Abstract
For most severe viral pandemics such as influenza and AIDS, the exact contribution of individual viral genes to pathogenicity is still largely unknown. A necessary step toward that understanding is a systematic comparison of different influenza virus strains at the level of transcriptional regulation in the host as a whole and interpretation of these complex genetic changes in the context of multifactorial clinical outcomes and pathology. We conducted a study by infecting pigtailed macaques (Macaca nemestrina) with a genetically reconstructed strain of human influenza H1N1 A/Texas/36/91 virus and hypothesized not only that these animals would respond to the virus similarly to humans, but that gene expression patterns in the lungs and tracheobronchial lymph nodes would fit into a coherent and complete picture of the host-virus interactions during infection. The disease observed in infected macaques simulated uncomplicated influenza in humans. Clinical signs and an antibody response appeared with induction of interferon and B-cell activation pathways, respectively. Transcriptional activation of inflammatory cells and apoptotic pathways coincided with gross and histopathological signs of inflammation, with tissue damage and concurrent signs of repair. Additionally, cDNA microarrays offered new evidence of the importance of cytotoxic T cells and natural killer cells throughout infection. With this experiment, we confirmed the suitability of the nonhuman primate model in the quest for understanding the individual and joint contributions of viral genes to influenza virus pathogenesis by using cDNA microarray technology and a reverse genetics approach.
Much progress has been made in the understanding of the structure and molecular functions of influenza virus (19, 35, 46, 54) and in the sequencing of the virus genome, most notably that of the 1918 pandemic strain (7, 57, 58, 72). New formulations for vaccines are being developed (55, 79), and new prophylactic and therapeutic drugs have provided the human population with a degree of protection and shown promise in mice, even against viruses recombined with the hemagglutinin and neuraminidase genes from the 1918 pandemic flu (69, 73). However, the genetic instability of influenza virus requires a new vaccine every year, and resistance to some of the drugs that prevent uncoating of the virus inside cells (M2 blockers) has already developed (24). Two great enigmas of pandemic influenza, particularly of the 1918 strain, are, apart from their much greater virulence, the reversal in patterns of age-specific mortality and the relatively larger proportion of deaths due to overwhelming primary viral pneumonia rather than secondary bacterial pneumonias (51, 71). The key to these features distinguishing pandemic from ordinary strains lies in the intricacies of host-virus interactions in addition to differences in existing immunity (45).
The present study was aimed at establishing a model system in nonhuman primates for the study of host-virus interactions at the genetic level and through the use of cDNA microarrays. A thorough understanding of a successful response to infection with “ordinary” influenza virus, in this case A/Texas/36/91, a strain used to infect humans in experimental settings (30, 33), is essential to elucidate how responses to more virulent strains differ and ultimately influence the outcome of infection. In the analysis of the cDNA array data, attention was given to general gene regulation patterns but with a particular focus on genes related to the innate immune response, which is believed to be a key factor in the clinical course and pathology of the disease, particularly during infection with more virulent strains (77). Other functions studied at the transcriptional level included immune cell chemotaxis and tissue transmigration, antigen presentation, T- and B-cell proliferation and activation, and apoptotic pathways. With this approach we were able to show that cDNA microarray data from relevant tissues yielded a picture that was consistent with clinical presentation, immune response, and pathology. These data will therefore be a suitable foundation for using transcriptional profiling to shed new light on the mechanisms of virulence of different influenza virus strains and subtypes.
MATERIALS AND METHODS
Animals.
Four female pigtailed macaques (Macaca nemestrina), ranging in age from 4 to 11.5 years and in weight from 5.8 to 9.8 kg, were provided by the Washington National Primate Research Center for the purpose of this study. None of these animals had received any major treatments in the past. In addition, a preliminary physical exam 2 weeks before scheduled inoculation, as well as a complete blood count, serum chemistry (both processed at the University of Washington Medical Center), and influenza virus A/B serum antibody titers (by complement fixation at Focus Technologies, Cypress, Calif.), ruled out any major health problems or prior exposure to influenza virus that would have rendered an animal unsuitable for the present study. All animals were moved into the room assigned to the study several days prior to the start of the experiment to ensure acclimation. During the study, the room was maintained at biosafety level 2 plus, and all procedures were performed according to guidelines approved by the University of Washington Environmental Health and Safety Committee, Occupational Health Administration, Primate Center Research Review Committee, and Institutional Animal Use and Care Committee.
Protocol.
The protocol was adapted from that of Rimmelzwaan et al. (44, 61, 62). The four animals were designated as either control (sham-inoculated) animals or experimental animals and matched for age and weight. One control and one experimental animal were assigned an endpoint of 4 days postinoculation, and the remaining control and experimental animals were assigned an endpoint of 7 days postinoculation. The control phase of the study was performed first, to avoid cross-infection by infected animals, and the timeline for both phases is illustrated in Fig. 1. Food consumption (Fiber Plus Monkey Diet 5049; LabDiet, PMI Nutrition International, St Louis, Mo.) was monitored for several days prior to the day of inoculation, and average daily biscuit consumption was calculated for each animal. Animals had ample amounts of body fat prior to the start of the study and continued to consume measured amounts of fruits and produce provided as part of the enrichment program.
FIG. 1.
Timeline of study. Two animals (one experimental and one control) had a 4-day endpoint, and two others had a 7-day endpoint.
All examinations and procedures were performed on the animals under chemical restraint (tiletamine-zolazepam at 2 to 10 mg/kg intramuscularly). Experimental animals were inoculated with 107 50% tissue culture infectious doses of reconstructed influenza A/Texas/36/91 virus. This infectious dose was admittedly more than that used on human subjects for this virus, which is on the order of 106 50% tissue culture infectious doses administered intranasally (30, 33), but this particular protocol, used by Rimmelzwaan et al., had proven to result in the desired level of infection in nonhuman primates.
The virus that we used was made by plasmid-based reverse genetics techniques with cDNAs corresponding to the eight RNA segments of the wild-type virus (26). The plasmid-derived recombinant virus had a replicative phenotype comparable to that of the wild-type virus in vitro and in mice. The viral suspension was diluted into 5 ml of standard bronchoalveolar lavage fluid (10) and distributed as 4 ml directly into the trachea by way of a gavage tube fitted inside an endotracheal tube; 0.5 ml was deposited onto the tonsils, and 0.25 ml was deposited onto each conjunctiva. Control animals were inoculated in the same manner but with bronchoalveolar lavage fluid alone. After the initial blood work, complete blood count and complete chemistry measurements were repeated at days 1, 3, and 4, and, for the two remaining animals, day 7 after inoculation. Influenza virus antibody titer determinations were repeated at days 3, 4, and 7 after inoculation. Additionally, bronchoalveolar lavage fluid samples were obtained by injecting and aspirating 2 ml of lavage fluid into the trachea as described above on days 3, 4, and 7 for cytology, immunohistochemistry, and virus isolation. At the same times, nasal and pharyngeal swabs were taken for virus isolation. Experimental euthanasia was achieved with an overdose of pentobarbital administered intravenously into a femoral vein. Tissue harvest at days 4 and 7 included lungs, trachea, tracheobronchial lymph nodes, heart, liver, spleen, kidneys, cerebrum, and cerebellum. Tissue samples were either saved in RNAlater (Ambion, Austin, Tex.), snap-frozen for cDNA arrays and viral isolation, or fixed in 10% formalin for histology and immunohistochemistry.
Viral isolation.
Samples set aside for viral isolation were weighed and homogenized in brain heart infusion medium (BHI) (Sigma, St. Louis, Mo.). Clarified homogenates and mucosal samples were titrated for virus infectivity from initial dilutions of 1:2 in tissues. Fifty percent egg infectious dose titers were determined by serial titration of viruses in eggs calculated by the method of Reed and Muench (56). Additionally, all samples were tested for infectious virus in a standard 48-h plaque assay performed on Madin-Darby canine kidney (MDCK) cells as described (32). The limit of virus detection was 100.8 50% egg infectious dose/g or PFU/g.
Histopathology and immunohistochemistry.
Tissues fixed in 10% neutral buffered formaldehyde were routinely processed for histopathological study. For immunohistochemical detection of virus-infected cells, deparaffinized and rehydrated sections were treated with 0.02% H2O2 in methanol for 10 min, followed by a rinse in Tris-buffered saline (TBS) and incubation in 0.1 M glycine in TBS for 15 min. Following a TBS rinse, the sections were blocked with 1% normal horse serum in TBS with Tween 20 and 0.5% bovine serum albumin. This was followed by incubation with mouse monoclonal antibody to the nucleoprotein of influenza A virus (A/Texas strain; Fitzgerald Industries Intern, Inc. Concord, Mass.) diluted 1:100 in blocking buffer and biotinylated horse anti-mouse immunoglobulin G (Vector Laboratories, Burlingame, Calif.). Bound antibody was visualized by the streptavidin-biotin-peroxidase complex procedure with diaminobenzidine or 3-amino-9-ethylcarbazole as the chromogen (70). Sections were counterstained with Harry's hematoxylin and examined in a Nikon Eclipse E600 microscope. Microphotographs were taken with a Nikon FDX-35 camera and Velvia RVP 135 film (Fujichrome). The photos were scanned with Adobe Photoshop 7.0.
Cytology.
Cells retrieved from bronchoalveolar lavage fluid were pelleted at 1,200 rpm, resuspended in phosphate-buffered saline, and either applied to glass slides with a cytospin centrifuge (Thermo Electron Corp., Waltham, Mass.) or spotted onto Teflon-coated spot slides (11, 23). Following thorough drying, the cells were fixed in 10% neutral buffered formaldehyde, rinsed in water, and stored dried until stained for virus antigen as described above.
cDNA microarrays.
Total RNA was isolated from areas of the lungs where lesions could be identified either grossly or microscopically in experimental animals and from tracheobronchial lymph nodes, the only two tissues showing pathology relevant to influenza virus infection in this experiment. After purification, reverse transcription, and amplification, the resulting cDNAs from each experimental tissue were labeled with two different dyes (indocarbocyanine and indodicarbocyanine), and the cDNAs from the corresponding controls were labeled in this manner as well. Complementary dye combinations of labeled experimental and control cDNA were then cohybridized to two replicate slides, each of which contained PCR products spotted in duplicate. On one slide, one sample was labeled with indodicarbocyanine and the other with indocarbocyanine; on the other slide, the labeling scheme was reversed. This hybridization scheme resulted in four separate measurements for each gene on the array, two in one labeling scheme and two in the other. This allowed calculation of a mean ratio and a standard deviation of the ratio, which was used as an error estimate. In addition, if one of the four measurements was an extreme outlier or was judged to have inferior spot quality by the image analysis software, it could be rejected and a standard deviation could still be calculated. The human cDNA arrays that were used in these studies were purchased from Agilent Technologies (Palo Alto, Calif.). These arrays contain duplicate spots of 13,026 unique cDNA clones (97% of which have been mapped to named human genes) and have been used extensively in our laboratory (27, 67, 75).
In addition to cohybridizing each experimental tissue with the corresponding time-matched control, cDNA of the pulmonary sample from the control animal at day 4 was cohybridized with that of the control animal at day 7 in an attempt to assess individual variability in these two healthy animals. Total gene lists of signature genes (P ≤ 0.05) in all arrays were generated in Resolver (Rosetta Inpharmatics, Kirkland, Wash.) and exported to a spreadsheet. Two main biosets were made through mining of Biocarta pathways (http://www.biocarta.com) and through addition of genes relevant to the innate immune response, immune cell migration, T- and B-cell activation and proliferation and genes relevant to apoptosis, either virus or immune cell induced. Theses sets were refined by gathering more specific information in Genecards (http://bioinformatics.weizmann.ac.il/cards/) and several immunology texts. The cDNA array comparing the two control lungs was used to eliminate the most variable genes from consideration [genes with a ≥1.5-fold change in the control versus the control array (P ≤ 0.05)], and a hierarchical cluster of each resulting bioset was generated (with mention of all genes in the biosets, for which a change of ≥1.5-fold and a P of ≤0.05 were obtained in at least two experiments). The final cluster figures were created in Spotfire Decision Site 7.1.1 by exporting the clustered data from Resolver. All primary data as well as a complete description of relevant laboratory protocols and of the Resolver System Error model are available at http://expression.microslu.washington.edu.
RESULTS
Clinical signs and ancillary findings.
The clinical signs observed in experimental animals are summarized in Fig. 2 and were overall consistent with those of human influenza. All animals maintained adequate hydration, as verified during physical exams and at necropsy, and the observed weight loss was thus due to loss of fat and possibly muscle tissue. Blood work (complete blood count and total chemistry) in experimental animals was typically nonspecific (14, 16) and complicated by the presence of a stress leukogram (also seen in control animals), but there was nevertheless lymphopenia and monocytosis in the infected macaques which was not noted in control animals.
FIG. 2.
cDNA microarray of selected genes pertaining to interferon pathways and upregulated concurrently with appearance of clinical signs in experimental animals. Lane L4, experimental versus control lungs at day 4; lane L7, experimental versus control lungs at day 7. Genes in this bioset were extracted from the cluster in appendix A, i.e., they were differentially regulated by 1.5-fold or more in at least two experiments (P ≤ 0.05) and were clustered according to a hierarchical algorithm. Red bars represent genes that were induced by the experiment; green bars represent genes that were repressed by the experiment; and darker colors represent lesser differential expression.
Live influenza A/Texas/36/91 virus was isolated from the lungs of the experimentally infected animal sacrificed at day 4 [2.37 log10 (50% egg infectious dose)/g of tissue] but not from the lungs of the experimentally infected animal sacrificed at day 7 or from any other tissues or samples at either time point. This confirmed the relatively low virulence of the Texas strain in the macaque model and suggested, at least in that animal, considerable clearance of the virus by day 7, also the time of detection of influenza virus-specific antibody in serum (titer = 1:8), performed by complement fixation with an antibody against influenza virus A soluble antigen. These results indicated that both animals had been successfully infected.
Gross pathology, histopathology, cytology, and immunohistochemistry.
The lungs and airways of the experimental macaque sacrificed at day 4 showed little gross pathology, but the animal had a mild suppurative tracheitis and mild tracheobronchial lymphadenopathy. Gross necropsy showed multifocal (on frontal and accessory lobes) to coalescing (on caudal lobes) vascular congestion, edema, and mild to moderate consolidation of the lung parenchyma of the virus-challenged macaque terminated on day 7 postinoculation. This animal also had moderate swelling of the tonsils, moderate amounts of foamy mucus in trachea and airways, and moderate tracheobronchial lymphadenopathy.
Histopathology of lung tissue is shown in Fig. 3 (day 4 postinoculation) and Fig. 4 (day 7 postinoculation), and that of tracheobronchial lymph nodes in Fig. 5. Immunohistochemistry of these tissues is shown in Fig. 6. All were consistent with progressive primary viral pneumonia as seen in humans (9, 39). There were no indications of secondary bacterial infection. The bronchoalveolar lavage fluid samples (not illustrated) on day 3 postinoculation showed disproportionate numbers of neutrophils relative to other cells, such as macrophages and epithelial cells, which normally predominate in samples from healthy animals, as they did in the controls. Influenza virus-specific labeling of cells in bronchoalveolar lavage fluid did not occur until day 4, when 80 to 90% of cells were neutrophils and the rest were a mixture of macrophages-monocytes and sloughed epithelial cells (not illustrated). That labeling was present mostly in immune cells and was both nuclear and cytoplasmic, suggestive of active phagocytosis of epithelial cells infected with the virus (15, 21). At day 7, however, cytology of the bronchoalveolar lavage fluid from the remaining animal consisted mainly of desquamated epithelial cells with only rare neutrophils and macrophages, and immunohistochemistry was negative for viral antigen, suggesting that the virus had largely been cleared (not illustrated).
FIG. 3.
Histopathology of lung tissue at day 4 postinoculation and cDNA microarrays of selected genes relevant to neutrophil and monocyte-macrophage function. The left slide shows uninfected lungs from the control animal; the right slide shows the lungs of the infected animal at day 4 postinoculation, showing pneumonia with mild alveolar septal and luminal infiltration of lymphocytes, neutrophils (black arrows), eosinophils, and macrophages (blue arrows), hypertrophy (red arrow), and hyperplasia (mitotic figures) in many pneumocytes with dark basophilic nuclei and cytoplasm but little or no apparent transudation to the alveoli. Magnification, ×70. The genes in these biosets were extracted from the cluster in appendix A, i.e., they were differentially regulated by 1.5-fold or more in at least two experiments (P ≤ 0.05) and were clustered according to a hierarchical algorithm. Red bars represent genes that were induced by the experiment; green bars represent genes that were repressed by the experiment; and darker colors represent lesser differential expression. The genes in the heat maps whose border color corresponds to a label on the histology slides are highly relevant to but not necessarily exclusive to the function of the labeled cells.
FIG. 4.
Histopathology of lung tissue at day 7 postinoculation and cDNA microarrays of selected genes relevant to neutrophil and monocyte-macrophage function. The left slide shows uninfected lungs from the control animal; the right slide (both sections) shows the lungs of the infected animal at day 7 postinoculation, showing pneumonia with alveolar flooding, hypertrophy, and hyperplasia of pneumocytes (red arrow), interstitial and alveolar infiltration by macrophages (blue arrows), lymphocytes, and a few neutrophils (black arrows), alveolar-septal necrosis (N), and areas of alveolar occlusion due to severe fibrin transudation and intra-alveolar cell migration. Hematoxylin and eosin stain. Magnification, ×70. The genes in these biosets were extracted from the cluster in appendix A, i.e., they were differentially regulated by 1.5-fold or more in at least two experiments (P ≤ 0.05) and were clustered according to a hierarchical algorithm. Red bars represent genes that were induced by the experiment; green bars represent genes that were repressed by the experiment; and darker colors represent lesser differential expression. The genes in the heat maps whose border color corresponds to a label on the histology slides are highly relevant to but not necessarily exclusive to the function of the labeled cells.
FIG. 5.
Histopathology of tracheobronchial lymph nodes. The left slide shows a quiescent node from the control animal; the middle panel shows the node from the infected animal at day 4 postinoculation, showing high cell density, dominance of small dense lymphocytes, macrophages with abundant cytoplasm (blue arrows), and golden to brown-black pigment granules and prominent high endothelial venules (green arrows); the right panel shows the node from the infected animal at day 7 postinoculation, showing diffuse edema, many activated macrophages (blue arrows) characterized by basophilic cytoplasm and nuclei with finely dispersed chromatin and a large central, magenta nucleolus (not visible at this magnification), dendritic cells (purple arrows), and hypertrophy of endothelial cells with large vesiculated nuclei (not visible at this magnification) in blood vessels (green arrows). Hematoxylin and eosin stain. Magnification, ×70.
FIG. 6.
Immunohistochemistry of lung and tracheal tissue. The left top panel shows lungs from the control animal at day 4 postinoculation, showing no influenza virus-positive cells; the left middle panel shows lungs from the infected animal at day 4 postinoculation, showing scattered influenza virus-positive pneumocytes (red arrows) and macrophages (blue arrows); the left lower panel shows lungs from the infected animal at day 7 postinfection showing scattered weak signals in macrophages (blue arrows) representing phagocytosed material from influenza virus-infected cells rather than productive infection and pneumocytes (red arrows); the right upper panel shows the trachea of the control animal at day 7 post-sham inoculation, showing no influenza virus-positive cells; the right middle panel shows the trachea from the infected animal at day 4 postinoculation, showing influenza virus-positive cells (red arrows) in addition to nonspecific binding of antibody to the basement membrane, a common occurrence enhanced by plasma transudation; note the collapse or disintegration of mucus goblet cells and atrophy of cilia due to inflammation; the right lower panel shows the trachea from the infected animal at day 7 postinoculation, showing influenza virus-positive cells (red arrows) present in a patchy pattern; note the collapse or disintegration of mucus goblet cells and the atrophy of cilia due to inflammation. Magnifications, ×100.
In both experimentally infected animals, necropsy showed livers with fatty changes and varying distention of the gall bladder, findings typical of protracted anorexia, especially in overweight animals. No pathology consistent with a viral respiratory infection or anorexia was found in either of the control animals, which were sacrificed at the same endpoints as the experimental animals (day 4 and day 7). In conclusion, histopathology and immunohistochemistry findings in virus-challenged animals indicated successful infection and progressive pathology consistent with ongoing immune-mediated clearance of the virus and with lesions seen in human patients previously naïve for the disease (16).
Regulation of immune response in lungs and tracheobronchial lymph nodes of infected animals.
Genes relevant to the immune response were clustered in Resolver (Appendix A in the supplemental material), and that cluster was organized into functional categories to illustrate associations between function and regulation (Appendix B in the supplemental material). The percentages of upregulated genes in each functional category are shown in Fig. 7.
FIG. 7.
Percentages of upregulated genes in functional categories. The percentages of clustered genes [differentially regulated by 1.5-fold or more in at least two experiments (P ≤ 0.05)] involved in general innate immune response, interferon pathways, immune cell migration, antigen presentation on MHC complexes, and presence and/or activation of neutrophils, monocytes-macrophages, dendritic cells, NK cells, T cells, and B cells and upregulated at days 4 and 7 in either lungs (upper panel) or lymph nodes (lower panel).
General innate immune response and interferon pathways.
Respiratory epithelial cells and leukocytes infected with influenza virus A (37) respond to the insult in part by secreting interferons, whose production starts about 2 h after infection and peaks 3 to 4 days later (48, 64), thereby activating antiviral defenses in neighboring cells (alpha/beta interferon response), limiting viral spread and eliminating a large proportion of the original viral load (3). The antiviral state in the lungs was evident in particular through regulation of the majority of genes enhancing or indicating the presence of an alpha/beta interferon or gamma interferon response (Fig. 2, Appendices A and B). Examples of activated genes included the human MX [homologue of murine MX1 or myxovirus (influenza virus) resistance 1], a cellular protein induced by alpha/beta interferon that inhibits influenza virus replication at the level of primary transcription (43) and gamma interferon itself. Interleukin-13, produced by activated T cells and known to inhibit gamma interferon production in monocytes (47), was consistently downregulated at day 4 in lung tissue (Appendices A and B). Likewise IRF2, a transcriptional repressor of alpha/beta interferon and of major histocompatibility complex (MHC) class I genes (31), was downregulated in lung tissue and at day 4 in tracheobronchial lymph nodes (Appendices A and B).
Induction of the interferon pathways at day 4 in the lungs also coincided with the onset of systemic signs, typically occurring 2 to 4 days after infection, and with successful isolation of live virus from lung tissue (Fig. 2). Alpha/beta interferon and gamma responses still showed signs of activation at day 7 (Fig. 2 and 7), although induction of other noncellular mediators of the innate immune response (interleukin-1RAP and certain elements of the complement pathways) tended to decrease in the two experimental animals from day 4 to 7 (Appendices A and B). Finally, both interferon responses seemed induced, albeit through different genes, in tracheobronchial lymph nodes, where gamma interferon-producing NK cells and T cells abound (Fig. 7, Appendices A and B) (65).
Immune cell migration and neutrophil and monocyte-macrophage response.
Many pneumocytes in infected lungs at day 4 were hypertrophic and had dark basophilic nuclei and cytoplasm (Fig. 3), typical of high synthetic activity of cells infected with influenza A virus and resulting in secretion of cytokines that attract and activate leukocytes (65). Light microscopy of lymph nodes showed that the endothelial cells of most blood vessels at day 7 were hypertrophic and had large vesiculated nuclei, both evidence of high synthetic activity (Fig. 5). Genes for mediators of chemotaxis, adhesion, and transmigration of immune cells through tissues, such as the chemokines CXCR3 and CXCR6, interleukin-8, CXCL11 and CXCL12, adhesion proteins AOC3 and PECAM-1, and integrins 2b, β1, and αL, were alternatively induced in lungs and tracheobronchial lymph nodes and suggested heavy traffic of immune cells into both of these tissues. Chemoattractants and other functional genes for neutrophils (interleukin-8, CCL3, AOAH, C3AR1, CSF2RB, EBI2, MNDA, etc.) tended to be most activated in the lungs at day 4 (Fig. 3 and 4 and Appendix B). Interestingly, an almost entirely different set of genes equally relevant to neutrophils and monocytes-macrophages was upregulated at day 7, particularly in tracheobronchial lymph nodes (Appendix B). Consequently, histopathology and cDNA microarrays concurred on the degree of abundance and activation of neutrophils and monocytes-macrophages in the lungs and lymph nodes (Fig. 7).
Antigen presentation (induction of MHC complexes), and T-cell, NK cell, and B-cell response.
There was overall prevalence of gene expression relevant to processing of antigens on MHC class I complexes, particularly in lung tissue (Appendix B). Even in lymph nodes, where most MHC class II-positive cells are found (12), a number of heat shock proteins were upregulated, which may be attributed to processing of antigen on MHC class I complexes, particularly calreticulin and hsp70, known to be actively involved in processing of these complexes through the endoplasmic reticulum (8). General upregulation of heat shock proteins in lymph nodes could also be caused by high mitotic activity in dividing immune cells. Tc cells and NK cells are classically affected by interaction with MHC class I complexes on the surface of infected cells. Tc cells are invariably activated by such interactions, and consistently, many genes coding for proteins relevant to T-cell function were induced at one time or another (GZMA and GZMB, TNFSF7, CD7, CCL28, FASTK, LAK, etc.) (Fig. 7 and Appendix B).
In the lungs at day 4, however, genes related to cytotoxicity were also likely to be expressed by NK cells, usually considered more active than Tc cells early on (65). Other genes almost exclusively expressed by NK cells were upregulated in lung tissue at day 4, including several inhibitory lectin-like receptors (KLRC1, KLRC3, KLRC4, and KLRD1) and several killer immunoglobulin-like receptors (KIR3DL2 and KIR2DL3) (Appendix B). These receptors interact with different components of MHC class I complexes that are normally expressed in larger numbers on the surface of uninfected cells (2, 49), and most of them were also induced at day 7 in lung tissue and day 4 in lymph nodes. Other genes of interest to NK cells were most active at day 7 in lymph nodes, such as NKG2D, which belongs to the C-type lectin family and also transmits an inhibitory signal through interactions with MHC class I complexes (5, 68).
In summary, although no genes coding for activating receptors of NK cells were included on the arrays, it is likely that both NK cells and Tc cells were present and active as early as day 4 in lung tissue, likewise in tracheobronchial lymph nodes, particularly at day 7, when cytotoxicity had to be enhanced by production of antibodies detected in serum. Gene regulation in the two experimental animals also suggested dominance of a T helper 1 response in lung tissue (IFNAR2 and gamma interferon), particularly early on. In the lymph nodes of these animals, a T helper 1 response prevailed at day 4 but appeared to give way to a T helper 2 response at day 7 (CMKBR3, interleukin-13). General T-cell attraction and activation were both suggested by the array and corroborated by lymphocyte infiltration seen on lung and lymph node histology.
Certain genes playing a role in B-cell function and proliferation (TCF3, TNFRSF17, PTPN7, NFKB1, etc), were only active or most active in lymph nodes at day 7, where and when B cells proliferate and may become activated into producing antibody as part of a T helper 2 response.
In conclusion, cDNA microarrays showed evidence of an innate immune response, characterized by infiltration, involvement, and proliferation of neutrophils, monocytes-macrophages, and NK and cytotoxic T cells as well as activation of interferon pathways and dominance of a T helper 1 over a T helper 2 response. These events were completed by development of an adaptive immune response in the form of increasing induction of a T helper 2 response, resulting in B-cell activation and production of antibodies.
Regulation of oxidative stress and apoptosis in lungs and tracheobronchial lymph nodes of infected animals.
Since oxidative and apoptotic pathways during infection can at the same time worsen and limit the inflammatory process and resultant tissue damage, regulation of related genes was assayed for in a separate bioset (Fig. 8). A number of genes relevant to apoptosis were preferentially upregulated in the lungs at day 4 (GZMA and GZMB, TNFRSF6, TNFSF6, etc.) and to a lesser extent in the lungs at day 7 and lymph nodes at day 4. For instance, several genes specific for cytotoxicity, including that for aldehyde oxidase (AOX1), an indirect catalyst for superoxide production, were induced. Although oxidative pathways play a role in antimicrobial defense, they are also a source of collateral damage to tissues. Moreover, these pathways are apparently used by viruses, particularly influenza A virus, to depress mitochondrial functions (66), although in our study the mitochondrial ribosomal protein S30 was upregulated throughout.
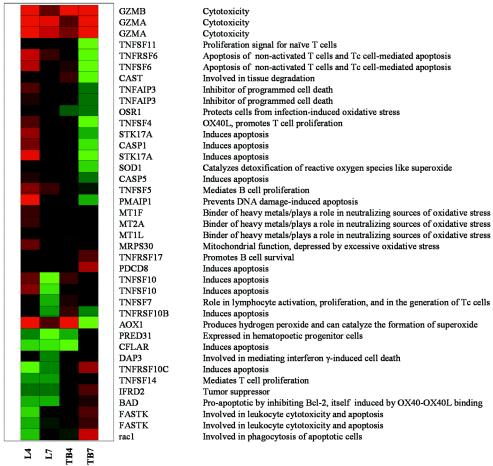
FIG. 8.
Regulation of genes relevant to apoptosis and oxidative stress. Lane L4, experimental versus control lungs at day 4; lane L7, experimental versus control lungs at day 7; lane TB4, experimental versus control tracheobronchial lymph nodes at day 4; lane TB7, experimental versus control tracheobronchial lymph nodes at day 7. The genes in this bioset [differentially regulated by 1.5-fold or more in at least two experiments (P ≤ 0.05)] were clustered according to a hierarchical algorithm; green bars represent genes that were repressed by the experiment, and darker colors represent lesser differential expression.
Regarding more general apoptosis, caspases 1, 3, and 8 are known to be activated during infection with influenza A virus, possibly through alpha/beta interferon (6). Consistently, both caspase 1, also involved in enhancing the T helper 1 response through processing of interleukin-1β and interleukin-18 (37; http://www.biocarta.com/pathfiles/h_il18Pathway.asp), and caspase 5 were upregulated, particularly at day 4 in lung tissue. In contrast, a number of genes coding for protective enzymes against oxidative damage were activated (OSR1, superoxide dismutase, and metallothioneins), as were a number of tumor necrosis factor-associated genes actually inhibiting apoptosis and promoting growth, particularly in naïve T and B cells (TNFSF5 and TNFSF14), either in lungs and lymph nodes. Several apoptosis inducers (CFLAR, DAP-3, TFRSF10C, IFRD2, FASTK, BAD, and RAC1) were downregulated in lungs but not in lymph nodes. Overall, growth and proliferation signals, in agreement with reactivity of lymph nodes and presence of mitotic figures in pneumocytes, coexisted with and even slightly dominated induction of proapoptotic and cytotoxic pathways, materialized through extensive necrosis of alveolar walls observed in consolidated areas of lung tissues at day 7 (Fig. 4, N).
DISCUSSION
With this study, we have taken steps to show that pigtailed macaques could be successfully infected with human influenza A/Texas/36/91 virus and that the resulting clinical, pathological, and gene expression pictures were likely consistent with a mild case of human influenza virus. We therefore propose that nonhuman primates will be an excellent model to assess the contributions of specific genes or motifs from different strains of influenza virus to the infectious and pathological process. In vitro studies have yielded useful information about the response of lung cells to influenza virus infection (28), but the artificial environment of cell cultures and the lack of critical interaction with components of the immune system limit their utility. Mouse models have allowed us to work with larger numbers of animals and have shown that individual genes from the 1918 influenza pandemic virus strain do in fact influence gene expression patterns and pathology in lungs infected with a mouse-adapted influenza virus (38a) and therefore are all the more likely to do so within the backbone of a human strain in a nonhuman primate model. Ferrets are also considered excellent models of human influenza (1, 60, 80), but there is a lack of ferret-specific genomic data, cDNA microarray slides, and immunological reagents which precludes their use in our studies. cDNA microarrays spotted with human genes are readily available, and nonhuman primate cDNA hybridizes satisfactorily to human cDNAs spotted on these arrays (59, 63, 74, 78).
Clinical signs, pathology, and gene expression data agreed with and completed each other.
Interferon production at day 4 postinfection, inferred from the array, is known to correlate with peak concentration of live viral particles isolated from lung tissue at that time and with maximal desquamation of infected cells (12). Desquamation of respiratory epithelial cells, confirmed by bronchoalveolar lavage, is responsible for the local symptoms of influenza virus infection that were observed in our experimental animals (9). The alpha/beta interferon and gamma pathways favor an initial T helper 1 response. Early T helper 1 responses are associated with a better prognosis for influenza virus infection than T helper 2 responses (20). Interleukin-13 is normally produced during a T helper 2 response, so its downregulation early in lung tissue and upregulation later, particularly in lymph nodes, was consistent with a protective T helper 1 response during the initial phase of infection and with a predominant T helper 2 response leading to the observed antibody production at day 7 (20).
The presumable clearance of the virus from the lungs observed at day 7, the presence of an acute titer at day 7, and its absence at day 4 also matched the course of human disease when recovery is expected (12). Therefore, the overall patterns of expression were remarkably consistent with the clinical signs, ancillary diagnostics, and pathology observed. The arrays also concurred with the body of literature supporting a critical role of Tc cells, NK cells, and neutrophils during infection with influenza viruses, in terms of cytotoxicity-induced viral clearance and pathology. Interestingly, cytotoxicity-related oxidative pathways were most intensely induced in mice infected with an adapted influenza virus (WSN) recombined with the hemagglutinin and neuraminidase genes of the highly virulent 1918 strain of influenza virus (38a).
Role of cytotoxic T cells.
The arrays pointed to the likely presence and functionality of Tc cells early on in lung tissue. Tc cells are known to be important for clearance of influenza virus (12, 25, 65), but they are believed to require activation in lymph nodes against viral nucleoproteins, matrix proteins, and polymerase proteins, usually conserved from one strain to the next (13, 29, 76), before gaining the ability to migrate to infected tissues and be effective against the virus (25). That differentiation may have taken place later, as gene regulation at day 7 suggested the presence and activation within lymph nodes of both Tc cells and NK cells, known to concentrate in T-cell areas of lymph nodes and to be required for the differentiation of influenza virus-specific Tc cells (42). This is important because Tc cells also play a role in the pathology associated with influenza virus infection that is dependent not only on the direct cytopathic effect of the virus on respiratory epithelial cells but also on the host's inflammatory response (20).
Of note, mice which failed to express OX40 (CD134) (41) or were treated with an antibody against OX40 (36) had much reduced morbidity and lung pathology but intact ability to clear virus infection. OX40 is a membrane protein on T helper and Tc cells that is normally upregulated after antigen presentation-driven activation and facilitates further stimulatory interactions between T cells and antigen-presenting cells. The cytotoxic action of NK cells and effective activation of Tc cells early in infection were credited for viral clearance in these mice, and later enhancement of the T-cell response appeared to be superfluous and harmful. OX40L (TNFSF4), the ligand for OX40 on antigen-presenting cells that becomes upregulated after interaction with T cells, was induced in our study at day 4 in lung tissue, and BAD, a protein which inhibits the antiapoptotic action of Bcl-2 induced by OX40-OX40L binding, was downregulated in the lung tissue (Appendices A and B).
We performed a Western blot confirming induction at days 4 and 7 postinfection of OX40 itself because it was not present on the array, as well a series of reverse transcription-PCRs, which confirmed the array data for tumor necrosis factor alpha, gamma interferon, interleukin-5, and Bcl-2, all of which are known to be positively regulated during induction of the OX40 pathway (34, 36, 52) (data not shown). A relative increase in expression of OX40 or OX40L during infections with more pathogenic influenza viruses will therefore be most significant.
Role of NK cells.
The intricate functioning of NK cells in this process will also need to be characterized further. Their role early in infection, which takes place within local lymph nodes as much as in tissues, is largely beneficial through cytotoxicity, production of interferon, which prolongs the protective T helper 1 response, and contribution to Tc-cell maturation (22). MHC class I complexes, expressed more easily by healthy cells, inhibit NK cells as a protective mechanism to avoid excessive tissue damage. However, it was recently established that influenza virus has evolved ways to use that mechanism to its advantage by causing infected cells to express MHC class I complexes that bind and inhibit NK cells more easily (2). MHC class I complexes appeared to be highly expressed in the tissues of our infected macaques, but this did not necessarily signify a dampening of NK cell cytotoxicity, since NK cells possess at least one activating receptor that specifically binds the viral hemagglutinin protein presented by infected cells (42). Therefore, not all interactions with antigens presented by MHC class I complexes result in inhibition of NK cells, and NK cells participate in a more specific immune response than just antibody-mediated cytotoxicity. NK cells are evidently under tight regulatory control which, if deregulated, presents the potential to significantly contribute to excessive tissue damage during infections with highly pathogenic strains of influenza virus.
Role of neutrophils.
Neutrophils are another highly significant source of tissue injury during the innate immune response (40). Consistently, mice infected with WSN expressing pandemic strain HA and NA genes were affected with more severe pneumonitis, characterized by more prominent neutrophilic pulmonary infiltration than in their counterparts infected with the wild-type WSN or with WSN expressing the HA and NA genes from a mildly pathogenic virus (38a). On the other hand, the absence of CCL3, a chemotactic factor for neutrophils and monocytes-macrophages found in our studies to be upregulated in lung tissue at day 4 postinoculation, was associated with reduced pneumonitis and reduced viral clearance in mice infected with influenza virus A (18).
While neutrophils become apoptotic after phagocytosing and destroying infected cells, as a way to limit and control tissue-damaging inflammation, certain pathogens have the ability to hasten or delay neutrophil apoptosis to their advantage (38, 65). When apoptosis is hastened, this may facilitate spread of the virus or the occurrence of secondary bacterial infections (21), and when apoptosis is delayed, excessive oxidative tissue damage (4, 50) or even downregulation of the T helper 1 response may result (4). Influenza A virus was shown by Collamussi et al. (17) and Engelich et al. (21) to accelerate the apoptosis of infected neutrophils in vitro. However, the question remains whether highly pathogenic strains of influenza virus would in fact also accelerate neutrophil apoptosis or on the contrary delay it to the point of contributing to the severe tissue damage and inflammation leading to the development of pulmonary edema (a component of acute respiratory distress syndrome), the cause of death in the most acute cases of the 1918 pandemic and other pandemics (53). Such a delay may in addition exhaust antioxidative pathways induced in the lungs of our infected animals and likely instrumental in limiting tissue damage.
Collectively, our data showed that pigtailed macaques can be used successfully to study influenza virus infection at the genetic level and that influenza A/Texas/36/91 virus causes a human-like influenza syndrome in this animal model, making that strain a suitable control and backbone for reverse genetic studies of more-pathogenic influenza viruses.
Supplementary Material
Acknowledgments
This work was in part supported by the WaNRPC through core funding from the National Institutes of Health NPRC (grant no. P51RR00166), by NIH grants P51 RR00166 and R24 RR16354 (to M. G. Katze), USDA/ARS CRIS project number 6612-32000-022-93 (to T. M. Tumpey), and NIH grant P01 AI48204 (to A. Garcia-Sastre).
We express our gratitude to Ann Schmidt, Research Scientist at the WaNRPC, for invaluable assistance, to Richard Cadagan for technical assistance, and to Jeanne H. Schickli and Peter Palese for providing reagents.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org.
REFERENCES
- 1.Abed, Y., A. M. Bourgault, R. J. Fenton, P. J. Morley, D. Gower, I. J. Owens, M. Tisdale, and G. Boivin. 2002. Characterization of 2 influenza A(H3N2) clinical isolates with reduced susceptibility to neuraminidase inhibitors due to mutations in the hemagglutinin gene. J. Infect. Dis. 186:1074-1080. [DOI] [PubMed] [Google Scholar]
- 2.Achdout, H., T. I. Arnon, G. Markel, T. Gonen-Gross, G. Katz, N. Lieberman, R. Gazit, A. Joseph, E. Kedar, and O. Mandelboim. 2003. Enhanced recognition of human NK receptors after influenza virus infection. J. Immunol. 171:915-923. [DOI] [PubMed] [Google Scholar]
- 3.Ada, G. L., and P. D. Jones. 1986. The immune response to influenza infection. Curr. Top. Microbiol. Immunol. 128:1-54. [DOI] [PubMed] [Google Scholar]
- 4.Akaike, T. 2001. Role of free radicals in viral pathogenesis and mutation. Rev. Med. Virol. 11:87-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bahram, S., M. Bresnahan, D. E. Geraghty, and T. Spies. 1994. A second lineage of mammalian major histocompatibility complex class I genes. Proc. Natl. Acad. Sci. USA 91:6259-6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balachandran, S., P. C. Roberts, T. Kipperman, K. N. Bhalla, R. W. Compans, D. R. Archer, and G. N. Barber. 2000. Alpha/beta interferons potentiate virus-induced apoptosis through activation of the FADD/caspase-8 death signaling pathway. J. Virol. 74:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Basler, C. F., A. H. Reid, J. K. Dybing, T. A. Janczewski, T. G. Fanning, H. Zheng, M. Salvatore, M. L. Perdue, D. E. Swayne, A. Garcia-Sastre, P. Palese, and J. K. Taubenberger. 2001. Sequence of the 1918 pandemic influenza virus nonstructural gene (NS) segment and characterization of recombinant viruses bearing the 1918 NS genes. Proc. Natl. Acad. Sci. USA 98:2746-2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Basu, S., R. J. Binder, T. Ramalingam, and P. K. Srivastava. 2001. CD91 is a common receptor for heat shock proteins gp96, hsp90, hsp70, and calreticulin. Immunity 14:303-313. [DOI] [PubMed] [Google Scholar]
- 9.Bender, B. S., and P. A. Small, Jr. 1992. Influenza: pathogenesis and host defense. Semin. Respir. Infect. 7:38-45. [PubMed] [Google Scholar]
- 10.Bielefeldt Ohmann, H., and L. A. Babiuk. 1986. Bovine alveolar macrophages: phenotypic and functional properties of subpopulations obtained by Percoll density gradient centrifugation. J. Leukoc. Biol. 39:167-181. [DOI] [PubMed] [Google Scholar]
- 11.Bielefeldt-Ohmann, H., D. W. Beasley, D. R. Fitzpatrick, and J. G. Aaskov. 1997. Analysis of a recombinant dengue-2 virus-dengue-3 virus hybrid envelope protein expressed in a secretory baculovirus system. J. Gen. Virol. 78:2723-2733. [DOI] [PubMed] [Google Scholar]
- 12.Bocharov, G. A., and A. A. Romanyukha. 1994. Mathematical model of antiviral immune response. III. Influenza A virus infection. J. Theor. Biol. 167:323-360. [DOI] [PubMed] [Google Scholar]
- 13.Boon, A. C., G. de Mutsert, Y. M. Graus, R. A. Fouchier, K. Sintnicolaas, A. D. Osterhaus, and G. F. Rimmelzwaan. 2002. The magnitude and specificity of influenza A virus-specific cytotoxic T-lymphocyte responses in humans is related to HLA-A and -B phenotype. J. Virol. 76:582-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Braunwald, E. 1998. Harrison's principles of internal medicine. McGraw Hill, New York, N.Y.
- 15.Cassidy, L. F., D. S. Lyles, and J. S. Abramson. 1988. Synthesis of viral proteins in polymorphonuclear leukocytes infected with influenza A virus. J. Clin. Microbiol. 26:1267-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cate, T. R. 1987. Clinical manifestations and consequences of influenza. Am. J. Med. 82:15-19. [DOI] [PubMed] [Google Scholar]
- 17.Colamussi, M. L., M. R. White, E. Crouch, and K. L. Hartshorn. 1999. Influenza A virus accelerates neutrophil apoptosis and markedly potentiates apoptotic effects of bacteria. Blood 93:2395-2403. [PubMed] [Google Scholar]
- 18.Cook, D. N., M. A. Beck, T. M. Coffman, S. L. Kirby, J. F. Sheridan, I. B. Pragnell, and O. Smithies. 1995. Requirement of MIP-1 alpha for an inflammatory response to viral infection. Science 269:1583-1585. [DOI] [PubMed] [Google Scholar]
- 19.De Jong, J. C., G. F. Rimmelzwaan, R. A. Fouchier, and A. D. Osterhaus. 2000. Influenza virus: a master of metamorphosis. J. Infect. 40:218-228. [DOI] [PubMed] [Google Scholar]
- 20.Durbin, J. E., A. Fernandez-Sesma, C. K. Lee, T. D. Rao, A. B. Frey, T. M. Moran, S. Vukmanovic, A. Garcia-Sastre, and D. E. Levy. 2000. Type I IFN modulates innate and specific antiviral immunity. J. Immunol. 164:4220-4228. [DOI] [PubMed] [Google Scholar]
- 21.Engelich, G., M. White, and K. L. Hartshorn. 2002. Role of the respiratory burst in cooperative reduction in neutrophil survival by influenza A virus and Escherichia coli. J. Med. Microbiol. 51:484-490. [DOI] [PubMed] [Google Scholar]
- 22.Fehniger, T. A., M. A. Cooper, G. J. Nuovo, M. Cella, F. Facchetti, M. Colonna, and M. A. Caligiuri. 2003. CD56bright natural killer cells are present in human lymph nodes and are activated by T cell-derived IL-2: a potential new link between adaptive and innate immunity. Blood 101:3052-3057. [DOI] [PubMed] [Google Scholar]
- 23.Fitzpatrick, D. R., M. Snider, L. McDougall, T. Beskorwayne, L. A. Babiuk, T. J. Zamb, and H. Bielefeldt Ohmann. 1990. Molecular mimicry: a herpes virus glycoprotein antigenically related to a cell-surface glycoprotein expressed by macrophages, polymorphonuclear leucocytes, and platelets. Immunology 70:504-512. [PMC free article] [PubMed] [Google Scholar]
- 24.Fleming, D. M. 2003. Zanamivir in the treatment of influenza. Expert Opin. Pharmacother. 4:799-805. [DOI] [PubMed] [Google Scholar]
- 25.Flynn, K. J., G. T. Belz, J. D. Altman, R. Ahmed, D. L. Woodland, and P. C. Doherty. 1998. Virus-specific CD8+ T cells in primary and secondary influenza pneumonia. Immunity 8:683-691. [DOI] [PubMed] [Google Scholar]
- 26.Fodor, E., L. Devenish, O. G. Engelhardt, P. Palese, G. G. Brownlee, and A. Garcia-Sastre. 1999. Rescue of influenza A virus from recombinant DNA. J. Virol. 73:9679-9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geiss, G. K., V. S. Carter, Y. He, B. K. Kwieciszewski, T. Holzman, M. J. Korth, C. A. Lazaro, N. Fausto, R. E. Bumgarner, and M. G. Katze. 2003. Gene expression profiling of the cellular transcriptional network regulated by alpha/beta interferon and its partial attenuation by the hepatitis C virus nonstructural 5A protein. J. Virol. 77:6367-6375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Geiss, G. K., M. Salvatore, T. M. Tumpey, V. S. Carter, X. Wang, C. F. Basler, J. K. Taubenberger, R. E. Bumgarner, P. Palese, M. G. Katze, and A. Garcia-Sastre. 2002. Cellular transcriptional profiling in influenza A virus-infected lung epithelial cells: the role of the nonstructural NS1 protein in the evasion of the host innate defense and its potential contribution to pandemic influenza. Proc. Natl. Acad. Sci. USA 99:10736-10741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gog, J. R., G. F. Rimmelzwaan, A. D. Osterhaus, and B. T. Grenfell. 2003. Population dynamics of rapid fixation in cytotoxic T lymphocyte escape mutants of influenza A. Proc. Natl. Acad. Sci. USA 100:11143-11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gubareva, L. V., L. Kaiser, M. N. Matrosovich, Y. Soo-Hoo, and F. G. Hayden. 2001. Selection of influenza virus mutants in experimentally infected volunteers treated with oseltamivir. J Infect. Dis. 183:523-531. [DOI] [PubMed] [Google Scholar]
- 31.Harada, H., T. Fujita, M. Miyamoto, Y. Kimura, M. Maruyama, A. Furia, T. Miyata, and T. Taniguchi. 1989. Structurally similar but functionally distinct factors, IRF-1 and IRF-2, bind to the same regulatory elements of IFN and IFN-inducible genes. Cell 58:729-739. [DOI] [PubMed] [Google Scholar]
- 32.Hayden, F. G., K. M. Cote, and R. G. Douglas, Jr. 1980. Plaque inhibition assay for drug susceptibility testing of influenza viruses. Antimicrob. Agents Chemother. 17:865-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hayden, F. G., J. J. Treanor, R. S. Fritz, M. Lobo, R. F. Betts, M. Miller, N. Kinnersley, R. G. Mills, P. Ward, and S. E. Straus. 1999. Use of the oral neuraminidase inhibitor oseltamivir in experimental human influenza: randomized controlled trials for prevention and treatment. JAMA 282:1240-1246. [DOI] [PubMed] [Google Scholar]
- 34.Higgins, L. M., S. A. McDonald, N. Whittle, N. Crockett, J. G. Shields, and T. T. MacDonald. 1999. Regulation of T cell activation in vitro and in vivo by targeting the OX40-OX40 ligand interaction: amelioration of ongoing inflammatory bowel disease with an OX40-IgG fusion protein, but not with an OX40 ligand-IgG fusion protein. J. Immunol. 162:486-493. [PubMed] [Google Scholar]
- 35.Hilleman, M. R. 2002. Realities and enigmas of human viral influenza: pathogenesis, epidemiology and control. Vaccine 20:3068-3087. [DOI] [PubMed] [Google Scholar]
- 36.Humphreys, I. R., G. Walzl, L. Edwards, A. Rae, S. Hill, and T. Hussell. 2003. A critical role for OX40 in T cell-mediated immunopathology during lung viral infection. J. Exp. Med. 198:1237-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Julkunen, I., T. Sareneva, J. Pirhonen, T. Ronni, K. Melen, and S. Matikainen. 2001. Molecular pathogenesis of influenza A virus infection and virus-induced regulation of cytokine gene expression. Cytokine Growth Factor Rev. 12:171-180. [DOI] [PubMed] [Google Scholar]
- 38.Kaplanski, G., V. Marin, F. Montero-Julian, A. Mantovani, and C. Farnarier. 2003. IL-6: a regulator of the transition from neutrophil to monocyte recruitment during inflammation. Trends Immunol. 24:25-29. [DOI] [PubMed] [Google Scholar]
- 38a.Kash, J. C., C. F. Basler, A. García-Sastre, V. Carter, R. Billharz, D. E. Swayne, R. M. Przygodzki, and J. K. Taubenberger. 2004. Global host immune response: pathogenesis and transcriptional profiling of type A influenza viruses expressing the hemagglutinin and neuraminidase genes from the 1918 pandemic virus. J. Virol. 78:9499-9511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim, E. A., K. S. Lee, S. L. Primack, H. K. Yoon, H. S. Byun, T. S. Kim, G. Y. Suh, O. J. Kwon, and J. Han. 2002. Viral pneumonias in adults: radiologic and pathologic findings. Radiographics Spec No. 22:S137-S149. [DOI] [PubMed] [Google Scholar]
- 40.Kobayashi, S. D., K. R. Braughton, A. R. Whitney, J. M. Voyich, T. G. Schwan, J. M. Musser, and F. R. DeLeo. 2003. Bacterial pathogens modulate an apoptosis differentiation program in human neutrophils. Proc. Natl. Acad. Sci. USA 100:10948-10953. [DOI] [PMC free article] [PubMed]
- 41.Kopf, M., C. Ruedl, N. Schmitz, A. Gallimore, K. Lefrang, B. Ecabert, B. Odermatt, and M. F. Bachmann. 1999. OX40-deficient mice are defective in Th cell proliferation but are competent in generating B cell and CTL Responses after virus infection. Immunity 11:699-708. [DOI] [PubMed] [Google Scholar]
- 42.Kos, F. J., and E. G. Engleman. 1996. Role of natural killer cells in the generation of influenza virus-specific cytotoxic T cells. Cell. Immunol. 173:1-6. [DOI] [PubMed] [Google Scholar]
- 43.Krug, R. M., M. Shaw, B. Broni, G. Shapiro, and O. Haller. 1985. Inhibition of influenza viral mRNA synthesis in cells expressing the interferon-induced Mx gene product. J. Virol. 56:201-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuiken, T., G. F. Rimmelzwaan, G. Van Amerongen, and A. D. Osterhaus. 2003. Pathology of human influenza A (H5N1) virus infection in cynomolgus macaques (Macaca fascicularis). Vet. Pathol. 40:304-310. [DOI] [PubMed] [Google Scholar]
- 45.Meltzer, M. I., N. J. Cox, and K. Fukuda. 1999. The economic impact of pandemic influenza in the United States: priorities for intervention. Emerg. Infect. Dis. 5:659-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mikulasova, A., E. Vareckova, and E. Fodor. 2000. Transcription and replication of the influenza a virus genome. Acta Virol. 44:273-282. [PubMed] [Google Scholar]
- 47.Minty, A., P. Chalon, J. M. Derocq, X. Dumont, J. C. Guillemot, M. Kaghad, C. Labit, P. Leplatois, P. Liauzun, B. Miloux, et al. 1993. Interleukin-13 is a new human lymphokine regulating inflammatory and immune responses. Nature 362:248-250. [DOI] [PubMed] [Google Scholar]
- 48.Mogensen, S. C., and J. L. Virelizier. 1987. The interferon-macrophage alliance. Interferon 8:55-84. [PubMed] [Google Scholar]
- 49.Moretta, A., C. Bottino, S. Sivori, E. Marcenaro, R. Castriconi, M. Della Chiesa, S. Carlomagno, R. Augugliaro, M. Nanni, M. Vitale, and R. Millo. 2001. Natural killer lymphocytes: “null cells” no more. Ital. J. Anat. Embryol. 106:335-342. [PubMed] [Google Scholar]
- 50.Nathan, C. 2002. Points of control in inflammation. Nature 420:846-852. [DOI] [PubMed] [Google Scholar]
- 51.Nguyen-Van-Tam, J. S., and A. W. Hampson. 2003. The epidemiology and clinical impact of pandemic influenza. Vaccine 21:1762-1768. [DOI] [PubMed] [Google Scholar]
- 52.Obermeier, F., H. Schwarz, N. Dunger, U. G. Strauch, N. Grunwald, J. Scholmerich, and W. Falk. 2003. OX40/OX40L interaction induces the expression of CXCR5 and contributes to chronic colitis induced by dextran sulfate sodium in mice. Eur. J. Immunol. 33:3265-3274. [DOI] [PubMed] [Google Scholar]
- 53.Oxford, J. S. 2000. Influenza A pandemics of the 20th century with special reference to 1918: virology, pathology and epidemiology. Rev. Med. Virol. 10:119-133. [DOI] [PubMed] [Google Scholar]
- 54.Palese, P. 1977. The genes of influenza virus. Cell 10:1-10. [DOI] [PubMed] [Google Scholar]
- 55.Palese, P., and A. Garcia-Sastre. 2002. Influenza vaccines: present and future. J. Clin. Investig. 110:9-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reed, L., J., and H. Muench. 1938. A simple method of estimating fifty per cent endpoints. Am. J. Hyg. 27:493-497. [Google Scholar]
- 57.Reid, A. H., T. G. Fanning, J. V. Hultin, and J. K. Taubenberger. 1999. Origin and evolution of the 1918 “Spanish” influenza virus hemagglutinin gene. Proc. Natl. Acad. Sci. USA 96:1651-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reid, A. H., T. G. Fanning, T. A. Janczewski, and J. K. Taubenberger. 2000. Characterization of the 1918 “Spanish” influenza virus neuraminidase gene. Proc. Natl. Acad. Sci. USA 97:6785-6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reinhart, T. A., B. A. Fallert, M. E. Pfeifer, S. Sanghavi, S. Capuano, 3rd, P. Rajakumar, M. Murphey-Corb, R. Day, C. L. Fuller, and T. M. Schaefer. 2002. Increased expression of the inflammatory chemokine CXC chemokine ligand 9/monokine induced by interferon-gamma in lymphoid tissues of rhesus macaques during simian immunodeficiency virus infection and acquired immunodeficiency syndrome. Blood 99:3119-3128. [DOI] [PubMed] [Google Scholar]
- 60.Renegar, K. B. 1992. Influenza virus infections and immunity: a review of human and animal models. Lab. Anim. Sci. 42:222-232. [PubMed] [Google Scholar]
- 61.Rimmelzwaan, G. F., T. Kuiken, G. van Amerongen, T. M. Bestebroer, R. A. Fouchier, and A. D. Osterhaus. 2001. Pathogenesis of influenza A (H5N1) virus infection in a primate model. J. Virol. 75:6687-6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rimmelzwaan, G. F., T. Kuiken, G. van Amerongen, T. M. Bestebroer, R. A. Fouchier, and A. D. Osterhaus. 2003. A primate model to study the pathogenesis of influenza A (H5N1) virus infection. Avian Dis. 47:931-933. [DOI] [PubMed] [Google Scholar]
- 63.Roberts, E. S., M. A. Zandonatti, D. D. Watry, L. J. Madden, S. J. Henriksen, M. A. Taffe, and H. S. Fox. 2003. Induction of pathogenic sets of genes in macrophages and neurons in NeuroAIDS. Am. J. Pathol. 162:2041-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Roberts, N. J., Jr., R. G. Douglas, Jr., R. M. Simons, and M. E. Diamond. 1979. Virus-induced interferon production by human macrophages. J. Immunol. 123:365-369. [PubMed] [Google Scholar]
- 65.Roitt, I. M., J. Brostoff, and D. K. Male. 2001. Immunology. Mosby, New York, N.Y.
- 66.Schwarz, K. B. 1996. Oxidative stress during viral infection: a review. Free Radic. Biol. Med. 21:641-649. [DOI] [PubMed] [Google Scholar]
- 67.Smith, M. W., Z. N. Yue, G. K. Geiss, N. Y. Sadovnikova, V. S. Carter, L. Boix, C. A. Lazaro, G. B. Rosenberg, R. E. Bumgarner, N. Fausto, J. Bruix, and M. G. Katze. 2003. Identification of novel tumor markers in hepatitis C virus-associated hepatocellular carcinoma. Cancer Res. 63:859-864. [PubMed] [Google Scholar]
- 68.Stephens, H. A. 2001. MICA and MICB genes: can the enigma of their polymorphism be resolved? Trends Immunol. 22:378-385. [DOI] [PubMed] [Google Scholar]
- 69.Stiver, G. 2003. The treatment of influenza with antiviral drugs. Cmaj 168:49-56. [PMC free article] [PubMed] [Google Scholar]
- 70.Swasdipan, S., M. McGowan, N. Phillips, and H. Bielefeldt-Ohmann. 2002. Pathogenesis of transplacental virus infection: pestivirus replication in the placenta and fetus following respiratory infection. Microb. Pathog. 32:49-60. [DOI] [PubMed] [Google Scholar]
- 71.Taubenberger, J. K., A. H. Reid, and T. G. Fanning. 2000. The 1918 influenza virus: A killer comes into view. Virology 274:241-245. [DOI] [PubMed] [Google Scholar]
- 72.Taubenberger, J. K., A. H. Reid, A. E. Krafft, K. E. Bijwaard, and T. G. Fanning. 1997. Initial genetic characterization of the 1918 “Spanish” influenza virus. Science 275:1793-1796. [DOI] [PubMed] [Google Scholar]
- 73.Tumpey, T. M., A. Garcia-Sastre, A. Mikulasova, J. K. Taubenberger, D. E. Swayne, P. Palese, and C. F. Basler. 2002. Existing antivirals are effective against influenza viruses with genes from the 1918 pandemic virus. Proc. Natl. Acad. Sci. USA 99:13849-13854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vahey, M. T., M. E. Nau, M. Taubman, J. Yalley-Ogunro, P. Silvera, and M. G. Lewis. 2003. Patterns of gene expression in peripheral blood mononuclear cells of rhesus macaques infected with SIVmac251 and exhibiting differential rates of disease progression. AIDS Res. Hum. Retroviruses 19:369-387. [DOI] [PubMed] [Google Scholar]
- 75.van 't Wout, A. B., G. K. Lehrman, S. A. Mikheeva, G. C. O'Keeffe, M. G. Katze, R. E. Bumgarner, G. K. Geiss, and J. I. Mullins. 2003. Cellular gene expression upon human immunodeficiency virus type 1 infection of CD4+-T-cell lines. J. Virol. 77:1392-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Voeten, J. T., T. M. Bestebroer, N. J. Nieuwkoop, R. A. Fouchier, A. D. Osterhaus, and G. F. Rimmelzwaan. 2000. Antigenic drift in the influenza A virus (H3N2) nucleoprotein and escape from recognition by cytotoxic T lymphocytes. J. Virol. 74:6800-6807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yewdell, J., and A. Garcia-Sastre. 2002. Influenza virus still surprises. Curr. Opin. Microbiol. 5:414-418. [DOI] [PubMed] [Google Scholar]
- 78.Yokota, C., H. Inoue, Y. Kuge, T. Abumiya, M. Tagaya, Y. Hasegawa, N. Ejima, N. Tamaki, and K. Minematsu. 2003. Cyclooxygenase-2 expression associated with spreading depression in a primate model. J. Cereb. Blood Flow Metab. 23:395-398. [DOI] [PubMed] [Google Scholar]
- 79.Zangwill, K. M. 2003. Cold-adapted, live attenuated intranasal influenza virus vaccine. Pediatr. Infect. Dis. J. 22:273-274. [DOI] [PubMed] [Google Scholar]
- 80.Zitzow, L. A., T. Rowe, T. Morken, W. J. Shieh, S. Zaki, and J. M. Katz. 2002. Pathogenesis of avian influenza A (H5N1) viruses in ferrets. J. Virol. 76:4420-4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.