Abstract
The human cytomegalovirus tegument protein, pTRS1, appears to function at several discrete stages of the virus replication cycle. We previously demonstrated that pTRS1 acts during the late phase of infection to facilitate the production of infectious virions. We now have more precisely identified the late pTRS1 function by further study of a mutant virus lacking the TRS1 region, ADsubTRS1. We observed a significant reduction in the production of capsids, especially DNA-containing C-capsids, in mutant virus-infected cells. ADsubTRS1 exhibited normal cleavage of DNA concatemers, so the defect in C-capsid production must occur after DNA cleavage and before DNA is stably inserted into a capsid. Further, the normal virus-induced morphological reorganization of the nucleus did not occur after infection with the pTRS1-deficient mutant.
Cytomegalovirus, a member of the betaherpesvirus subfamily, is a large DNA virus with a narrow host range (19). Human cytomegalovirus (HCMV) infections are widespread and generally asymptomatic in healthy individuals. After infection, the virus can persist in a lifelong, latent state. Although relatively little is known about the events involved the establishment of HCMV latency, the cascade of gene expression and the events that occur during the lytic replication cycle of the virus have been studied in considerable detail (24).
The genome of HCMV contains two unique domains, one long and one short, each flanked by repeat sequences. Two open reading frames, TRS1 and IRS1, include sequence from both repeated and unique domains. The N-terminal two-thirds of pTRS1 is encoded in the c repeat region, and the remainder of the protein is coded within the unique short region. The related protein, pIRS1, is encoded in the internal or c′ repeat region plus the adjacent unique short region. Consequently, the N-terminal domains of pTRS1 and pIRS1 are nearly identical, and the two proteins have different C-terminal domains (37). Because their amino-terminal domains are encoded in the repeat region, the transcription of these genes is controlled by identical immediate-early promoters. Both pTRS1 and pIRS1 are packaged into the virion and therefore are delivered to the cell immediately upon infection (32); pTRS1 and pIRS1 can be detected in infected cells as early as 2 h postinfection (33, 36).
The first function ascribed to pTRS1 and pIRS1 was transcriptional activation. Both proteins were found to act in conjunction with the immediate-early transcriptional regulatory proteins, IE1 and IE2, but not on their own, to increase expression from the UL44 promoter in transient-transfection assays (36). Subsequent analysis identified TRS1/IRS1 as 1 of 11 loci that are required for transient complementation of HCMV oriLyt-dependent DNA replication (28). In this assay, pTRS1 and pIRS1 likely facilitate the accumulation of other proteins that function directly in the replication process (15). More recently, pTRS1 and pIRS1 have been shown to block the double-stranded RNA-dependent protein kinase R response pathway, an antiviral pathway that shuts down protein synthesis in infected cells (5). Finally, we have shown that pTRS1, but not pIRS1, acts late during the virus replication cycle to facilitate the production of virions (3).
Our analysis of pTRS1 function used a mutant virus, ADsubTRS1, that lacks the central portion of the TRS1 open reading frame (3). Although DNA replication was normal and there was no significant difference in the accumulation of six viral mRNAs in mutant compared to wild-type virus-infected fibroblasts, ADsubTRS1 produced ≥10-fold-reduced yields of both extracellular and intracellular virus. In addition, the localization of the pp65 and pp71 tegument proteins was found to be abnormal in mutant virus-infected cells. After infection with ADsubTRS1, these two tegument proteins localized exclusively to the nucleus and did not move to the cytoplasmic sites of virus assembly, as happens after infection with wild-type virus. Since this defect was not complemented by protein expressed from the corresponding open reading frame, IRS1, it was clear that this was a separate function from those exhibited by both proteins and, therefore, the activity was ascribed to the unique C-terminal one-third of pTRS1. Indeed, deletion of the pIRS1 coding region produces a viable virus that replicates with wild-type kinetics (3, 17).
Our earlier analysis localized the ADsubTRS1 defect to the late phase of the replication cycle. As viral DNA accumulates during the late phase of infection, structural proteins are produced, capsid shells are assembled, DNA is inserted into the shells, mature capsids associated with some tegument proteins move from the nucleus to the cytoplasm, additional tegument proteins are added, and the capsid with tegument proteins are enveloped in a virus-modified membrane to produce an infectious virion (reviewed in reference 7). Now we demonstrate that the production of DNA-filled C-capsids and nuclear reorganization induced by HCMV infection both require pTRS1 function.
MATERIALS AND METHODS
Cells and viruses.
Primary human foreskin fibroblasts were cultured in medium containing 10% fetal bovine serum or 10% newborn calf serum. The wild-type AD169 strain of HCMV (ADwt) and two substitution mutants derived from AD169, ADsubTRS1 and ADsubIRS1 (3), were propagated and studied in fibroblasts.
Microarray analysis.
At 72 h postinfection, polyadenylated RNA was isolated from cells by using the MicroPoly(A)Pure mRNA purification kit according to the manufacturer's protocol (Ambion). Cy3-dUTP-labeled cDNA was produced from 5 μg of RNA. Microarray slides were made as previously described (9). Each array contained PCR-amplified cDNA segments corresponding to the known HCMV open reading frames, as well as control cellular genes, and DNAs were printed in triplicate on amino-silane-coated glass slides. The cDNA (60 μl) was then hybridized to arrays at 42°C for 14 h (11). Arrays were washed and then scanned by using a GenePix 4000B scanner (Axon Instruments) at a 10-μm resolution. Analysis of the data was done with GenePix Pro 4.0 software (Axon). The Axon software was set to subtract both local and hybridization background values. After aberrant spots were flagged, the mean intensity of each spot was calculated, and the average values were normalized by using the cellular cDNAs. Values calculated for the cellular cDNAs were used for comparisons between slides.
Thin-section electron microscopy.
Fibroblasts were infected with ADwt, ADsubTRS1, or ADsubIRS1 at a multiplicity of infection (MOI) of 0.5 PFU/cell. At 72 h postinfection cells were washed once with phosphate-buffered saline (PBS) and fixed in 3% glutaraldehyde in sodium cacodylate buffer at room temperature for 1 h. Staining was performed by resuspending the cell pellet in an aqueous solution of 1% osmium tetroxide and 2% uranyl acetate. The stained cells were dehydrated through ethanol steps and then embedded in a blend of EmBed 812, dodecyl succinic anhydride, and nadic methyl anhydride resins (Electron Microscopy Sciences). Thin sections were prepared and post-sectioning staining was performed with aqueous 2% uranyl acetate and modified Sato's lead citrate (10) before sections were viewed at 80 kV with a Leo912a electron microscope.
Sucrose gradient analysis of capsids.
The basic protocol used was described previously (26), but several modification were used. Fibroblasts were infected with ADwt or ADsubTRS1 at an MOI of 0.5 PFU/cell. At 72 h postinfection the cells were washed in TNE buffer (20 mM Tris [pH 7.4], 500 mM NaCl, 1 mM EDTA) and then lysed in the same buffer with 1% Triton. Cellular membranes were disrupted by three freeze-thaw cycles, and particles in the lysate were pelleted through a 2-ml 35% sucrose cushion at 20,000 rpm in an SW41 rotor for 75 min. The pellet was resuspended in a small volume of TNE and treated with RQ1 DNase I (Promega), 8 mM dithiothreitol, and 4 mM MgCl2 to digest unencapsidated DNA. The lysates were sonicated to disperse aggregates and layered onto 20 to 65% sucrose gradients, and the gradients were spun at 40,000 rpm in an SW41 rotor for 20 min. When viewed with top illumination from a fiber optic lamp, bands corresponding to A-, B-, and C-capsids were visible. The gradient was fractionated, and a portion of each fraction was analyzed by Western blotting. After electrophoresis through a 10% sodium dodecyl sulfate (SDS)-containing gel, proteins were transferred to a Protran membrane (Schleicher & Schuell) and then blocked in 0.5% bovine serum albumin. Membranes were probed with antibodies to the major capsid protein (UL86; monoclonal antibody [MAb] 28-4) and minor capsid protein (UL85; MAb 85-13), both gifts from W. Gibson (The Johns Hopkins University School of Medicine). The secondary antibody was an 125I-conjugated goat anti-mouse immunoglobulin G (Perkin-Elmer). Membranes were exposed to a phosphor screen for accurate quantitation. A Storm PhosphorImager (Molecular Dynamics) was used for scanning; ImageQuant software was used for data analysis.
For analysis of the virion DNA content, 100-μl portions of each fraction were incubated for 2 h at 37°C in an equal volume of 2× capsid lysis buffer (200 mM NaCl, 20 mM Tris [pH 7.4], 50 mM EDTA, 1.2% SDS, 0.2 mg of proteinase K/ml). The DNA was denatured for 1 h at 65°C with one-tenth volume of 3 M NaOH and then adjusted to a final salt concentration of 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Precipitated material was removed by a 2-min centrifugation in a microfuge. The supernatant was analyzed by slot blot assay by using a probe to a 1,697-bp fragment containing the HCMV UL82 gene prepared by random priming with the Klenow fragment of DNA polymerase I in the presence of [32P]dCTP (6).
Immunofluoresence.
Fibroblasts were grown to near confluence on sterile glass coverslips in six-well plates. Cells were then infected with ADwt or ADsubTRS1 at an MOI of 0.5 PFU/cell. After 72 h cells were washed in PBS, fixed for 15 min in 2% paraformaldehyde in PBS, washed again with PBS, and permeabilized for 15 min in 0.1% Triton X-100 in PBS. After a wash with PBS containing 0.2% Tween 20, the cells were incubated for 1 h in PBS-blocking buffer containing 2% bovine serum albumin and 0.2% Tween 20. Cells were then incubated in PBS-blocking buffer for 1 h at room temperature with a mouse MAb to the major capsid protein (pUL86, clone MAb 28-4, diluted 1:5 [W. Gibson, The Johns Hopkins University School of Medicine]), minor capsid protein (pUL85, clone MAb 85-13, diluted 1:5 [W. Gibson]), smallest capsid protein (pUL48.5, clone 11-2-23, diluted 1:5 [W. Britt, University of Alabama School of Medicine]), polymerase accessory protein (pUL44, diluted 1:400 [Virosys]), replication compartment protein (pUL112-113, diluted 1:5 [E. Bogner, Universität Erlangen-Nürnberg]), or the virus-coded DNase (pUL98, diluted 1:5). After a further wash with PBS containing 0.2% Tween 20, slides were incubated for 30 min at room temperature with a goat anti-mouse- or anti-rabbit-Alexa 546 conjugate (diluted 1:1,000; Molecular Probes). The secondary antibody was supplemented with 1 ng of DAPI (4′,6′-diamidino-2-phenylindole; Molecular Probes)/ml to counterstain chromosomal DNA. After the final wash, the coverslips were mounted with Slow-Fade (Molecular Probes) to preserve samples on microscope slides. Microscopy and image acquisition was carried out with a Zeiss LSM510 confocal microscope.
FISH.
The fluorescence in situ hybridization (FISH) technique was described previously (20). Briefly, 72 h after fibroblasts were infected with ADwt, ADsubTRS1, or ADsubIRS1 at an MOI of 0.5 PFU/cell, cells were harvested, treated with 75 mM KCl for 20 min at 37°C, and fixed in methanol-acetic acid (3:1) at 4°C. Spread chromosomes and interphase nuclei were treated with DNase-free RNase A (100 mg/ml; Sigma) in 2× SSC prior to hybridization. DNA probes were labeled with biotin-16-dUTP (Roche) by nick translation (31). The DNA fragment length was assessed to be 200 to 500 bp. The denatured probe (50 to 100 ng) was hybridized to chromosomal DNA in 50% formamide-2× SSC-10% dextran sulfate-0.1% SDS at 37°C under a sealed coverslip. After a washing step, samples were preincubated for 30 min in a blocking solution (3% blocking reagent [Roche] in 4× SSC) and then incubated for 30 min at 37°C with avidin-fluorescein isothiocyanate (Vector) diluted 1:400 in blocking solution. Samples were washed and, for amplification of the fluorescent signal, treated sequentially with avidin-specific biotinylated antibody (Vector) diluted 1:40 in 4× SSC and with avidin-fluorescein isothiocyanate diluted 1:400 in blocking solution. Chromosomal DNA was counterstained with propidium iodide, and the samples were mounted in Slow-Fade. Images were captured on a Zeiss LSM510 confocal microscope.
DNA analysis.
To study cleavage of the concatemeric DNA, a protocol modified from (23) was used; fibroblasts were infected at an MOI of 0.5 PFU/cell, and total DNA was prepared. HpaI-restricted DNA fragments were electrophoretically separated on a 0.8% agarose gel and subjected to Southern blot analysis. A random-primed, 32P-labeled, 500-bp SacI-AatII fragment corresponding to the viral RL3 gene was used to probe HCMV genomic termini. This probe recognizes both the terminal repeat long (TRL) and the inverted repeat long (IRL) regions. As a control, ADwt DNA, purified from virions, was processed in the same way. Bands were quantified by using the Molecular Dynamics PhosphorImager system and ImageQuant software.
RESULTS
No alterations are detected in ADsubTRS1 gene expression.
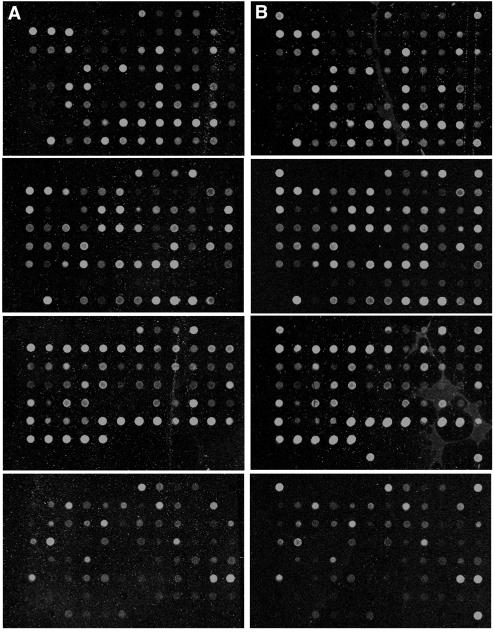
Earlier analysis of the ADsubTRS1 pTRS1-deficient mutant indicated that its defect occurs after viral DNA replication and the synthesis of late proteins but before infectious particles are assembled (3). Since pTRS1 is expressed throughout the entire replication cycle, it is possible that one or more viral proteins are not produced in cells infected with the ADsubTRS1 mutant as a result of the loss of pTRS1 transactivation activity. However, nine virus-coded proteins were tested by Western blotting, and they all accumulated to normal levels in cells infected with the mutant (3). To further test the possibility that the loss of pTRS1 transactivation activity inhibited the expression of viral genes, we performed a DNA array analysis. The array (9) contained DNAs corresponding to most viral open reading frames believed to encode proteins (25) plus a set of cellular gene controls. At 72 h after infection of fibroblasts at an MOI of 0.5 PFU/cell with ADsubTRS1 (Fig. 1B) or its wild-type parent, ADwt (Fig. 1A), more than 100 viral RNAs were detected at levels above background (Fig. 1, top three sets of panels), and more than 20 cellular RNAs were expressed at sufficiently high levels for detection (Fig. 1, bottom set of panels). A PCR-amplified cDNA segment corresponding to green fluorescent protein (GFP) was spotted on each panel in three of the four corners and once in position six of the bottom row of the third panel. The ADsubTRS1 mutant was constructed by substitution of a GFP cassette for the central portion of the TRS1 open reading frame, so these spots function as positive controls. No difference was evident in the total number of RNAs expressed at a detectable level, and no RNAs showed a >3-fold difference in the levels of their expression between ADsubTRS1 and ADwt. In fact, most transcripts showed <2-fold change. This level of variability is observed between independent infections with ADwt (reference 9 and data not shown). The array analysis and the earlier Western blot analysis (3) argue that the normal complement of viral RNAs and proteins accumulate in the absence of pTRS1.
FIG. 1.
Microarray analysis of the ADwt (A) and ADsubTRS1 (B) mutant reveals similar patterns of HCMV gene expression in infected fibroblasts. DNA arrays were probed with cDNA prepared from fibroblasts infected at an MOI of 0.5 PFU/cell and harvested at 72 h postinfection. The top three panels in each section have cDNAs that span the entire HCMV genome along with several controls. The bottom panel in each section contains a set of control cellular genes.
Reduced accumulation of capsids in ADsubTRS1-infected cells.
The pp65 (pUL83) and pp71 (pUL82) tegument proteins belong to the late kinetic class of virus proteins and were previously found to remain in the nucleus late after infection with the mutant virus (3). Normally, they are substantially localized to a region within the cytoplasm, where capsids acquire their tegument proteins and final envelope (reviewed in reference 35). It is possible that pp65 and pp71, which are localized to the nucleus early in infection, move to the cytoplasm in association with newly formed capsids. If this is the case, then their mislocalization in ADsubTRS1-infected cells could result from a failure to produce or assemble capsid constituents or from a failure to translocate capsids from the nucleus to cytoplasm.
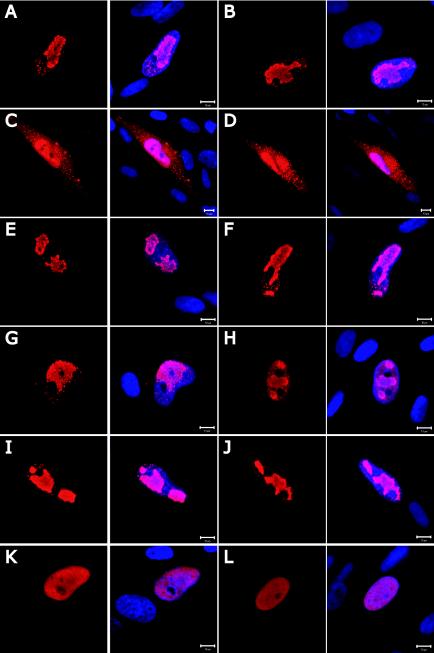
Five late virus-coded proteins, including the major capsid protein, were assayed previously by Western blotting and found to accumulate to normal levels after infection with ADsubTRS1 (3). To determine whether the defect in ADsubTRS1-infected cells results from the failure to properly localize the proteins that comprise the capsid, we monitored the location of the major capsid protein pUL86 (Fig. 2A and B), minor capsid protein pUL85 (Fig. 2C and D), and smallest capsid protein pUL48.5 (Fig. 2E and F) by immunofluorescence. Moreover, we localized several virus-coded proteins involved in DNA replication: replication compartment protein pUL112-113 (Fig. 2G and H), polymerase accessory protein pUL44 (Fig. 2I and J), and DNase pUL98 (Fig. 2K and L). No differences were evident in the locations of the proteins at 72 h postinfection with ADwt compared to ADsubTRS1. Thus, the principal capsid constituents, as well as several proteins involved in HCMV DNA replication, are produced and accumulate in the correct cellular compartment of ADsubTRS1-infected fibroblasts.
FIG.2.
Immunofluorescence analysis of the intracellular localization of HCMV proteins involved in capsid formation and DNA replication after infection with ADwt (A, C, E, G, I, and K) or ADsubTRS1 (B, D, F, H, J, and L). Fibroblasts were fixed at 72 h postinfection and probed with MAbs specific to the pUL86 major capsid protein (A and B), pUL85 minor capsid protein (C and D), pUL48.5 smallest capsid protein (E and F), pUL112/113 replication compartment protein (G and H), pUL44 polymerase accessory proteins (I and J), and pUL98 virus-coded DNase (K and L). The left panel in each pair shows the antibody-specific signal alone (red), and in the right panel of each pair the DNA is counterstained with DAPI (blue). Bars, 10 μm.
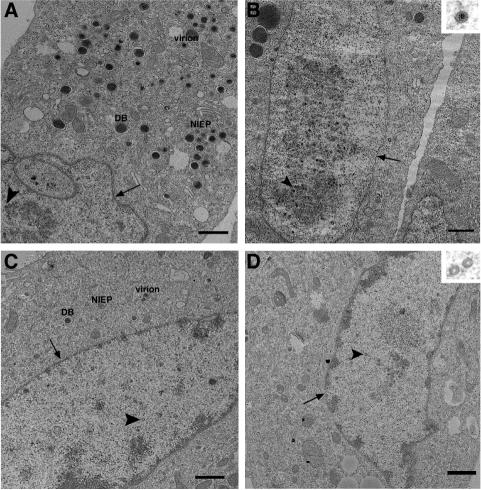
To monitor the formation of capsids, we initially analyzed infected fibroblasts by using thin-section transmission electron microscopy. Cells infected with ADwt showed an accumulation of virions, noninfectious enveloped particles (NIEPs) and dense bodies (DBs) in the cytoplasm, plus an abundance of capsids in the nucleus (Fig. 3A and B). Three capsid types (reviewed in reference 7), were evident: B-capsids, which are precursor capsids that have not yet had their internal scaffold protein removed (their cores appear gray in the uranyl acetate-stained sections); C-capsids, which are mature DNA-containing capsids that have lost their scaffold protein and contain DNA (their electron-dense cores are black); and A-capsids, which have lost the internal scaffolding protein, presumably during an abortive attempt to package DNA (their empty cores are white). ADsubTRS1-infected cells contained a reduced number of all types of virus particles (Fig. 3C and D) than were evident in ADwt-infected cells. Fewer virions, NIEPs and DBs were present in the cytoplasm, and there were reduced numbers of all capsid types in the nucleus. When capsids were viewed at higher magnification there appeared to be no significant defects in the morphology of capsids produced in mutant virus-infected cells compared to wild-type (Fig. 3B and D, insets).
FIG. 3.
The ADsubTRS1 mutant causes a decrease in capsid production and a disruption of viral replication centers. Thin-section transmission electron microscopy of fibroblasts infected at an MOI of 0.5 PFU/cell and fixed at 72 h postinfection. Fibroblasts were infected with ADwt (A and B) or ADsubTRS1 (C and D). The nuclear envelope is indicated by arrows, and the arrowheads point to capsids. NIEPs, virions, and DBs are labeled in the cytoplasm (A and C). All images (except for higher magnification insets) are taken at the same magnification. Bars, 1 μm.
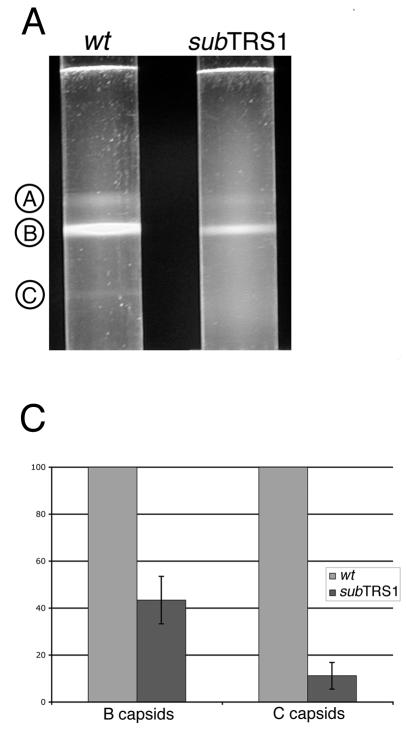
Since by electron microscopy there was a clear reduction in the number of capsids produced in ADsubTRS1-infected nuclei, we quantified the relative levels of the different capsid types by a second assay. The three types of capsids were separated by velocity sedimentation in sucrose gradients. Previous work has shown that more NIEPs than virions are produced in cells infected with the HCMV AD169 strain from which ADwt is derived; in fact, AD169 produces 10-fold more NIEPs than other HCMV strains (13). NIEPs are enveloped B-capsids (14), so it was anticipated that the B-capsid would be the most abundant form of capsid isolated from ADwt-infected cells.
Lysates were prepared at 72 h after infection with ADsubTRS1 or ADwt and then subjected to velocity sedimentation. Figure 4A displays a representative separation of the A-, B-, and C-capsids. All three capsid types were observed after separation of the particles in lysates of wild-type virus-infected cells and, as expected, B-capsids were the most abundant. A band comprised of C-capsids was seen after centrifugation of extracts from wild-type virus- but not mutant virus-infected cells. The numbers of C-capsids in the ADsubTRS1 mutant were too few to be visualized above the background in the gradients.
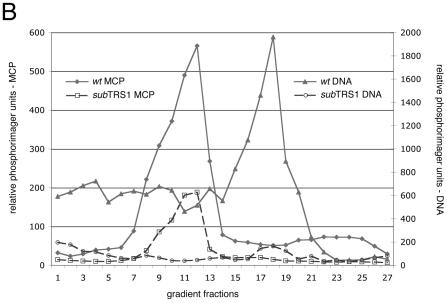
FIG. 4.
Rate velocity gradient separation of HCMV nucleocapsids. Lysates from ADwt- or ADsubTRS1-infected fibroblasts were layered onto 20 to 65% sucrose gradients. (A) Illumination from the top revealed three light-scattering bands in ADwt, indicated by circled letters. A-capsids are empty; B-capsids are precursors that contain the scaffolding protein, which is then released from the capsid to allow DNA insertion and the maturation to the C-capsid form. (B) Profile of gradient fractions probed for the presence of capsid proteins or HCMV DNA. This representative sample shows the distribution of both the major capsid protein (on the left axis) and viral DNA (on the right axis) along the fractions of the rate velocity gradient. (C) When the ADsubTRS1 mutant is compared to ADwt, the reduction in capsid numbers is different for the two different capsid types analyzed. Quantitation for both the B and C peaks shows the fold decrease for ADsubTRS1 compared to the ADwt strain.
To identify and quantify each capsid type, the gradient was fractionated and the fractions were analyzed for their capsid protein and viral DNA content. Western blot analysis with antibodies to the major capsid protein (Fig. 4B) or minor capsid protein (data not shown) identified the abundant B-capsids in lysates of both wild-type and mutant virus-infected cells. A-capsids formed a shoulder to the left of the B-capsid peak, and C-capsids could occasionally be detected as a minor protein peak in a later fraction. A slot blot assay using a probe for HCMV DNA conclusively identified the C-capsids (Fig. 4B). Radioactivity in each peak was quantified by phosphorimager analysis (Fig. 4C). The data from multiple experiments demonstrated a reduction of B- and C-capsids in lysates of mutant compared to wild-type virus. ADsubTRS1 produced ∼2-fold fewer B-capsids and 10-fold fewer C-capsids than did ADwt. The decrease in the number of C-capsids in ADsubTRS1-infected cells argues that pTRS1 is required for efficient DNA encapsidation. We were not able to reliably quantify A-capsids in gradient fractions since they overlapped the major B-capsid peak. However, visual inspection of the gradients (Fig. 4A) and electron microscopy images indicated that the mutant and wild-type viruses produced similar percentages of A-capsids.
Concatemeric ADsubTRS1 DNA is cleaved normally.
It is generally believed that, early in the HCMV replication cycle, the viral genome circularizes and begins rolling circle replication (24), although a recent report suggested that in herpes simplex virus type 1 (HSV-1) there is little or no circular viral DNA found during a productive infection (16). Previous work in HCMV has indicated that DNA is replicated from the ori-Lyt origin and concatemers of the viral genome are generated. The concatemers must then be cleaved into unit-length pieces as the DNA is packaged into preassembled procapsids. This cleavage has been shown to utilize ATP and proceed concomitantly with insertion of the DNA into the capsid (12). In HSV-1 infections, at least seven proteins function at this point, including the large UL28 and small UL15 terminase subunits and the UL6 portal protein (18, 38, 39). In HCMV, the protein products of UL56 and UL89 have been identified as the large and small terminase subunits (4, 34), but further details of the HCMV proteins involved in this process have not been investigated.
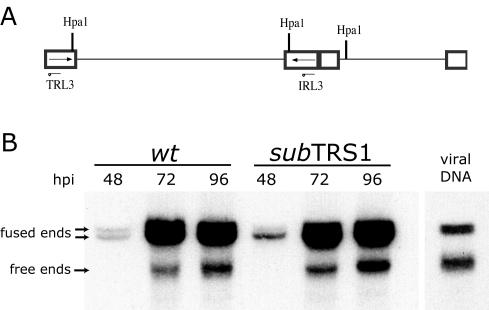
It has previously been shown that ADsubTRS1 DNA is replicated with kinetics similar to that of ADwt (3), and we have shown here that there is a defect in the production of DNA-containing C-capsids. Consequently, it appeared possible that pTRS1 is required for cleavage of concatemeric HCMV DNA during the packaging process. To investigate whether the mutant DNA is properly cleaved, we determined the ratio of free ends (cleaved genomes) to fused ends (genomes that are still part of the concatemer) in the mutant compared to wild-type virus-infected cells (22, 23). Total DNA was isolated from fibroblasts at several times after infection and genomic termini were monitored by Southern blotting with a probe to the IRL3/TRL3 region of the long repeats (diagramed in Fig. 5A). Since the probe corresponds to the repeat region, it will identify the free end (TRL3), as well as the internal fused region (IRL3), of the HCMV genome. HpaI-digested viral DNA generates free ends of 7 kbp and two fragments (10 and 13 kbp each) that span the long-short junction; genomic isomerization yields the two different sizes of junction fragments. The ratio of free to fused ends was not significantly different in DNA isolated from ADsubTRS1-infected compared to ADwt-infected cells (Fig. 5B and Table 1). The normal accumulation of free HCMV genome ends in ADsubTRS1-infected cells indicates that pTRS1 is not required for cleavage of viral DNA concatemers.
FIG. 5.
Processing of newly replicated viral DNA. Fibroblasts were infected with either ADwt or ADsubTRS1 virus at an MOI of 0.5 PFU/cell, and the total DNA was prepared at the indicated times. (A) Shows the relevant fragments of an HCMV genome digested with HpaI. (B) A representative Southern blot with a 32P-labeled probe specific for RL3. Purified cell-free viral DNA was used as a control for size. The indicated fused and free fragments were quantified by using a phosphorimager.
TABLE 1.
Percentage of total viral DNA that has been cleaved from the concatemer
| Time postinfection (h) | % Total viral DNA
|
|
|---|---|---|
| Wild type | subTRS1 | |
| 48 | 9.5 | 11.2 |
| 72 | 15.4 | 15.0 |
| 96 | 17.7 | 18.4 |
Altered nuclear organization in ADsubTRS1-infected cells.
In addition to the failure to produce normal numbers of C-capsids, a marked difference in nuclear morphology was evident in mutant compared to wild-type virus-infected cells. Nuclear morphology is profoundly reorganized after HCMV infection (1), as well as in HSV-1-infected cells (30). Large replication and capsid assembly centers with a diffuse electron-dense appearance are evident in the nucleus. These domains contain high concentrations of the viral components needed to form a DNA-filled capsid (see, for example, reference 29); including viral DNA replication proteins, terminase subunits, viral DNA, and capsid proteins. As seen by electron microscopy, ADwt-infected nuclei contained these large, diffuse, electron-dense domains with associated capsids (Fig. 3A, lower left, and 3B). ADsubTRS1-infected nuclei seldom contained these electron-dense structures, and the capsids that did form in the nucleus were generally not clustered in such regions (Fig. 3C and D).
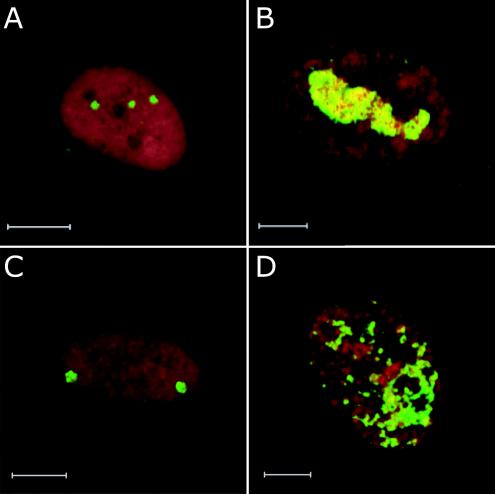
To further document the nuclear reorganization defect, we monitored the location of viral DNA in infected fibroblasts by FISH. Relatively early after infection (24 h), a few fluorescent dots of DNA were observed in both ADwt-infected (Fig. 6A) and ADsubTRS1-infected (Fig. 6C) nuclei. Later after infection (96 h), the numbers and sizes of fluorescent patches had increased and coalesced in the nuclei of wild-type virus-infected cells (Fig. 6B). In contrast, the DNA-containing dots and patches did not coalesce late after infection with the mutant virus. Rather, many small dispersed fluorescent puncta accumulated (Fig. 6D). The altered pattern of DNA accumulation in ADsubTRS1-infected nuclei could be the cause or effect of a failure to assemble DNA-containing capsids. As an additional control, viral DNA accumulation was monitored in cells infected with ADsubIRS1, and the pattern of fluorescence was indistinguishable from that seen for the wild-type virus (data not shown). We conclude that, although normal levels of ADsubTRS1 DNA are produced, it is not localized normally in the nucleus.
FIG. 6.
FISH of fibroblasts fixed infected with ADwt (A and B) or ADsubTRS1 (C and D) and then fixed at 24 h (A and C) or 96 h postinfection (B and D). HCMV DNA was visualized by FISH analysis and appears in green. Cellular DNA is counterstained with propidium iodide and appears in red.
DISCUSSION
Previous work has shown that a pTRS1-deficient HCMV mutant exhibits attenuated growth on human fibroblasts (3). Viral DNA replication proceeds at the same rate and to the same extent (3), and no difference is evident in the accumulation of viral RNAs (Fig. 1) and proteins (3) in ADsubTRS1- compared to ADwt-infected cells. Normal DNA replication argues that the ADsubTRS1 defect occurs late in the infectious cycle, and the failure to find a deficit in RNA or protein accumulation is consistent with the view that the mutant's defect occurs at the level of assembly.
Electron microscopic examination of infected cells revealed that the TRS1-deficient virus produced reduced numbers of virus particles in comparison to its wild-type parent (Fig. 3). Fewer virions, NIEPs, and DBs were observed in the cytoplasm, and fewer capsids were evident in the nucleus. The relative amounts of the different capsid types were quantified by velocity sedimentation in sucrose gradients. The mutant proved to generate 2-fold fewer B-capsids and 10-fold fewer C-capsids (Fig. 4B and C), whereas the amount of A-capsids appeared to be normal (Fig. 4A). The reduction in DNA-containing C-capsids without a concomitant increase in empty A-capsids, which are believed to arise from aborted DNA encapsidation events (reviewed in reference 7), in ADsubTRS1-infected cells suggests that a failure in the sequence of packaging events occurs before the scaffolding proteins exit the capsid.
We were able to further localize the defect by examining the cleavage of concatemeric viral DNA in mutant virus-infected cells, and cleavage occurred normally (Fig. 5). This further localized a defect to a point after DNA cleavage but before the exit of scaffolding proteins from the capsid.
Nuclear organization was also abnormal in ADsubTRS1- compared to ADwt-infected fibroblasts. Large, electron-dense centers of replication and assembly were evident in the nuclei of cells infected with the wild-type virus, but not the mutant (Fig. 3), and FISH analysis demonstrated that mutant DNA is compartmentalized differently than wild-type DNA in the nucleus (Fig. 6). Whereas the fluorescent signal for ADwt DNA coalesces to form one or two large domains late after infection, numerous individual fluorescent puncta of ADsubTRS1 DNA accumulate but do not coalesce. It is possible that pTRS1 functions to properly locate the viral DNA in the nucleus so that it can be efficiently packaged. Alternatively, the altered localization might result from a failure to efficiently package viral DNA.
We favor the view that pTRS1 is needed to produce a normal B-capsid that is competent to accept DNA, i.e., in the absence of pTRS1, only a portion of the B-capsids are competent for DNA packaging. This could explain the simultaneous 2-fold reduction in B-capsids and 10-fold reduction in C-capsids. B-capsids are complex structures. HCMV B-capsids were characterized some years ago to contain a major capsid protein, a minor capsid protein, the smallest capsid protein, and assembly protein (14), along with the minor capsid protein-binding protein (8). More recent work with HSV-1 has shown that its B-capsids include nine virus-coded polypeptides (reviewed in reference 21). Although pTRS1 is not homologous to any of these nine HSV-1 proteins, it is possible that pTRS1 functions as an additional constituent of HCMV B-capsids, and this would be consistent with its presence in virions (32). Further characterization of the B-capsids is needed to reveal whether pTRS1 associates with the particles produced in ADwt-infected cells and whether the B-capsids produced in ADsubTRS1-infected cells exhibit additional abnormalities in their composition.
Although pTRS1 doesn't exhibit homology to the HSV-1 UL25 protein, there are similarities in the phenotypes of ADsubTRS1 and HSV-1 mutants lacking the UL25 gene. The UL25 protein is found in virions (2), and it is required for the encapsidation of HSV-1 DNA but not for the cleavage of concatemers (21). Recent work has shown that the UL25 protein binds to the B-capsid, as well as to viral DNA (27).
Recently, Child et al. (5) have shown that pIRS1 and pTRS1 both complement a vaccinia virus mutant that lacks the E3L double-stranded RNA-binding protein. The complementation experiment argues that the HCMV proteins, like the E3L protein, block the shutoff of translation by host cell antiviral pathways. It is unlikely that this intriguing function is related to the assembly function that we have described for pTRS1. Since both pIRS1 and pTRS1 antagonize the host antiviral response, this function almost certainly resides in their common N-terminal domains. An assembly defect is evident only after infection with a pTRS1-deficient virus and not in the absence of pIRS1. Consequently, the pTRS1 assembly function must reside at least in part in its unique C-terminal domain.
Acknowledgments
We thank E. Bogner, W. Britt, and W. Gibson for generous gifts of antibodies, F. Goodrum for providing HCMV cDNA arrays, and M. Bisher for assistance with electron microscopy. We also thank M. Nevels for critical reading of the manuscript.
This study was supported by a grant from the National Cancer Institute (CA85786) to T.S. and a fellowship award from the Leukemia and Lymphoma Society to J.E.A.
REFERENCES
- 1.AbuBakar, S., W. W. Au, M. S. Legator, and T. Albrecht. 1988. Induction of chromosome aberrations and mitotic arrest by cytomegalovirus in human cells. Environ. Mol. Mutagen. 12:409-420. [DOI] [PubMed] [Google Scholar]
- 2.Ali, M. A., B. Forghani, and E. M. Cantin. 1996. Characterization of an essential HSV-1 protein encoded by the UL25 gene reported to be involved in virus penetration and capsid assembly. Virology 216:278-283. [DOI] [PubMed] [Google Scholar]
- 3.Blankenship, C. A., and T. Shenk. 2002. Mutant human cytomegalovirus lacking the immediate-early TRS1 coding region exhibits a late defect. J. Virol. 76:12290-12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bogner, E., K. Radsak, and M. F. Stinski. 1998. The gene product of human cytomegalovirus open reading frame UL56 binds the pac motif and has specific nuclease activity. J. Virol. 72:2259-2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Child, S. J., M. Hakki, K. L. De Niro, and A. P. Geballe. 2004. Evasion of cellular antiviral responses by human cytomegalovirus TRS1 and IRS1. J. Virol. 78:197-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feinberg, A. P., and B. Vogelstein. 1983. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 132:6-13. [DOI] [PubMed] [Google Scholar]
- 7.Gibson, W. 1996. Structure and assembly of the virion. Intervirology 39:389-400. [DOI] [PubMed] [Google Scholar]
- 8.Gibson, W., M. K. Baxter, and K. S. Clopper. 1996. Cytomegalovirus “missing” capsid protein identified as heat-aggregable product of human cytomegalovirus UL46. J. Virol. 70:7454-7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodrum, F. D., C. T. Jordan, K. High, and T. Shenk. 2002. Human cytomegalovirus gene expression during infection of primary hematopoietic progenitor cells: a model for latency. Proc. Natl. Acad. Sci. USA 99:16255-16260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayat, M. 2000. Principles and techniques of electron microscopy, 4th ed. Cambridge University Press, Cambridge, United Kingdom.
- 11.Hegde, P., R. Qi, K. Abernathy, C. Gay, S. Dharap, R. Gaspard, J. E. Hughes, E. Snesrud, N. Lee, and J. Quackenbush. 2000. A concise guide to cDNA microarray analysis. BioTechniques 29:548-556. [DOI] [PubMed] [Google Scholar]
- 12.Hwang, J. S., and E. Bogner. 2002. ATPase activity of the terminase subunit pUL56 of human cytomegalovirus. J. Biol. Chem. 277:6943-6948. [DOI] [PubMed] [Google Scholar]
- 13.Irmiere, A., and W. Gibson. 1983. Isolation and characterization of a noninfectious virion-like particle released from cells infected with human strains of cytomegalovirus. Virology 130:118-133. [DOI] [PubMed] [Google Scholar]
- 14.Irmiere, A., and W. Gibson. 1985. Isolation of human cytomegalovirus intranuclear capsids, characterization of their protein constituents, and demonstration that the B-capsid assembly protein is also abundant in noninfectious enveloped particles. J. Virol. 56:277-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iskenderian, A. C., L. Huang, A. Reilly, R. M. Stenberg, and D. G. Anders. 1996. Four of eleven loci required for transient complementation of human cytomegalovirus DNA replication cooperate to activate expression of replication genes. J. Virol. 70:383-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jackson, S. A., and N. A. DeLuca. 2003. Relationship of herpes simplex virus genome configuration to productive and persistent infections. Proc. Natl. Acad. Sci. USA 100:7428-7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones, T. R., and V. P. Muzithras. 1992. A cluster of dispensable genes within the human cytomegalovirus genome short component: IRS1, US1 through US5, and the US6 family. J. Virol. 66:2541-2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koslowski, K. M., P. R. Shaver, J. T. Casey, 2nd, T. Wilson, G. Yamanaka, A. K. Sheaffer, D. J. Tenney, and N. E. Pederson. 1999. Physical and functional interactions between the herpes simplex virus UL15 and UL28 DNA cleavage and packaging proteins. J. Virol. 73:1704-1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lafemina, R. L., and G. S. Hayward. 1988. Differences in cell-type-specific blocks to immediate-early gene expression and DNA replication of human, simian and murine cytomegalovirus. J. Gen. Virol. 69(Pt. 2):355-374. [DOI] [PubMed] [Google Scholar]
- 20.Lichter, P., C. J. Tang, K. Call, G. Hermanson, G. A. Evans, D. Housman, and D. C. Ward. 1990. High-resolution mapping of human chromosome 11 by in situ hybridization with cosmid clones. Science 247:64-69. [DOI] [PubMed] [Google Scholar]
- 21.McNab, A. R., P. Desai, S. Person, L. L. Roof, D. R. Thomsen, W. W. Newcomb, J. C. Brown, and F. L. Homa. 1998. The product of the herpes simplex virus type 1 UL25 gene is required for encapsidation but not for cleavage of replicated viral DNA. J. Virol. 72:1060-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McVoy, M. A., and S. P. Adler. 1994. Human cytomegalovirus DNA replicates after early circularization by concatemer formation, and inversion occurs within the concatemer. J. Virol. 68:1040-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meier, J. L., and J. A. Pruessner. 2000. The human cytomegalovirus major immediate-early distal enhancer region is required for efficient viral replication and immediate-early gene expression. J. Virol. 74:1602-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mocarski, E. S., and C. T. Courcelle. 2001. Cytomegaloviruses and their replication, p. 2629-2673. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott/The Williams & Wilkins Co., Philadelphia, Pa.
- 25.Murphy, E., I. Rigoutsos, T. Shibuya, and T. E. Shenk. 2003. Reevaluation of human cytomegalovirus coding potential. Proc. Natl. Acad. Sci. USA 100:13585-13590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Newcomb, W. W., and J. C. Brown. 1991. Structure of the herpes simplex virus capsid: effects of extraction with guanidine hydrochloride and partial reconstitution of extracted capsids. J. Virol. 65:613-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ogasawara, M., T. Suzutani, I. Yoshida, and M. Azuma. 2001. Role of the UL25 gene product in packaging DNA into the herpes simplex virus capsid: location of UL25 product in the capsid and demonstration that it binds DNA. J. Virol. 75:1427-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pari, G. S., and D. G. Anders. 1993. Eleven loci encoding trans-acting factors are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA replication. J. Virol. 67:6979-6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Penfold, M. E., and E. S. Mocarski. 1997. Formation of cytomegalovirus DNA replication compartments defined by localization of viral proteins and DNA synthesis. Virology 239:46-61. [DOI] [PubMed] [Google Scholar]
- 30.Puvion-Dutilleul, F., L. Venturini, M. C. Guillemin, H. de The, and E. Puvion. 1995. Sequestration of PML and Sp100 proteins in an intranuclear viral structure during herpes simplex virus type 1 infection. Exp. Cell Res. 221:448-461. [DOI] [PubMed] [Google Scholar]
- 31.Rigby, P. W., M. Dieckmann, C. Rhodes, and P. Berg. 1977. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J. Mol. Biol. 113:237-251. [DOI] [PubMed] [Google Scholar]
- 32.Romanowski, M. J., E. Garrido-Guerrero, and T. Shenk. 1997. pIRS1 and pTRS1 are present in human cytomegalovirus virions. J. Virol. 71:5703-5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Romanowski, M. J., and T. Shenk. 1997. Characterization of the human cytomegalovirus irs1 and trs1 genes: a second immediate-early transcription unit within irs1 whose product antagonizes transcriptional activation. J. Virol. 71:1485-1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scheffczik, H., C. G. Savva, A. Holzenburg, L. Kolesnikova, and E. Bogner. 2002. The terminase subunits pUL56 and pUL89 of human cytomegalovirus are DNA-metabolizing proteins with toroidal structure. Nucleic Acids Res. 30:1695-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silva, M. C., Q. C. Yu, L. Enquist, and T. Shenk. 2003. Human cytomegalovirus UL99-encoded pp28 is required for the cytoplasmic envelopment of tegument-associated capsids. J. Virol. 77:10594-10605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stasiak, P. C., and E. S. Mocarski. 1992. Transactivation of the cytomegalovirus ICP36 gene promoter requires the alpha gene product TRS1 in addition to IE1 and IE2. J. Virol. 66:1050-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weston, K., and B. G. Barrell. 1986. Sequence of the short unique region, short repeats, and part of the long repeats of human cytomegalovirus. J. Mol. Biol. 192:177-208. [DOI] [PubMed] [Google Scholar]
- 38.White, C. A., N. D. Stow, A. H. Patel, M. Hughes, and V. G. Preston. 2003. Herpes simplex virus type 1 portal protein UL6 interacts with the putative terminase subunits UL15 and UL28. J. Virol. 77:6351-6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu, D., and S. K. Weller. 1998. Genetic analysis of the UL 15 gene locus for the putative terminase of herpes simplex virus type 1. Virology 243:32-44. [DOI] [PubMed] [Google Scholar]