weight/volume dilutions). These dilutions (10−8 for TSV and 4 × 10−8 to 6 × 10−8 for WSSV) were chosen as doses that consistently resulted in mortality rates of between 60 and 90% over a large number of experiments and titration trials. Negative controls were injected with extracts of specific-pathogen-free shrimp at equivalent dilutions. Injection volumes were 20 μl, unless otherwise specified. Mortality was recorded daily, and water exchange and feeding regimes were as described above.
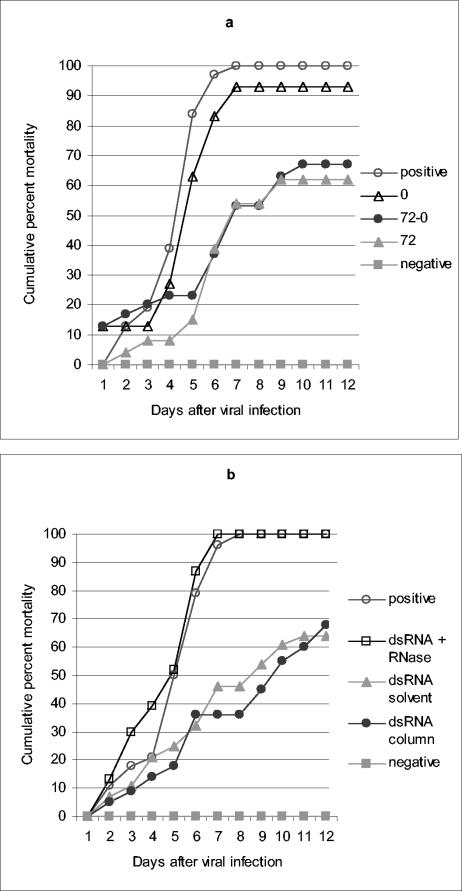
FIG. 2.
Induction of the antiviral state is due to RNA. Shrimp (2 to 3 g) were injected intramuscularly with either saline (positive [○] and negative [░⃞] controls) or dsRNA. At 72 h after this initial injection, animals were infected with WSSV either alone or mixed with dsRNA. (a) Animals (n = 26 to 31) were injected each time with 50 μl of a solution containing ca. 10 to 15 μg of dsRNA for duck υ and challenged with WSSV-positive extract. The 72-0 group (•) received dsRNA both 72 and 0 h (coinjection) before viral infection, the 72 group (gray triangles) received dsRNA only 72 h prior to infection, and the 0 group (▵) received dsRNA only mixed with the WSSV inoculum. (b) Shrimp (n = 20 to 28) were kept in a recirculation system (see Materials and Methods), injected with dsRNA 72 h before viral challenge, and then reinjected with dsRNA mixed with WSSV-positive extract. Positive (○) and negative (░⃞) controls were as described for panel a. dsRNA for the duck υ was used in every case and applied at 10 to 15 μg per injection. dsRNA was purified with phenol and chloroform (gray triangles), RNeasy columns (Qiagen) (•), or purified with RNeasy columns and treated with a cocktail of RNases (RNase A, RNase T1, and RNase V1 from Ambion) (□). The Fisher exact test was used to assess the significance of the observed antiviral protection by comparing the final cumulative mortality in dsRNA-treated groups with that of their respective positive controls: (a) 72-0 treatment (P = 0.0003) and 72 treatment (P = 0.0001) and (b) dsRNA column (P = 0.0017) and dsRNA solvent (P = 0.0003).
dsRNA.
Single-stranded RNA (ssRNA) was transcribed in vitro from linearized plasmid constructs by using T3 and T7 phage RNA polymerases (Promega), and the DNA template was then degraded by addition of DNase I (Promega) at a ratio of 1 U/μg of template. The transcripts were then purified by organic solvent extraction by standard methods or by silica matrix absorption (RNeasy; Qiagen). cRNA strands were mixed in the presence of 400 mM NaCl-10 mM Tris-Cl (pH 7.4) and annealed by incubation at 75°C for 15 min, 65°C for 15 min, and room temperature for 15 min. The formation of dsRNA was monitored by determining the size shift in agarose gel electrophoresis, and the concentration of dsRNA was measured spectrophotometrically. The DNA templates used for in vitro transcription were pBluescript vector (Stratagene) hosting either a 309-bp portion of the immunoglobulin (Ig) υ H chain of the duck, Anas platyrhynchos (accession no. AJ312200); a 1,316-bp genomic noncoding region of clone BAC6 from the catfish Ictalurus punctatus IgH locus (accession no. CC936713); a 1,079-bp portion of pig Sus scrofa IgG cDNA (accession no. U03778); or a 1,184-bp fragment of the bacterial artificial chromosome (BAC) cloning vector pBeloBAC11. Poly(C-G), poly(I-C), and poly(C) are commercial dsRNA and ssRNA analogs (Sigma-Aldrich).
Histology and immunohistochemistry.
Whole shrimp were fixed in Davidson's solution (33% ethanol, 22% formalin, 11.5% acetic acid) for 24 h and then transferred to 70% ethanol and stored until histological analysis by hematoxylin-eosin staining (33) and immunohistochemistry by using an anti-WSSV monoclonal antibody detection system (Diagxotics).
RESULTS
dsRNA induces an antiviral state in shrimp.
We first observed that injection of shrimp with dsRNA afforded antiviral protection while testing the effects of RNAi-mediated downregulation of signal transduction pathways on the outcome of challenge with TSV. Our experimental approach was to inject purified cognate dsRNA and then challenge animals with TSV to explore whether animals in which genes of interest were downregulated would display increased susceptibility to virus. Surprisingly, injection of any dsRNA, whether representing shrimp genes or nonspecific dsRNA controls, resulted in increased survival to TSV challenge (not shown). This initial observation prompted us to address the possibility that, as in vertebrates, dsRNA can induce an antiviral program in shrimp.
To explore whether dsRNA induces a general antiviral response in the shrimp, we analyzed the effect of dsRNA treatment on infection with TSV, an ssRNA virus (8), and on infection with WSSV, a complex enveloped dsDNA virus (45). Reductions in cumulative mortality of 50 to 75% were observed in animals treated with dsRNA, compared to mock-treated animals (Fig. 1a and b). The level of antiviral protection was reduced (to ca. 12%) when shrimp were subjected to a significantly higher dose of TSV (Fig. 1a), suggesting that, like any immune response, dsRNA-induced antiviral protection in shrimp can be overwhelmed by increasing the load of infectious agent. The dsRNA used to protect against both TSV and WSSV in the experiments summarized in Fig. 1 was transcribed from the gene for the duck Ig υ heavy chain. This sequence has no similarity to any known shrimp gene or to the genomes of WSSV (accession no. NC_003225) or TSV (accession no. NC_003005). Thus, the observed antiviral response induced by duck υ dsRNA is unlikely to involve the sequence-specific RNAi pathway and therefore may represent a more general antiviral mechanism active against two unrelated viruses.
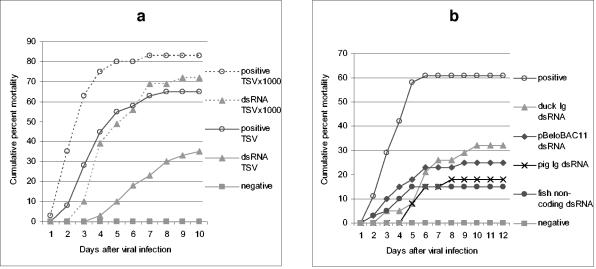
FIG. 1.
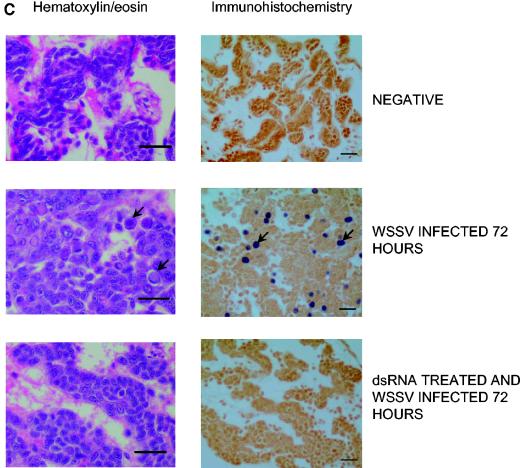
dsRNA induces an antiviral state in shrimp. Shrimp (1 to 2 g) were injected intramuscularly with either saline (positive [○] and negative [░⃞] controls) or 8 μg of dsRNA as indicated. At 72 h after dsRNA injection, animals (n = 38 to 42) were infected by intramuscular injection with TSV (a) or WSSV (b and c) as described in Materials and Methods. In panel a, the effect of a 1,000-fold increase in the dose of TSV (from 10−8 to 10−5 [wt/vol] dilutions) in both positive controls and dsRNA-treated shrimp is also shown in the dotted mortality curves. The dsRNA preparations were a 309-bp portion of duck Igυ ( ; accession no. AJ312200), a 1,316-bp genomic noncoding region of clone BAC6 from the catfish IgH locus (•; accession no. CC936713), a 1,079-bp portion of pig IgG cDNA (✽; accession no. U03778), and a 1,184-bp fragment of the BAC cloning vector pBeloBAC11 (26) (♦). The chi-square statistic was used to assess the significance of the observed antiviral protection by comparing the final cumulative mortality in dsRNA-treated groups with that of their respective positive controls: (a) duck Igυ dsRNA (χ2 = 6.05 [0.01 < P < 0.025]) and duck Igυ dsRNA TSVx1000 (χ2 = 0.75 [not significant]) and (b) duck Igυ dsRNA (χ2 = 5.29 [0.01 < P < 0.025]), pig Ig dsRNA (χ2 = 12.93 [P < 0.001]), pBeloBAC11 dsRNA (χ2 = 8.67 [0.001 < P < 0.005]), and fish noncoding dsRNA (χ2 = 15.39 [P < 0.001]). Panel c shows eosin-hematoxylin staining and anti-WSSV immunostaining of hemopoietic tissue sections from a control infected shrimp in comparison to a dsRNA-treated/WSSV-infected shrimp. The sections shown were obtained 72 h after viral infection. Arrows indicate the presence of intranuclear inclusions characteristic of WSSV infection and WSSV-positive immunoreactivity. Scale bars, 20 μm.
Preliminary experiments (data not shown) suggested that 1 μg of duck υ dsRNA was close to the minimal effective dose required to induce protection against WSSV infection and that as much as 100 μg of dsRNA could be injected into a shrimp of 1 to 2 g without causing obvious signs of toxicity. Thus, intermediate doses of 7 to 15 μg were subsequently used in the studies reported here to investigate the characteristics of the dsRNA-induced antiviral response in shrimp.
Confirmation that the induction of an antiviral state by dsRNA was sequence independent was sought by challenging shrimp with WSSV after treatment with four different types of unrelated dsRNA sequences. We used sequences derived from vertebrate immunoglobulin genes (duck υ and pig γ), fish noncoding genomic DNA, and bacterial vector sequence (pBeloBAC11 vector). Each one of these sequences induced protection against WSSV infection (Fig. 1b). To determine whether dsRNA treatment inhibits viral infection or results in attenuation of WSSV-induced mortality without affecting viral accumulation in the host, histological analysis was performed on shrimp challenged with WSSV with or without pretreatment with dsRNA. Dramatic differences were observed when we tested the accumulation of WSSV particles and the occurrence of classical tissue damage in control and dsRNA-treated shrimp 36 and 72 h after infection (data not shown and Fig. 1c). Histological sections of control infected animals revealed large basophilic granular viral inclusions in nuclei of gastric and cuticular epithelial tissues (not shown), as well as hemopoietic tissues (Fig. 1c). These features were absent from animals protected by dsRNA treatment. These observations suggest that WSSV fails to accumulate significantly in dsRNA-treated shrimp, probably due to the induction of an antiviral protection system. Collectively, these data support the conclusion that dsRNA induces, in a sequence-independent manner, an antiviral program in shrimp.
Induction of the antiviral state is due to RNA.
Some potent inducers of innate immunity can exert strong effects on sensitive cells at relatively low concentrations. We considered the possibility that the relative resistance of dsRNA-treated animals to viral challenge was due to contaminants in the dsRNA preparations that could copurify with dsRNA during organic solvent extraction (e.g., LPS and DNA). In addition, we considered the possibility that compounds present in the RNA preparations could directly inactivate the viruses used in our studies, even when 72 h were allowed to pass between dsRNA treatment and viral injections. To address these issues, we conducted a series of experiments in which we subjected the animals to a two-step dsRNA application regime. First, we injected animals with dsRNA 72 h before viral challenge as done before and then applied an additional dose of dsRNA mixed with the viral preparation. We reasoned that, if contaminants in dsRNA preparations were responsible for direct inactivation of the virus, we would observe high survival rates in shrimp injected with viral inoculum that has been mixed with the dsRNA preparations. Figure 2a shows that a single dsRNA dose administered 72 h before viral challenge protects shrimp against WSSV to a similar extent as two applications of dsRNA (72 h before infection and injection of virus-dsRNA mixtures). Furthermore, when dsRNA was injected only as a mixture with the viral inoculum, poor protection (<10% reduction in mortality) was observed relative to the 72-h pretreatment or the double treatment with dsRNA. These results demonstrate that direct inactivation of the virus by dsRNA or by putative contaminants cannot account for the observed antiviral protection. Furthermore, Fig. 2b shows that RNase treatment destroyed the antiviral protection afforded by our preparations and that dsRNA purified from in vitro transcription reactions by two different methods (organic solvent extraction or silica matrix adsorption) have similar abilities to induce the antiviral response. Taken together, these results demonstrate that intact RNA (and not other compounds present in the dsRNA preparations) is the inducer of the observed antiviral immunity in shrimp.
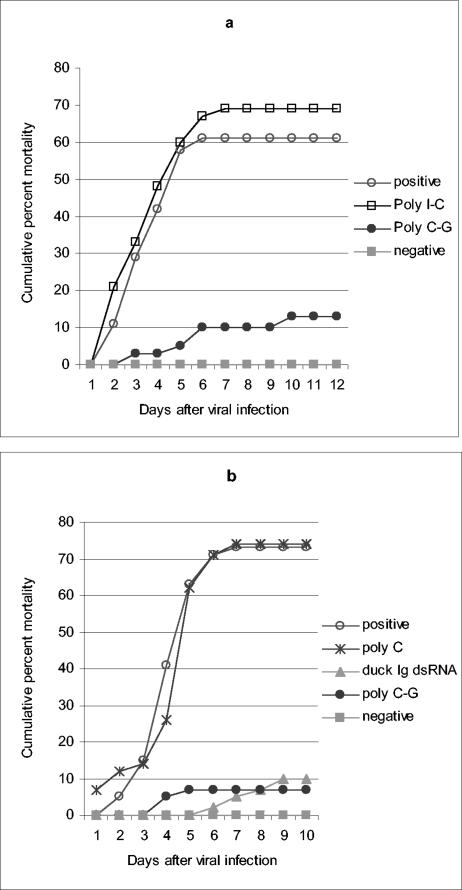
Poly(C-G), but not poly(I-C) or poly(C), induces the antiviral state.
Poly(I-C) is a potent inducer of the innate antiviral response in vertebrates and a popular experimental tool for studying the biology of dsRNA in these systems. We explored the possibility that this and other synthetic commercial RNAs would also induce the antiviral response in shrimp. Figure 3a shows that poly(I-C) failed to induce protection against WSSV infection, whereas another dsRNA analogue, poly(C-G) was a highly effective inducer of antiviral protection. This result suggests differences in the mechanisms for recognition of dsRNA between vertebrates and shrimp, and further supports the notion that dsRNA can induce the antiviral response in shrimp independently of the method by which it is prepared or purified. Importantly, the synthetic ssRNA poly(C) failed to induce antiviral protection (Fig. 3b), suggesting that, like its vertebrate counterpart, the antiviral system of the shrimp is capable of discriminating between ssRNA and dsRNA.
FIG. 3.
Poly(C-G), but not poly(I-C) or poly(C), induces the antiviral state. Shrimp (1 to 2 g, n = 38 to 42) were injected with 8 μg (a) or 7 μg (b) of synthetic dsRNA analogues as indicated and infected 72 h later with WSSV. All RNA analogues were purchased from Sigma-Aldrich, reconstituted in 400 mM NaCl-10 mM Tris-Cl (pH 7.4), and annealed and quantified as described in Materials and Methods. dsRNA for the duck υ (7 μg) was included in the experiment on panel b for comparison. The chi-square statistic was used to assess the significance of the observed antiviral protection relative to positive controls: (a) poly CG (χ2 = 16.92 [P < 0.001]) and (b) duck Igυ dsRNA (χ2 = 32.17 [P < 0.001]) and poly(C-G) (χ2 = 35.05 [P < 0.001]).
DISCUSSION
The data presented here show that dsRNA evokes, in a marine invertebrate, protection against viral infection. This provides the first evidence that an inducible, general antiviral immune response may be expressed in an invertebrate. Other studies have suggested that microbial products (e.g., LPS and β-glucans), as well as certain viral proteins, can enhance antiviral resistance in shrimp (11, 24, 41, 43, 44). Our results, however, further advance our knowledge on antiviral immunity in crustaceans in three ways. First, the results presented here demonstrate an antiviral immune response that is active against two unrelated viruses, TSV and WSSV. The working hypothesis that we propose on the basis of these results is that dsRNA induces a general antiviral response in shrimp. Second, our findings show that the antiviral immune system of the shrimp responds to dsRNA as a molecular pattern regardless of its sequence and base composition [except in the case of poly(I-C)]. Third, these results suggest a possible evolutionary link (recognition of dsRNA) between innate antiviral immunity in vertebrates and invertebrates.
Although our data demonstrate that dsRNA can protect shrimp from lethal infection by TSV and WSSV under controlled experimental conditions, it is important to note that animals were infected with viral doses intended to kill <100% of the shrimp. The protection induced by dsRNA can be overwhelmed by a high dose of infectious virus. For instance, we found that viral doses eightfold higher than the WSSV inoculum used here cause almost 100% mortality in both control and dsRNA-treated shrimp (data not shown) and that a dramatic increase in viral dose can minimize the protective effect of dsRNA upon TSV infection (Fig. 1a).
Although much is known of the molecular basis for antibacterial and antifungal responses in invertebrates, especially in insects (reviewed in reference 21), there is no information concerning immune mechanisms directed against virus infections in any invertebrate. It seems clear that invertebrates lack antiviral systems homologous to vertebrate IFN, based on the analysis of four complete invertebrate genomes (i.e., Drosophila melanogaster, Anopheles gambiae, Caenorhabditis elegans, and Ciona intestinalis). However, the absence of homologous genes does not rule out the existence of invertebrate immune systems analogous to those present in vertebrates. For instance, antigen-specific immune memory has recently been demonstrated for a crustacean in the context of parasitic infection (27). This is surprising, since a bona fide adaptive immune system homologous to the vertebrate system, which is based on T and B lymphocytes and rearranging antigen receptor genes, is clearly absent from invertebrates. Genetic diversification and the acquisition of novel gene function during the evolution of the metazoa may have resulted in convergent evolution of antiviral immunity. Thus, focusing on function rather than sequence homology may hold the key to understanding the evolution of antiviral immunity at the molecular level.
Our demonstration that dsRNA induces an antiviral program in an invertebrate opens the possibility of finding novel molecular mechanisms of innate immunity. It would be surprising if shrimp expressed genes homologous to IFNs or to IFN-induced effectors of antiviral immunity, such as PKR or Mx, since these genes are not present in the genomes of other arthropods. Interestingly, immune-responsive oligoadenylate synthetases have been identified in an invertebrate group, the sponges (17, 28), and the possibility that these genes exist in crustaceans and play a role in antiviral immunity cannot be excluded. On the other hand, signal transduction pathways homologous to those known to regulate the vertebrate IFN response do exist in arthropods. In Drosophila, the JAK/STAT system regulates hemopoiesis and sexual determination (reviewed in reference 46) and also controls expression of some immune function genes (e.g., thiol ester-containing proteins and Turandot A) (2, 9). In A. gambiae, two homologues of STAT have been described (12), one of which has been shown to translocate to the nucleus in response to immune challenge (5), and our own work has identified at least one homologue of STAT in shrimp (NCBI accession no. CA991435). The discovery that mammalian TLR-3 recognizes dsRNA, leading to activation of the IFN response, opens the possibility that a TLR/NF-κB cassette is also involved in invertebrate antiviral immunity, and specifically in the dsRNA-induced response reported here.
The widespread existence of dsRNA-induced immune responses in other invertebrate taxa is an important issue that remains to be explored. For instance, understanding the antiviral defense mechanisms of invertebrate vectors of human disease (e.g., mosquitoes) will have important implications for public health. Furthermore, dsRNA-mediated genetic interference (RNAi) is a widely used reverse genetics tool in many invertebrate models. The possible induction of an immune response by dsRNA may need to be taken into consideration in interpreting studies with RNAi.
Acknowledgments
This study was supported by awards from the National Science Foundation (MCB-0315393), the South Carolina Sea Grant Consortium (R/MT-6), the USDA-CSREES US Marine Shrimp Farming Program, and the South Carolina Department of Natural Resources (03030650). J.R. is supported by Escuela Superior Politécnica del Litoral (Ecuador) and a scholarship from the Fundación para la Ciencia y Tecnología (Ecuador).
We thank Robert Chapman for support and fruitful discussions.
REFERENCES
- 1.Adams, M. D., S. E. Celniker, R. A. Holt, C. A. Evans, J. D. Gocayne, P. G. Amanatides, S. E. Scherer, P. W. Li, R. A. Hoskins, R. F. Galle, R. A. George, S. E. Lewis, S. Richards, M. Ashburner, S. N. Henderson, G. G. Sutton, J. R. Wortman, M. D. Yandell, Q. Zhang, L. X. Chen, R. C. Brandon, Y. H. Rogers, R. G. Blazej, M. Champe, B. D. Pfeiffer, K. H. Wan, C. Doyle, E. G. Baxter, G. Helt, C. R. Nelson, G. L. Gabor, J. F. Abril, A. Agbayani, H. J. An, C. Andrews-Pfannkoch, D. Baldwin, R. M. Ballew, A. Basu, J. Baxendale, L. Bayraktaroglu, E. M. Beasley, K. Y. Beeson, P. V. Benos, B. P. Berman, D. Bhandari, S. Bolshakov, D. Borkova, M. R. Botchan, J. Bouck, P. Brokstein, P. Brottier, K. C. Burtis, D. A. Busam, H. Butler, E. Cadieu, A. Center, I. Chandra, J. M. Cherry, S. Cawley, C. Dahlke, L. B. Davenport, P. Davies, B. de Pablos, A. Delcher, Z. Deng, A. D. Mays, I. Dew, S. M. Dietz, K. Dodson, L. E. Doup, M. Downes, S. Dugan-Rocha, B. C. Dunkov, P. Dunn, K. J. Durbin, C. C. Evangelista, C. Ferraz, S. Ferriera, W. Fleischmann, C. Fosler, A. E. Gabrielian, N. S. Garg, W. M. Gelbart, K. Glasser, A. Glodek, F. Gong, J. H. Gorrell, Z. Gu, P. Guan, M. Harris, N. L. Harris, D. Harvey, T. J. Heiman, J. R. Hernandez, J. Houck, D. Hostin, K. A. Houston, T. J. Howland, M. H. Wei, C. Ibegwam, et al. 2000. The genome sequence of Drosophila melanogaster. Science 287:2185-2195. [DOI] [PubMed] [Google Scholar]
- 2.Agaisse, H., U. M. Petersen, M. Boutros, B. Mathey-Prevot, and N. Perrimon. 2003. Signaling role of hemocytes in Drosophila JAK/STAT-dependent response to septic injury. Dev. Cell 5:441-450. [DOI] [PubMed] [Google Scholar]
- 3.Akira, S., and H. Hemmi. 2003. Recognition of pathogen-associated molecular patterns by TLR family. Immunol. Lett. 85:85-95. [DOI] [PubMed] [Google Scholar]
- 4.Alexopoulou, L., A. C. Holt, R. Medzhitov, and R. A. Flavell. 2001. Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature 413:732-738. [DOI] [PubMed] [Google Scholar]
- 5.Barillas-Mury, C., Y. S. Han, D. Seeley, and F. C. Kafatos. 1999. Anopheles gambiae Ag-STAT, a new insect member of the STAT family, is activated in response to bacterial infection. EMBO J. 18:959-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bauer, S., C. J. Kirschning, H. Hacker, V. Redecke, S. Hausmann, S. Akira, H. Wagner, and G. B. Lipford. 2001. Human TLR9 confers responsiveness to bacterial DNA via species-specific CpG motif recognition. Proc. Natl. Acad. Sci. USA 98:9237-9242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernstein, E., A. A. Caudy, S. M. Hammond, and G. J. Hannon. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409:363-366. [DOI] [PubMed] [Google Scholar]
- 8.Bonami, J. R., K. W. Hasson, J. Mari, B. T. Poulos, and D. V. Lightner. 1997. Taura syndrome of marine penaeid shrimp: characterization of the viral agent. J. Gen. Virol. 78(Pt. 2):313-319. [DOI] [PubMed] [Google Scholar]
- 9.Boutros, M., H. Agaisse, and N. Perrimon. 2002. Sequential activation of signaling pathways during innate immune responses in Drosophila. Dev. Cell 3:711-722. [DOI] [PubMed] [Google Scholar]
- 10.Caenorhabditis elegans Sequencing Consortium. 1998. Genome sequence of the nematode Caenorhabditis elegans: a platform for investigating biology. Science 282:2012-2018. [DOI] [PubMed] [Google Scholar]
- 11.Chang, C. F., M. S. Su, H. Y. Chen, and I. C. Liao. 2003. Dietary β-1,3-glucan effectively improves immunity and survival of Penaeus monodon challenged with white spot syndrome virus. Fish Shellfish Immunol. 15:297-310. [DOI] [PubMed] [Google Scholar]
- 12.Christophides, G. K., E. Zdobnov, C. Barillas-Mury, E. Birney, S. Blandin, C. Blass, P. T. Brey, F. H. Collins, A. Danielli, G. Dimopoulos, C. Hetru, N. T. Hoa, J. A. Hoffmann, S. M. Kanzok, I. Letunic, E. A. Levashina, T. G. Loukeris, G. Lycett, S. Meister, K. Michel, L. F. Moita, H. M. Muller, M. A. Osta, S. M. Paskewitz, J. M. Reichhart, A. Rzhetsky, L. Troxler, K. D. Vernick, D. Vlachou, J. Volz, C. von Mering, J. Xu, L. Zheng, P. Bork, and F. C. Kafatos. 2002. Immunity-related genes and gene families in Anopheles gambiae. Science 298:159-165. [DOI] [PubMed] [Google Scholar]
- 13.Dehal, P., Y. Satou, R. K. Campbell, J. Chapman, B. Degnan, A. De Tomaso, B. Davidson, A. Di Gregorio, M. Gelpke, D. M. Goodstein, N. Harafuji, K. E. Hastings, I. Ho, K. Hotta, W. Huang, T. Kawashima, P. Lemaire, D. Martinez, I. A. Meinertzhagen, S. Necula, M. Nonaka, N. Putnam, S. Rash, H. Saiga, M. Satake, A. Terry, L. Yamada, H. G. Wang, S. Awazu, K. Azumi, J. Boore, M. Branno, S. Chin-Bow, R. DeSantis, S. Doyle, P. Francino, D. N. Keys, S. Haga, H. Hayashi, K. Hino, K. S. Imai, K. Inaba, S. Kano, K. Kobayashi, M. Kobayashi, B. I. Lee, K. W. Makabe, C. Manohar, G. Matassi, M. Medina, Y. Mochizuki, S. Mount, T. Morishita, S. Miura, A. Nakayama, S. Nishizaka, H. Nomoto, F. Ohta, K. Oishi, I. Rigoutsos, M. Sano, A. Sasaki, Y. Sasakura, E. Shoguchi, T. Shin-i, A. Spagnuolo, D. Stainier, M. M. Suzuki, O. Tassy, N. Takatori, M. Tokuoka, K. Yagi, F. Yoshizaki, S. Wada, C. Zhang, P. D. Hyatt, F. Larimer, C. Detter, N. Doggett, T. Glavina, T. Hawkins, P. Richardson, S. Lucas, Y. Kohara, M. Levine, N. Satoh, and D. S. Rokhsar. 2002. The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science 298:2157-2167. [DOI] [PubMed] [Google Scholar]
- 14.de Veer, M. J., M. Holko, M. Frevel, E. Walker, S. Der, J. M. Paranjape, R. H. Silverman, and B. R. Williams. 2001. Functional classification of interferon-stimulated genes identified using microarrays. J. Leukoc. Biol. 69:912-920. [PubMed] [Google Scholar]
- 15.Ehrt, S., D. Schnappinger, S. Bekiranov, J. Drenkow, S. Shi, T. R. Gingeras, T. Gaasterland, G. Schoolnik, and C. Nathan. 2001. Reprogramming of the macrophage transcriptome in response to interferon-gamma and Mycobacterium tuberculosis: signaling roles of nitric oxide synthase-2 and phagocyte oxidase. J. Exp. Med. 194:1123-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fire, A., S. Xu, M. K. Montgomery, S. A. Kostas, S. E. Driver, and C. C. Mello. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391:806-811. [DOI] [PubMed] [Google Scholar]
- 17.Grebenjuk, V. A., A. Kuusksalu, M. Kelve, J. Schutze, H. C. Schroder, and W. E. Muller. 2002. Induction of (2′-5′)oligoadenylate synthetase in the marine sponges Suberites domuncula and Geodia cydonium by the bacterial endotoxin lipopolysaccharide. Eur. J. Biochem. 269:1382-1392. [DOI] [PubMed] [Google Scholar]
- 18.Hammond, S. M., E. Bernstein, D. Beach, and G. J. Hannon. 2000. An RNA-directed nuclease mediates posttranscriptional gene silencing in Drosophila cells. Nature 404:293-296. [DOI] [PubMed] [Google Scholar]
- 19.Hammond, S. M., S. Boettcher, A. A. Caudy, R. Kobayashi, and G. J. Hannon. 2001. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293:1146-1150. [DOI] [PubMed] [Google Scholar]
- 20.Hayashi, F., K. D. Smith, A. Ozinsky, T. R. Hawn, E. C. Yi, D. R. Goodlett, J. K. Eng, S. Akira, D. M. Underhill, and A. Aderem. 2001. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 410:1099-1103. [DOI] [PubMed] [Google Scholar]
- 21.Hetru, C., L. Troxler, and J. A. Hoffmann. 2003. Drosophila melanogaster antimicrobial defense. J. Infect. Dis. 187(Suppl. 2):S327-S334. [DOI] [PubMed] [Google Scholar]
- 22.Holt, R. A., G. M. Subramanian, A. Halpern, G. G. Sutton, R. Charlab, D. R. Nusskern, P. Wincker, A. G. Clark, J. M. Ribeiro, R. Wides, S. L. Salzberg, B. Loftus, M. Yandell, W. H. Majoros, D. B. Rusch, Z. Lai, C. L. Kraft, J. F. Abril, V. Anthouard, P. Arensburger, P. W. Atkinson, H. Baden, V. de Berardinis, D. Baldwin, V. Benes, J. Biedler, C. Blass, R. Bolanos, D. Boscus, M. Barnstead, S. Cai, A. Center, K. Chatuverdi, G. K. Christophides, M. A. Chrystal, M. Clamp, A. Cravchik, V. Curwen, A. Dana, A. Delcher, I. Dew, C. A. Evans, M. Flanigan, A. Grundschober-Freimoser, L. Friedli, Z. Gu, P. Guan, R. Guigo, M. E. Hillenmeyer, S. L. Hladun, J. R. Hogan, Y. S. Hong, J. Hoover, O. Jaillon, Z. Ke, C. Kodira, E. Kokoza, A. Koutsos, I. Letunic, A. Levitsky, Y. Liang, J. J. Lin, N. F. Lobo, J. R. Lopez, J. A. Malek, T. C. McIntosh, S. Meister, J. Miller, C. Mobarry, E. Mongin, S. D. Murphy, D. A. O'Brochta, C. Pfannkoch, R. Qi, M. A. Regier, K. Remington, H. Shao, M. V. Sharakhova, C. D. Sitter, J. Shetty, T. J. Smith, R. Strong, J. Sun, D. Thomasova, L. Q. Ton, P. Topalis, Z. Tu, M. F. Unger, B. Walenz, A. Wang, J. Wang, M. Wang, X. Wang, K. J. Woodford, J. R. Wortman, M. Wu, A. Yao, E. M. Zdobnov, H. Zhang, Q. Zhao, et al. 2002. The genome sequence of the malaria mosquito Anopheles gambiae. Science 298:129-149. [DOI] [PubMed] [Google Scholar]
- 23.Hoshino, K., O. Takeuchi, T. Kawai, H. Sanjo, T. Ogawa, Y. Takeda, K. Takeda, and S. Akira. 1999. Cutting edge: Toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide: evidence for TLR4 as the Lps gene product. J. Immunol. 162:3749-3752. [PubMed] [Google Scholar]
- 24.Huang, C. C., and Y. L. Song. 1999. Maternal transmission of immunity to white spot syndrome associated virus (WSSV) in shrimp (Penaeus monodon). Dev. Comp. Immunol. 23:545-552. [DOI] [PubMed] [Google Scholar]
- 25.Jacobs, B. L., and J. O. Langland. 1996. When two strands are better than one: the mediators and modulators of the cellular responses to double-stranded RNA. Virology 219:339-349. [DOI] [PubMed] [Google Scholar]
- 26.Kim, U. J., B. W. Birren, T. Slepak, V. Mancino, C. Boysen, H. L. Kang, M. I. Simon, and H. Shizuya. 1996. Construction and characterization of a human bacterial artificial chromosome library. Genomics 34:213-218. [DOI] [PubMed] [Google Scholar]
- 27.Kurtz, J., and K. Franz. 2003. Innate defense: evidence for memory in invertebrate immunity. Nature 425:37-38. [DOI] [PubMed] [Google Scholar]
- 28.Kuusksalu, A., A. Pihlak, W. E. Muller, and M. Kelve. 1995. The (2′-5′)oligoadenylate synthetase is present in the lowest multicellular organisms, the marine sponges. Demonstration of the existence and identification of its reaction products. Eur. J. Biochem. 232:351-357. [PubMed] [Google Scholar]
- 29.Leulier, F., C. Parquet, S. Pili-Floury, J. H. Ryu, M. Caroff, W. J. Lee, D. Mengin-Lecreulx, and B. Lemaitre. 2003. The Drosophila immune system detects bacteria through specific peptidoglycan recognition. Nat. Immunol. 4:478-484. [DOI] [PubMed] [Google Scholar]
- 30.Levy, D. E., and A. Garcia-Sastre. 2001. The virus battles: IFN induction of the antiviral state and mechanisms of viral evasion. Cytokine Growth Factor Rev. 12:143-156. [DOI] [PubMed] [Google Scholar]
- 31.Li, H., W. X. Li, and S. W. Ding. 2002. Induction and suppression of RNA silencing by an animal virus. Science 296:1319-1321. [DOI] [PubMed] [Google Scholar]
- 32.Li, W. X., H. Li, R. Lu, F. Li, M. Dus, P. Atkinson, E. W. Brydon, K. L. Johnson, A. Garcia-Sastre, L. A. Ball, P. Palese, and S. W. Ding. 2004. Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing. Proc. Natl. Acad. Sci. USA 101:1350-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lightner, D. V. 1996. A handbook of shrimp pathology and diagnostic procedures for diseases of cultured penaeid shrimp. World Aquaculture Society, Baton Rouge, La.
- 34.Meurs, E., K. Chong, J. Galabru, N. S. Thomas, I. M. Kerr, B. R. Williams, and A. G. Hovanessian. 1990. Molecular cloning and characterization of the human double-stranded RNA-activated protein kinase induced by interferon. Cell 62:379-390. [DOI] [PubMed] [Google Scholar]
- 35.Misquitta, L., and B. M. Paterson. 1999. Targeted disruption of gene function in Drosophila by RNA interference (RNA-i): a role for nautilus in embryonic somatic muscle formation. Proc. Natl. Acad. Sci. USA 96:1451-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oshiumi, H., M. Matsumoto, K. Funami, T. Akazawa, and T. Seya. 2003. TICAM-1, an adaptor molecule that participates in Toll-like receptor 3-mediated interferon-beta induction. Nat. Immunol. 4:161-167. [DOI] [PubMed] [Google Scholar]
- 37.Poltorak, A., X. He, I. Smirnova, M. Y. Liu, C. Van Huffel, X. Du, D. Birdwell, E. Alejos, M. Silva, C. Galanos, M. Freudenberg, P. Ricciardi-Castagnoli, B. Layton, and B. Beutler. 1998. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science 282:2085-2088. [DOI] [PubMed] [Google Scholar]
- 38.Prior, S., C. L. Browdy, E. F. Shepard, R. Laramore, and P. G. Parnell. 2003. Controlled bioassay systems for determination of lethal infective doses of tissue homogenates containing Taura syndrome or white spot syndrome virus. Dis. Aquat. Organ. 54:89-96. [DOI] [PubMed] [Google Scholar]
- 39.Schwandner, R., R. Dziarski, H. Wesche, M. Rothe, and C. J. Kirschning. 1999. Peptidoglycan- and lipoteichoic acid-induced cell activation is mediated by Toll-like receptor 2. J. Biol. Chem. 274:17406-17409. [DOI] [PubMed] [Google Scholar]
- 40.Silhavy, D., A. Molnar, A. Lucioli, G. Szittya, C. Hornyik, M. Tavazza, and J. Burgyan. 2002. A viral protein suppresses RNA silencing and binds silencing-generated, 21- to 25-nucleotide double-stranded RNAs. EMBO J. 21:3070-3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Song, Y. L., J. J. Liu, L. C. Chan, and H. H. Sung. 1997. Glucan-induced disease resistance in tiger shrimp (Penaeus monodon). Dev. Biol. Stand. 90:413-421. [PubMed] [Google Scholar]
- 42.Svoboda, P., P. Stein, H. Hayashi, and R. M. Schultz. 2000. Selective reduction of dormant maternal mRNAs in mouse oocytes by RNA interference. Development 127:4147-4156. [DOI] [PubMed] [Google Scholar]
- 43.Takahashi, Y., M. Kondo, T. Itami, T. Honda, H. Inagawa, T. Nishizawa, G. I. Soma, and Y. Yokomizo. 2000. Enhancement of disease resistance against penaeid acute viraemia and induction of virus-inactivating activity in haemolymph of kuruma shrimp, Penaeus japonicus, by oral administration of Pantoea agglomerans lipopolysaccharide (LPS). Fish Shellfish Immunol. 10:555-558. [DOI] [PubMed] [Google Scholar]
- 44.Witteveldt, J., C. C. Cifuentes, J. M. Vlak, and M. C. van Hulten. 2004. Protection of Penaeus monodon against white spot syndrome virus by oral vaccination. J. Virol. 78:2057-2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang, F., J. He, X. Lin, Q. Li, D. Pan, X. Zhang, and X. Xu. 2001. Complete genome sequence of the shrimp white spot bacilliform virus. J. Virol. 75:11811-11820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zeidler, M. P., E. A. Bach, and N. Perrimon. 2000. The roles of the Drosophila JAK/STAT pathway. Oncogene 19:2598-2606. [DOI] [PubMed] [Google Scholar]