Abstract
Reovirus replication and assembly are thought to occur within cytoplasmic inclusion bodies, which we call viral factories. A strain-dependent difference in the morphology of these structures reflects more effective microtubule association by the μ2 core proteins of some viral strains, which form filamentous factories, than by those of others, which form globular factories. For this report, we identified and characterized another strain-dependent attribute of the factories, namely, the extent to which they colocalized with conjugated ubiquitin (cUb). Among 16 laboratory strains and field isolates, the extent of factory costaining for cUb paralleled factory morphology, with globular strains exhibiting higher levels by far. In reassortant viruses, factory costaining for cUb mapped primarily to the μ2-encoding M1 genome segment, although contributions by the λ3- and λ2-encoding L1 and L2 genome segments were also evident. Immunoprecipitations revealed that cells infected with globular strains contained higher levels of ubiquitinated μ2 (Ub-μ2). In M1-transfected cells, cUb commonly colocalized with aggregates formed by μ2 from globular strains but not with microtubules coated by μ2 from filamentous strains, and immunoprecipitations revealed that μ2 from globular strains displayed higher levels of Ub-μ2. Allelic changes at μ2 residue 208 determined these differences. Nocodazole treatment of cells infected with filamentous strains resulted in globular factories that still showed low levels of costaining for cUb, indicating that higher levels of costaining were not a direct result of decreased microtubule association. The factories of globular strains, or their μ2 proteins expressed in transfected cells, were furthermore shown to gain microtubule association and to lose colocalization with cUb when cells were grown at reduced temperature. From the sum of these findings, we propose that μ2 from globular strains is more prone to temperature-dependent misfolding and as a result displays increased aggregation, increased levels of Ub-μ2, and decreased association with microtubules. Because so few of the viral strains formed factories that were regularly associated with ubiquitinated proteins, we conclude that reovirus factories are generally distinct from cellular aggresomes.
Many of the processes that constitute genome replication and assembly of the mature virions of orthoreoviruses (viruses with 10-segmented, double-stranded RNA genomes from the family Reoviridae, genus Orthoreovirus) are thought to occur in specialized regions of the infected cytoplasm, which have been variably called viroplasms, viral inclusions, or viral factories (see references 36 and 48 for general reviews about these viruses). The factories of nonfusogenic mammalian orthoreoviruses (reoviruses) form early in infection as punctate, phase-dense inclusions, which grow larger and move towards the perinuclear region as infection proceeds (1, 21, 31, 38, 42). Viral and cellular proteins, double-stranded RNA, and partially and fully assembled viral particles, but not ribosomes, all localize to factories during infection (1, 2, 7, 8, 13, 32, 38, 41, 42, 44, 47). Recent studies have suggested that the viral nonstructural protein μNS, which forms inclusions morphologically similar to some viral factories when expressed in the absence of other viral proteins, constitutes the main structural matrix of the factories (2, 8, 47). In addition, μNS appears to play a key role in recruiting other components to the factories. To date, viral nonstructural protein σNS (2, 32, 47); viral core proteins σ2, μ2, λ2, and λ1 (7, 8); and intact viral core particles (7) have been shown to localize to factory-like inclusions through association with μNS. Association of μNS with the μ2 protein in particular plays a key role in determining the morphology of viral factories (8, 38).
The μ2 protein is a structurally minor component of the viral inner capsid particle, or core, being present in only 20 to 24 copies per particle (11, 34). In vitro μ2 binds RNA (5) and exhibits nucleoside and RNA triphosphatase activities (25, 37) that are likely important for its proposed role as a cofactor for the viral polymerase, λ3 (50). In addition to these roles, μ2 from certain reovirus strains associates with and stabilizes cellular microtubules when expressed in transfected cells in the absence of other viral proteins (38). Differences in the microtubule-associating capacity of μ2 correlate with striking differences in the morphologies of reovirus factories among strains examined to date. Factories from strains in which μ2 effectively associates with microtubules have a filamentous morphology (filamentous strains), whereas factories from strains in which μ2 does not effectively associate with microtubules have a globular morphology (globular strains) (38). When expressed in the absence of other viral proteins, μ2 from globular strains shows little or no association with microtubules and instead forms angular aggregates in most cells, suggesting that it may have a tendency to misfold (38).
Aggresomes are cellular inclusion bodies that accumulate in cells in certain pathological conditions, including a number of neurodegenerative diseases (reviewed in references 19 and 26). Aggresomes form in the pericentrosomal region of cells as a programmed response to protein misfolding and consist of aggregated proteins that have been specifically transported to the inclusions by a microtubule-dependent mechanism. Many of the aggregated proteins that localize in aggresomes have been tagged for destruction by the ubiquitin (Ub)-proteasome system, and thus immunostaining for multiubiquitinated proteins can often be used to identify these structures (19). Reovirus factories, especially those with globular morphology, have been noted to share several similarities with cellular aggresomes (38).
Based on the preceding results, we hypothesized that the μ2 protein from globular strains might be more prone to misfolding and then targeted for degradation by the Ub-proteasome system (reviewed in references 20 and 39). In this study, we showed there was indeed a strong correlation between the morphologies of reovirus factories formed by 16 different viral laboratory strains or field isolates and their costaining for conjugated Ub (cUb), with globular strains exhibiting higher levels by far. The viral M1 genome segment, which encodes μ2, was the primary genetic determinant of this difference between strains type 3 Dearing (T3D) and type 1 Lang (T1L), and the μ2 protein from globular strains displayed much higher levels of ubiquitinated μ2 (Ub-μ2) in both infected and transfected cells. We also discovered that strains that form globular factories at 37°C showed increased formation of filamentous factories and decreased colocalization with cUb at 31°C. From the sum of the results, we propose that μ2 from globular strains is more prone to temperature-dependent misfolding as a primary defect that results in increased aggregation, increased levels of Ub-μ2, and decreased association with microtubules as secondary effects. Because so few of the field strains formed factories that costained for ubiquitinated proteins, we conclude that reovirus factories are generally distinct from cellular aggresomes.
MATERIALS AND METHODS
Cells and viruses.
CV-1 and HeLa cells were maintained in Dulbecco's modified Eagle's medium (Invitrogen Life Technologies) containing 10% fetal bovine serum (HyClone Laboratories) and 10 μg of gentamicin (Invitrogen Life Technologies) per ml. L929 cells adapted to spinner culture were maintained in Joklik's minimal essential medium (Irvine Scientific) containing 2% calf serum and 2% fetal bovine serum (HyClone Laboratories) as well as 2 mM l-glutamine, 100 μg of streptomycin per ml, and 100 U of penicillin G (Irvine Scientific) per ml. Reoviruses T3DN and T1L were our laboratory stocks for the isolates formally identified as T3/human/Ohio/Dearing/1955 and T1/human/Ohio/Lang/1953, respectively, using the following nomenclature: serotype/species of origin/place of origin/strain designation/year of isolation (22). The superscript N in T3DN differentiates our laboratory's clones of T3D from a clone obtained from the laboratory of L. W. Cashdollar (Medical College of Wisconsin), for which our designation is T3DC. T3DN and T3DC have been shown to differ in both M1 nucleotide sequence and viral factory morphology (38). T1L × T3D reassortant viruses were previously described (9, 12, 14, 35). The 13 additional reovirus isolates were also previously described (22, 24, 27, 38) and are formally identified as follow: T3C12, T3/human/Washington,DC/1/1957; T1C50, T1/bovine/Maryland/clone50/1960; T2N84, T2/human/Netherlands/1/1984; T1N84, T1/human/Netherlands/1/1984; T3A, T3/human/Washington,DC/Abney/1957; T2J, T2/human/Ohio/Jones/1955; T3C8, T3/human/Tahiti/clone8/1960; T1C39, T1/bovine/Maryland/clone39/1960; T2S59, T2/simian/Maryland/SV59/1958; T1N85, T1/human/Netherlands/1/1985; T1C23, T1/bovine/Maryland/clone23/1959; T3N83, T3/human/Netherlands/1/1983; and T3C9, T3/murine/France/clone9/1961. Unless otherwise indicated, second- or third-passage stocks of doubly or triply plaque-purified clones of viruses were used for infections.
Antibodies and other reagents.
Rabbit polyclonal antibodies against the viral proteins μ2 and μNS have been described previously (6, 38). Monoclonal antibodies (MAbs) that recognize multiubiquitinated proteins (FK1) or both multi- and monoubiquitinated proteins (FK2), but not free Ub in either case (15), were obtained from Affiniti Research Products. MAb HA.11, specific for the influenza virus hemagglutinin (HA) epitope described below, was obtained from Covance Research Products. Goat anti-rabbit immunoglobulin G (IgG) conjugated to Alexa 594 and goat anti-mouse IgG conjugated to Alexa 488 were obtained from Molecular Probes. Donkey anti-rabbit or donkey anti-mouse IgG conjugated to horseradish peroxidase (HRP) was obtained from Pierce. Proteasome inhibitor MG132 was obtained from Calbiochem and used at a final concentration of 10 μM. The microtubule-depolymerizing drug nocodazole was obtained from Sigma-Aldrich and used at a final concentration of 10 μM.
Plasmid constructs.
All of the plasmid constructs used to express μ2 in mammalian cells [pCI-M1(T3DN), pCI-M1(T1L), pCI-M1(T3DC), pCI-M1(T3DN-S208P), and pCI-M1(T1L-P208S)] have been previously described (38). In each case, the M1 gene is cloned behind the immediate-early human cytomegalovirus promoter of the pCI-neo vector (Promega). pHAUb expresses a version of Ub tagged with an influenza virus HA epitope as previously described (33).
Transfections and infections.
For immunostaining, 1.5 × 105 CV-1, 3 × 105 Mv1Lu, or 3 × 105 L929 cells were seeded onto six-well (9.6-cm2) dishes containing 18-mm-diameter round coverslips on the day before infection or transfection. To improve cell adherence in some experiments, acid-washed poly-l-lysine-treated coverslips were used. For transfections, a total of 2 μg of plasmid DNA was mixed with 7 μl of Lipofectamine 2000 in Optimem medium (Invitrogen Life Technologies), added to cells, and incubated for 4 h. The cells were then overlaid with Dulbecco's modified Eagle's medium and incubated at 31 or 37°C for 14 h. Transfections for immunoprecipitation were done essentially the same way, but with 3 × 105 CV-1 cells seeded onto 60-mm-diameter dishes, 4 μg of DNA, and 12 μl of Lipofectamine 2000. Infections for either immunostaining or immunoprecipitation were begun by adsorbing virus stocks to cells at a multiplicity of infection (MOI) of 0.5 to 50 PFU/cell, as indicated, for 1 h at room temperature in phosphate-buffered saline (PBS) (137 mM NaCl, 3 mM KCl, 8 mM Na2HPO4, 1 mM KH2PO4 [pH 7.5]) containing 2 mM MgCl2. Cells were then overlaid with growth medium and incubated at 31°C or 37°C for 8 to 44 h as indicated. For experiments involving both transfection and infection, cells were first transfected as described above, incubated at 37°C for 6 h, and then infected as described above except that intermediate subvirion particles were prepared from purified virions (16) and used in place of virus stocks. Following infection, cells were further incubated for 14 to 18 h before harvesting.
Immunostaining and immunofluorescence (IF) microscopy.
Infected or transfected cells were fixed at room temperature for 20 min with 2% paraformaldehyde in PBS and then washed three times with PBS. Fixed cells were permeabilized by incubation with 0.2% Triton X-100 in PBS for 5 min and blocked by a 5-min incubation with 1% bovine serum albumin in PBS. Primary and secondary antibodies were diluted in 1% bovine serum albumin in PBS. After blocking, cells were incubated for 45 to 60 min with primary antibody, washed three times with PBS, and then incubated for a further 45 to 60 min with secondary antibody. Immunostained cells were washed a final three times with PBS, incubated for 5 min in 300 nM 4,6-diamidino-2-phenylindole (DAPI), and mounted on slides with Prolong reagent (Molecular Probes). Immunostained samples were examined with a Nikon TE-300 inverted microscope equipped with fluorescence optics, and images were collected as described previously (38). Images were prepared for publication by using Photoshop and Illustrator software (Adobe Systems).
Quantitation of infected cells containing viral factories that costained for cUb.
Following immunostaining as described above, infected CV-1, HeLa, or L929 cells or transfected CV-1 cells were analyzed by using stringent criteria to score percentages of cells with filamentous distributions of viral proteins or colocalization of Ub with viral proteins. A particular infected or transfected cell was scored as positive for filaments if any indication of filamentous features was seen. Similarly, a particular infected or transfected cell was scored as positive for colocalization with cUb if any indication of costaining, either intense or faint, was visible. Over 100 such cells were scored from each experimental sample, and unless otherwise indicated, a minimum of three independent experiments were performed and scored for each reported virus strain or protein expressed after transfection. The InStat3 program (GraphPad Software) was used for performing Wilcoxon-Mann-Whitney analyses on the data for reassortant viruses.
Immunoprecipitations.
Transfected or infected cells were washed with PBS and harvested by being scraped from plates. In some experiments, the proteasome inhibitor MG132 was added 6 h before cells were harvested. Cells were collected by centrifugation at 13,000 × g and then lysed in nondenaturing buffer A (10 mM Tris [pH 8.0], 150 mM NaCl, 10 mM EDTA, 1% NP-40) and analyzed for total protein concentration by use of a Bradford assay (Bio-Rad Laboratories). Equivalent amounts of total protein were then added to buffer A containing 2% sodium dodecyl sulfate (SDS) for denaturation. Cells were incubated at 95°C for 5 min and then diluted to a final concentration of 0.125% SDS with buffer A. Antibodies that were preincubated for 2 h with protein A-conjugated magnetic beads (Dynal Biotech) were washed three times with buffer A containing 0.125% SDS, added to the prepared cell lysates, and incubated rotating overnight at 4°C. Magnetic beads containing antibody and antigen were then washed four to six times with buffer A containing 0.125% SDS and resuspended in sample buffer (125 mM Tris [pH 6.8], 10% sucrose, 1% SDS, 0.02% β-mercaptoethanol, 0.01% bromophenol blue).
Immunoblotting.
Immunoprecipitated proteins obtained as described above were boiled for 3 min and separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE). Proteins were transferred by electroblotting to nitrocellulose in transfer buffer (25 mM Tris, 192 mM glycine, 20% methanol [pH 8.3]). The nitrocellulose containing the transferred proteins was blocked for 15 min with 5% milk in Tris-buffered saline (20 mM Tris, 137 mM NaCl [pH 7.6]) containing 1% Tween (TBS-T) and then incubated overnight with primary antibodies in TBS-T containing 1% milk. Blots were washed three times for 15 min each with TBS-T, followed by a 4-h incubation with HRP-conjugated secondary antibodies in TBS-T containing 1% milk. Bound antibodies were detected with Supersignal chemiluminescence reagent (Pierce), and Supersignal-treated immunoblots were exposed to film to visualize the bound HRP conjugates.
RESULTS
Extent of factory costaining for cUb correlates with factory morphology.
To establish whether reovirus factories contain ubiquitinated proteins, we used IF microscopy to examine the factories in CV-1 cells infected at 37°C with any of 16 different viral strains, including the common laboratory prototypes and other field isolates (22, 24, 27, 38). Samples were analyzed at 19 to 24 h postinfection (p.i.), by which time reovirus has completed the exponential phase of growth (45). To detect cUb, we used MAb FK2, which recognizes both multi- and monoubiquitinated proteins but not free Ub (15). To identify factories, we used polyclonal antiserum against viral nonstructural protein μNS or core protein μ2 (6, 38). In cells infected with either of the strains that have been previously shown to form globular factories (T3DN and T3C12) (38), cUb costained with factories in a large percentage of cells (95 and 76%, respectively) (Fig. 1, top row; see Table S1 in the supplemental material). In cells infected with any of the other 14 strains (T1L, T3DC, etc.), all of which form filamentous factories (38), cUb costained with factories in many fewer cells (≤16%) (Fig. 1, middle row; see Table S1 in the supplemental material). Thus, among these strains, the extent of factory costaining for cUb correlated with factory morphology.
FIG. 1.
Distribution of cUb in infected cells with globular or filamentous viral factories. CV-1 cells were mock infected (bottom row) or infected with T3DN (top row) or T1L (middle row) at an MOI of 5 PFU/cell and then incubated at 37°C. Cells were fixed at 24 h p.i. and immunostained with rabbit anti-μ2 polyclonal serum followed by anti-rabbit IgG conjugated to Alexa 594 (α-μ2) (left column) and with mouse MAb FK2 against cUb followed by anti-mouse IgG conjugated to Alexa 488 (α-cUb) (right column). Bars, 10 μm. The brightly stained region at bottom left in the middle right panel is the nucleus of an uninfected cell in this field.
We performed several additional experiments with strains T3DN and T1L to determine the effects of other variables on viral factories formed at 37°C in CV-1 cells. MOIs of between 0.5 and 50 PFU/cell had minimal effects on the percentage of cells in which cUb and factories colocalized at 24 h p.i. (data not shown). At increasing times p.i., the percentage of cells showing factory costaining for cUb remained largely unchanged: high for T3DN and low for T1L (Table 1). We also tested MAb FK1, which recognizes multiubiquitinated proteins but neither monoubiquitinated proteins nor free Ub (15), and obtained results for colocalization of cUb with viral factories at 24 h p.i. that were similar to those obtained with FK2 (data not shown). Effects of growth temperature are described in a later section. In addition, T3DN and T1L showed differences in the extents of factory costaining for cUb in L929 and HeLa cells that were similar to those in CV-1 cells (Table 1).
TABLE 1.
Effect of cell type and time p.i. on viral factory costaining for cUb
| Virus strain | Cell type | Factory morphologya | % of infected cells with factories that costained with cUb at the following time (h) p.i.b:
|
|||
|---|---|---|---|---|---|---|
| 12 | 24 | 36 | 48 | |||
| T3DN | CV-1 | Globular | 90 ± 2 | 90 ± 5 | 91 ± 5 | 88 ± 3 |
| HeLa | Globular | NDc | 49 ± 18 | ND | 85 ± 6 | |
| L929 | Globular | ND | 85 ± 5 | ND | 83 ± 3 | |
| T1L | CV-1 | Filamentous | 6 ± 1 | 8 ± 3 | 9 ± 2 | 8 ± 3 |
| HeLa | Filamentous | ND | 2 ± 2 | ND | 14 ± 6 | |
| L929 | Filamentous | ND | 4 ± 3 | ND | 3 ± 1 | |
For CV-1 cells, in which counts were performed, filamentous indicates that ≥96% of cells displayed obvious filaments, and globular indicates that ≤1% of cells displayed obvious filaments.
Results are means of three determinations ± standard deviations; the MOI was 5 PFU/cell. Factories were identified by staining with anti-μ2 or anti-μNS serum. cUb was identified by staining with MAb FK2.
ND, not determined.
Viral M1 genome segment determines factory costaining for cUb.
To ascertain the genetic basis of the different levels of cUb costaining with T3DN and T1L factories, we infected CV-1 cells with a panel of T1L × T3D reassortants previously generated in the laboratory of B. N. Fields (9, 12, 14, 35). T3D clones traceable to that lab, including our own T3DN clones, form globular factories (51). The factories of the T1L × T3D reassortants were then examined for colocalization with cUb by IF microscopy. After ranking the viruses by the percentage of cells with factories that costained for cUb, we found that reassortants with genome segment L1 or M1 from T3D had the highest percentages of cells with this phenotype, whereas reassortants with both L1 and M1 from T1L had the lowest percentages (Table 2; Fig. 2). Statistical analysis of the full data set by using the nonparametric Wilcoxon-Mann-Whitney test (4, 43, 52) revealed that the parental origins of both L1 and M1 segregated with the phenotype to significant degrees (L1, P = 0.03; M1, P < 0.0001; other segments, P > 0.05).
TABLE 2.
Genetic analysis of factory costaining for cUb by using T1L × T3D reassortant viruses
| Virus straina | Genotypeb
|
% cUbc
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L1 | L2 | L3 | M1 | M2 | M3 | S1 | S2 | S3 | S4 | Mean ± SD | Rank | |
| EB86 | L | D | D | D | D | L | D | D | D | L | 95 ± 1 | 1 |
| T3DN | D | D | D | D | D | D | D | D | D | D | 92 ± 6 | 2 |
| EB137 | D | D | D | D | L | L | D | L | L | L | 90 ± 4 | 3 |
| EB138 | D | L | L | D | D | L | D | D | L | L | 89 ± 7 | 4 |
| EB113 | L | L | L | D | L | L | L | L | D | L | 88 ± 13 | 5 |
| EB134 | L | D | L | D | L | L | L | L | D | L | 86 ± 9 | 6 |
| EB108 | L | D | L | D | L | L | L | L | D | D | 84 ± 11 | 7 |
| EB136 | D | D | D | L | D | L | D | D | D | D | 80 ± 8 | 8 |
| EB18 | D | D | L | D | D | D | L | L | D | L | 79 ± 6 | 9 |
| EB31 | L | L | L | D | L | L | L | D | D | L | 77 ± 16 | 10 |
| G16 | L | L | L | D | L | L | L | D | L | L | 75 ± 20 | 11 |
| EB127 | D | D | L | L | D | L | L | D | D | L | 73 ± 14 | 12 |
| EB13 | D | D | D | D | D | D | D | D | D | L | 72 ± 4 | 13 |
| EB143 | D | L | L | L | L | L | D | L | L | L | 70 ± 3 | 14 |
| EB146 | L | L | L | D | L | L | L | L | L | D | 69 ± 20 | 15 |
| H60 | D | D | L | L | D | D | D | D | D | L | 64 ± 24 | 16 |
| EB140 | D | D | L | L | L | L | L | L | D | L | 61 ± 14 | 17.5 |
| H41 | D | D | L | L | L | D | L | L | D | L | 61 ± 13 | 17.5 |
| H15 | L | D | D | L | D | D | D | D | D | L | 56 ± 18 | 19 |
| EB98 | L | D | L | L | L | L | L | D | L | D | 52 ± 9 | 20 |
| EB39 | L | D | D | L | D | D | D | D | D | D | 38 ± 14 | 21 |
| EB87 | L | D | L | L | D | L | L | D | L | L | 36 ± 7 | 22 |
| EB67 | L | D | L | L | L | D | L | L | L | L | 20 ± 12 | 23 |
| EB144 | L | L | L | L | D | D | L | L | D | L | 14 ± 8 | 24 |
| T1L | L | L | L | L | L | L | L | L | L | L | 13 ± 3 | 25 |
| EB85 | L | L | L | L | L | D | L | D | L | L | 12 ± 9 | 26.5 |
| H24 | L | L | L | L | L | L | L | L | L | D | 12 ± 10 | 26.5 |
| KC19 | L | L | L | L | D | L | D | L | D | L | 8 ± 5 | 28 |
| H14 | L | L | D | L | L | L | L | D | D | L | 7 ± 5 | 29 |
Reassortants are described in Materials and Methods. Strains are listed in order of decreasing frequencies of factory costaining for cUb.
Parental origins of genome segments as indicated: D, T3D; L, T1L.
Percentage of infected CV-1 cells with factories that costained with cUb at 24 h p.i. Results are for three to six determinations; the MOI was 5 PFU/cell. Factories were identified by staining with anti-μ2 or anti-μNS serum. cUb was identified by staining with MAb FK2. The rank was assigned in preparation for the Wilcoxon-Mann-Whitney test.
FIG. 2.
Factory costaining for cUb in cells infected with T1L × T3D reassortant viruses. Mean values from Table 2 are plotted with respect to the parental origins of genome segments L1, L2, and M1. Filled symbols, reassortants containing T3D M1 (in boldface at bottom); open symbols, reassortants containing T1L M1. Three nonoverlapping groups are defined by the parental origin of genome segments L1 and L2 among the reassortants containing T1L M1: (i) L1 from T3D and L2 from either T3D or T1L, (ii) L1 from T1L and L2 from T3D, and (iii) both L1 and L2 from T1L.
Based on the preceding results, we further analyzed the reassortant data by separating them into two sets defined by the parental origin of M1 and again applying the Wilcoxon-Mann-Whitney test. For reassortants containing T3D M1, no other segment was significantly associated with the phenotype (for each segment, P > 0.05). For reassortants containing T1L M1, however, both L1 and L2 were significantly associated with the phenotype (L1, P = 0.0002; L2, P = 0.007; other segments, P > 0.05). In fact, the extents of factory costaining for cUb among the reassortants containing T1L M1 divided into three nonoverlapping groups according to the parental origins of L1 and L2, with reassortants containing T3D L1 having the highest values (Fig. 2).
From these experiments, we draw the following conclusions. (i) The μ2-encoding M1 genome segment of T3D is associated with higher levels of factory costaining for cUb and can demonstrate this effect in the absence of any other T3D genome segment. In the remainder of this report, we focus attention on defining the basis of this effect. (ii) In the presence of the μ2-encoding M1 genome segment of reovirus T1L, associations of two other T3D genome segments with higher levels of factory costaining for cUb become apparent: the λ3-encoding L1 genome segment and the λ2-encoding L2 genome segment. (iii) The effect of T3D L1 on levels of factory costaining for cUb is greater than that of T3D L2. The basis of the L1/λ3 and L2/λ2 effects in the presence of T1L M1/μ2 is discussed below but requires further studies to clarify.
T3DN μ2 is itself ubiquitinated in infected cells.
Ubiquitinated viral or cellular proteins may account for the colocalization of cUb with viral factories. Because the extent of factory costaining for cUb genetically mapped to the μ2-encoding M1 segment, we hypothesized that ubiquitinated forms of μ2 may accumulate to different levels during infections by the different viral strains. Because M1/μ2 also determines factory morphology (38), we looked for Ub-μ2 in cells infected with strains that form either globular factories (T3DN) or filamentous factories (T1L or T3DC). To improve detection of Ub-μ2, CV-1 cells were first transfected with a plasmid expressing HA-tagged Ub (HAUb) (33). Cell lysates were harvested at 18 h p.i., and the μ2 proteins were immunoprecipitated under denaturing conditions. After SDS-PAGE and blotting to nitrocellulose, immunoprecipitated proteins were probed with appropriate antibodies to visualize either μ2 (Fig. 3, top panels) or HAUb (Fig. 3, bottom panels). Strikingly, T3DN-infected cells displayed clear evidence for Ub-μ2, i.e., a characteristic ladder of HAUb-tagged μ2 species of higher molecular weight in immunoblots (Fig. 3, bottom panels, left). In contrast, T1L- or T3DC-infected cells displayed limited evidence for Ub-μ2 (Fig. 3, bottom panels, middle and right). These results suggest that ubiquitinated T3DN μ2 contributes to the colocalization of cUb with T3DN factories.
FIG. 3.
Ubiquitination of μ2 in T3DN-, T1L-, or T3DC-infected cells. CV-1 cells were transfected with 3 μg of pHAUb per 60-mm-diameter dish and 6 h later were infected with T3DN, T1L, or T3DC intermediate subvirion particles, as indicated, at an MOI of 50 PFU/cell and then incubated at 37°C. At 18 h p.i., cells were harvested and equilibrated for total protein concentration. Following lysis in denaturating buffer, samples were immunoprecipitated with rabbit anti-μ2 polyclonal serum (α-μ2 IP). Protein A-Sepharose beads alone (BA) were used as a control. Immunoprecipitated proteins were separated by SDS-PAGE, transferred to nitrocellulose, and immunoblotted with the anti-μ2 serum followed by HRP-conjugated anti-rabbit IgG (α-μ2 Blot) (upper row) or with mouse anti-HA MAb followed by HRP-conjugated anti-mouse IgG (α-HA Blot) (bottom row). Bound HRP conjugates were detected by chemiluminescence. Asterisks, cross-reaction of HRP-conjugated antibodies with IgG and protein A used in immunoprecipitations.
T3DN μ2 is also ubiquitinated in transfected cells and forms aggregates that colocalize with cUb.
To determine whether μ2 is ubiquitinated in the absence of infection, we transfected CV-1 cells with plasmids expressing T3DN or T1L μ2 and examined the cells for colocalization of μ2 and cUb by IF microscopy. In some experiments, endogenous cUb was visualized by staining with MAb FK2 (Fig. 4A). In other experiments, we cotransfected the cells with a plasmid expressing HAUb and examined them for cUb by staining with a MAb specific for the HA tag (Fig. 4B). In both types of experiment, T3DN μ2 formed aggregates that strongly colocalized with cUb in a majority of the transfected cells, whereas T1L μ2 associated with microtubules and only rarely colocalized with cUb (Fig. 4A and B; for representative quantitations, see Table 4). The different morphological patterns of these two allelic forms of μ2 (in aggregates or on microtubules) have been previously noted (38). Since the M1 segment of globular strain T3C12 encodes a μ2 protein with a sequence identical to that of T3DN (51), these (and subsequent) transfection results for T3DN μ2 represent those for T3C12 μ2 as well.
FIG. 4.
Distribution of cUb and ubiquitination of μ2 in transfected cells expressing T3DN or T1L μ2. (A) IF microscopy of CV-1 cells transfected with 2 μg of pCI-M1(T3DN) (top row) or pCI-M1(T1L) (bottom row) per well. Cells were fixed at 18 h p.t. and immunostained with rabbit anti-μ2 polyclonal serum followed by anti-rabbit IgG conjugated to Alexa 594 (α-μ2) (left column) (red in merge) and with mouse MAb FK2 against cUb followed by anti-mouse IgG conjugated to Alexa 488 (α-cUb) (middle column) (green in merge). Nuclei were counterstained with DAPI (blue in merge). Bars, 10 μm. (B) IF microscopy of CV-1 cells cotransfected with 1 μg of pHAUb and 1 μg of either pCI-M1(T3DN) (top row) or pCI-M1(T1L) (bottom row) per well. Cells were fixed at 18 h p.t. and immunostained with the anti-μ2 serum followed by anti-rabbit IgG conjugated to Alexa 594 (α-μ2) (left column) (red in merge) and with mouse anti-HA MAb followed by anti-mouse IgG conjugated to Alexa 488 (α-cUb) (middle column) (green in merge). Nuclei were counterstained with DAPI (blue). Bars, 10 μm. (C) CV-1 cells transfected with 2 μg of pHAUb and 2 μg of pCI-M1(T3DN), pCI-M1(T1L), or pCI-neo (Vector) per plate as indicated. Half of the samples as indicated were treated with MG132 at 12 h p.t. Cells were harvested at 18 h p.t. and equilibrated for total protein concentration. Following lysis in denaturing buffer, samples were immunoprecipitated with the anti-μ2 serum (α-μ2 IP). Immunoprecipitated proteins were separated by SDS-PAGE, transferred to nitrocellulose, and immunoblotted with the anti-μ2 serum followed by HRP-conjugated anti-rabbit IgG (α-μ2 Blot) (left panel) or with the anti-HA MAb followed by HRP-conjugated anti-mouse IgG (α-HA Blot) (right panel). Bound HRP conjugates were visualized by chemiluminescence. Asterisks, cross-reaction of HRP-conjugated antibody with IgG and protein A used in immunoprecipitations. Arrowhead, suspected cleavage product of T3DN μ2 (left panel, lanes 1 and 2).
TABLE 4.
Effect of temperature on μ2 colocalization with cUb
| M1 gene | Temp (°C) | % of transfected CV-1 cells showing μ2 colocalization with cUba |
|---|---|---|
| T3DN | 37 | 86 ± 6 |
| 31 | 50 ± 1 | |
| T1L | 37 | 22 ± 1 |
| 31 | 16 ± 6 | |
| T3DN-S208P | 37 | 38 ± 3 |
| 31 | 42 ± 3 | |
| T1L-P208S | 37 | 85 ± 4 |
| 31 | 57 ± 1 |
Results are means of three determinations ± standard deviations. CV-1 cells were transfected with 2 μg of pCI-M1 (T3DN), pCI-M1 (T1L), pCI-M1(T3DN-S208P), or pCI-M1 (T1L-P208S) per well and incubated at 37 or 31°C. At 18 h p.t., cells were fixed and immunostained for μ2 and cUb as described for Fig. 7.
To extend the preceding results, we cotransfected CV-1 cells with plasmids expressing T3DN or T1L μ2 and HAUb and examined them for Ub-μ2 by immunoprecipitation from cell lysates. In some samples, we optimized for detection of ubiquitinated proteins by treating the cells with MG132, which blocks proteasomal degradation of such proteins and thereby causes them to accumulate (reviewed in reference 28). The μ2 proteins were immunoprecipitated from lysates under denaturing conditions. After SDS-PAGE and blotting to nitrocellulose, immunoprecipitated proteins were probed with appropriate antibodies to visualize either μ2 (Fig. 4C, left panel) or HAUb (Fig. 4C, right panel). Cells expressing T1L μ2 displayed levels of Ub-μ2 that were higher in the presence than in the absence of MG132 (Fig. 4C, right panel), identifying a role for the Ub-proteasome system in degradation of this protein. The higher levels of total T1L μ2 protein obtained in the presence of MG132 (Fig. 4C, left panel) are consistent with this conclusion. In contrast, cells expressing T3DN μ2 displayed high levels of Ub-μ2 even in the absence of MG132, and little change in the levels of either Ub-μ2 or total μ2 was evident in the presence of inhibitor (Fig. 4C, left and right panels).
In sum, the results in this section suggest that ubiquitinated T3DN protein accumulates to high levels in transfected cells. Although the T1L μ2 protein was also ubiquitinated in these experiments, this was detectable only when its degradation was disrupted by MG132; moreover, the levels to which ubiquitinated T1L μ2 accumulated did not approach those of T3DN μ2 even in the presence of the inhibitor, and T1L μ2 did not commonly form aggregates. The limited effect of MG132 on T3DN μ2 possibly reflects that this protein accumulates in aggregates, where it is resistant to degradation even when the inhibitor is absent. Notably, the results obtained with transfected cells in the absence of MG132 paralleled those obtained with infected cells, according to the origin of M1/μ2.
Allelic variation at μ2 residue 208 affects levels of Ub-μ2 in transfected cells.
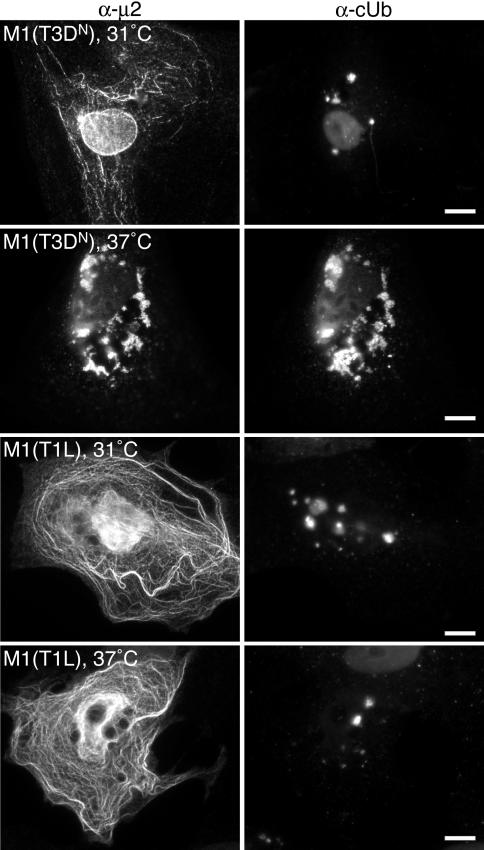
Viral strain-dependent differences in μ2 association with microtubules are determined by variation at μ2 residue 208: Pro208, as in T1L and T3DC μ2, causes effective microtubule association, while Ser208, as in T3DN and T3C12 μ2, causes ineffective association (38). Because the levels of Ub-μ2 appeared to correlate with the microtubule association capacities of μ2, we hypothesized that variation at position 208 may also influence the ubiquitination phenotype of μ2. We therefore transfected CV-1 cells with plasmids to express either T3DN-S208P or T1L-P208S μ2, each of which has the opposite microtubule association phenotype from its respective parent protein (38), and examined them for μ2 costaining with cUb by IF microscopy. We found that each of these reciprocal changes caused at least partial reversal of the ubiquitination phenotype: T1L-P208S μ2, which formed aggregates in most cells (38), colocalized with cUb in most cells (Fig. 5A, bottom row), whereas T3DN-S208P μ2, which associated with microtubules in most cells (38), colocalized with cUb in many fewer cells than did T3DN μ2 (Fig. 5A, top row [compare with Fig. 4A, top row; for representative quantitations, see Table 4]).
FIG. 5.
Distribution of cUb and ubiquitination of μ2 in transfected cells expressing T3DN or T1L μ2 with allelic variation at amino acid 208. (A) IF microscopy of CV-1 cells transfected with 2 μg of either pCI-M1(T3DN-S208P) (top row) or pCI-M1(T1L-P208S) (bottom row) per well. Cells were fixed at 18 h p.t. and immunostained with rabbit anti-μ2 polyclonal serum followed by anti-rabbit IgG conjugated to Alexa 594 (α-μ2) (left column) and with mouse MAb FK2 against cUb followed by anti-mouse IgG conjugated to Alexa 488 (α-cUb) (right column). Bars, 10 μm. (B) CV-1 cells transfected with 2 μg of pHAUb and 2 μg of pCI-M1(T3DN-S208P), pCI-M1(T1L-P208S), pCI-M1(T3DN), pCI-M1(T1L), or pCI-neo (Vector) per plate as indicated. Half of the samples as indicated were treated with MG132 at 12 h p.t. Cells were harvested at 18 h p.t. and equilibrated for total protein concentration. Following lysis in denaturing buffer, samples were immunoprecipitated with the anti-μ2 serum (α-μ2 IP). Immunoprecipitated proteins were separated by SDS-PAGE, transferred to nitrocellulose, and immunoblotted with the anti-μ2 serum followed by HRP-conjugated anti-rabbit IgG (α-μ2 Blot) (top panel) or with mouse anti-HA MAb followed by HRP-conjugated anti-mouse IgG (α-HA Blot) (bottom panel). Bound HRP conjugates were visualized by chemiluminescence. Asterisks, cross-reaction of HRP-conjugated antibody with IgG and protein A used in immunoprecipitations. Arrowhead, suspected cleavage product of T3DN or T3DN-derived μ2 (upper panel, lanes 1, 2, 5, and 6).
We also used immunoprecipitation to look for Ub-μ2 in cells expressing the T1L or T3DN μ2 protein with changes at residue 208. CV-1 cells were cotransfected with expression plasmids encoding HAUb and T3DN, T1L, T3DN-S208P, or T1L-P208S μ2. MG132 was added to some of the samples to optimize detection of ubiquitinated proteins. The μ2 proteins were immunoprecipitated from the cell lysates under denaturing conditions. After SDS-PAGE and blotting to nitrocellulose, immunoprecipitated proteins were probed with appropriate antibodies to visualize either μ2 (Fig. 5B, top panel) or HAUb (Fig. 5B, bottom panel). Cells expressing T1L-P208S μ2 displayed much higher levels of Ub-μ2 than cells expressing T1L μ2, in the absence of MG132 as well as in its presence (Fig. 5B, bottom panel). Additionally, based on the levels of total μ2 protein obtained in the absence or presence of MG132, T1L-P208S μ2 appeared to be less efficiently degraded than T1L μ2 (Fig. 5B, top panel), consistent with its presence in aggregates (Fig. 5A, bottom row). Cells expressing T3DN-S208P μ2, in contrast, displayed lower levels of Ub-μ2 than cells expressing T3DN μ2 in the absence of MG132 (Fig. 5B, bottom panel). Moreover, again based on the overall levels of μ2 obtained in the absence or presence of MG132, T3DN-S208P μ2 appeared to be more efficiently degraded than T3DN μ2 (Fig. 5B, top panel), consistent with its nonaggregated state (Fig. 5A, top row).
In sum, the findings in this section provide further evidence for a key role of μ2 residue Ser208 or Pro208 in influencing the aggregation, ubiquitination, degradation, and microtubule association phenotypes of the μ2 protein in transfected cells.
Factory costaining for cUb is not affected by microtubule disruption.
Disruption of microtubules by nocodazole treatment early in infection results in both T3DN and T1L forming small globular factories that remain dispersed through the cytoplasm (38). To test whether microtubule association is necessary for the more limited colocalization of cUb with T1L factories, we infected cells with T3DN or T1L, treated them with nocodazole starting at 6 h p.i., and then analyzed them for ubiquitinated proteins in viral factories by IF microscopy at 24 h p.i. As expected, we found that the factories of both strains were now globular in nature (see Fig. S2 in the supplemental material). Nonetheless, the percentages of infected cells exhibiting factory costaining for cUb were similar to those in the absence of nocodazole: high for T3DN but low for T1L (see Fig. S2 and Table S3 in the supplemental material). These results provide evidence that the levels of ubiquitinated proteins in viral factories are not directly determined by whether or not the factories are anchored to microtubules.
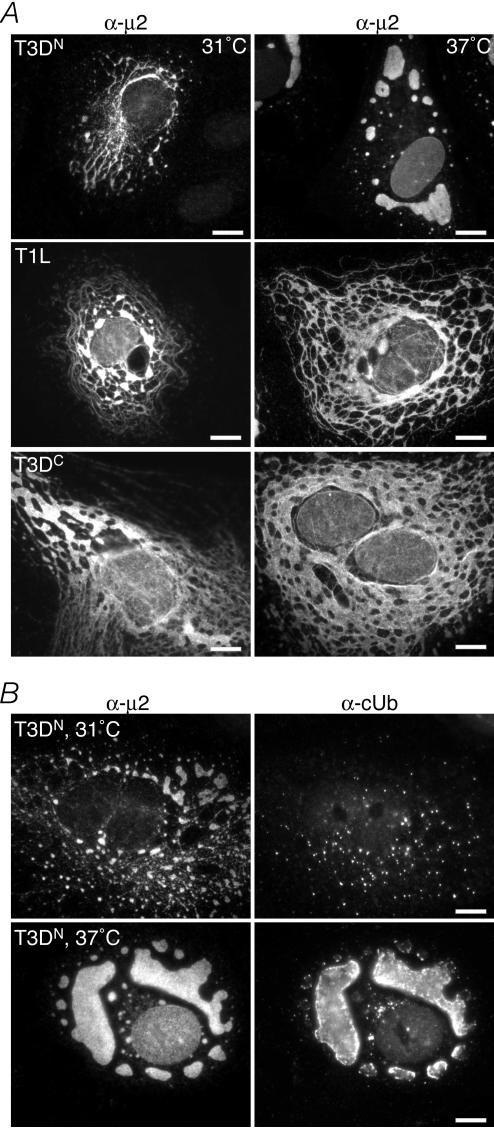
In infected cells, T3DN factory morphology and costaining for cUb are temperature dependent.
The preceding results in this report were generated with cells grown at 37°C. In an effort to learn more about the basis of the strain-dependent differences displayed by μ2 in this study, we tested whether any of these phenotypes may be temperature dependent. To this end, we infected CV-1 cells with T3DN, T1L, or T3DC at 37 or 31°C and examined factory morphology by IF microscopy. We found that although there were no obvious differences in the factories of T1L or T3DC at the two temperatures, the factories of T3DN at 31°C were much more commonly filamentous than were those at 37°C (Fig. 6A; Table 3). When we costained for cUb, we saw an associated decrease in the colocalization of cUb with the factories of T3DN at 31°C (Fig. 6B; Table 3). Results at 31°C that were similar to these with T3DN were obtained with T3C12, which also forms globular factories at 37°C (data not shown). These findings indicate that factory morphology and costaining for cUb, both of which phenotypes have been mapped to M1/μ2, are sensitive to the temperature at which T3DN or T3C12 is grown. The findings also suggest that the μ2 proteins of these strains have a temperature-dependent defect.
FIG. 6.
Viral factory morphology and cUb distribution in infected cells at 31 or 37°C. (A) IF microscopy of CV-1 cells infected with T3DN (top row), T1L (middle row), or T3DC (bottom row) at an MOI of 5 PFU/cell and incubated for 48 h at 31°C (left column) or 37°C (right column). Following incubation, cells were fixed and immunostained with rabbit anti-μ2 polyclonal serum followed by anti-rabbit IgG conjugated to Alexa 594 (α-μ2). Bars, 10 μm. (B) IF microscopy of CV-1 cells infected with T3DN at 31°C (top row) or 37°C (bottom row). At 48 h p.i., cells were fixed and immunostained with the anti-μ2 serum followed by anti-rabbit IgG conjugated to Alexa 594 (α-μ2) (left column) and with mouse MAb FK2 against cUb followed by anti-mouse IgG conjugated to Alexa 488 (α-cUb) (right column). Bars, 10 μm.
TABLE 3.
Effect of temperature on viral factory morphology and costaining for cUb
| Virus strain | Temp (°C) | % of infected CV-1 cells with factories that area:
|
|
|---|---|---|---|
| Filamentous | Costained for cUb | ||
| T3DN | 37 | 2 ± 3 | 90 ± 1 |
| 31 | 32 ± 5 | 71 ± 3 | |
| T1L | 37 | 99 ± 0 | 7 ± 1 |
| 31 | 99 ± 0 | 22 ± 9 | |
Results are means of three or four determinations ± standard deviations; the MOI was 5 PFU/cell. Factories were identified by staining with anti-μ2 serum. cUb was identified by staining with MAb FK2.
In transfected cells, microtubule association and colocalization with cUb by T3DN μ2 are temperature dependent and are affected by allelic variation at μ2 residue 208.
To address whether the shift from globular to filamentous factories in T3DN-infected cells at 31°C reflects increased microtubule association by T3DN μ2, we transfected CV-1 cells with expression plasmids encoding the T3DN or T1L μ2 protein and maintained the cells at 37 or 31°C. At 18 h posttransfection (p.t.), we examined the cells for μ2 and ubiquitinated proteins by IF microscopy. The properties of T1L μ2 (i.e., association with microtubules and less common colocalization with cUb) appeared similar at both temperatures (Fig. 7, bottom rows; Table 4). In contrast, T3DN μ2, which formed aggregates that colocalized with cUb in most cells at 37°C, was shifted to a more filamentous distribution and associated with cUb in many fewer cells at 31°C (Fig. 7, top rows; Table 4). These results support our hypothesis that microtubule association by T3DN μ2 is temperature dependent and that at the lower temperature, more of the protein is competent for microtubule association while less is subject to aggregation and accumulation in ubiquitinated forms.
FIG. 7.
Distribution of cUb and T3DN or T1L μ2 in transfected cells at 31 or 37°C. CV-1 cells were transfected with 2 μg of pCI-M1(T3DN) (top two rows) or 2 μg of pCI-M1(T1L) (bottom two rows) per well. Transfected cells were incubated at 31 or 37°C as indicated. At 18 h p.t., cells were fixed and immunostained with rabbit anti-μ2 polyclonal serum followed by anti-rabbit IgG conjugated to Alexa 594 (α-μ2) (left column) and with mouse MAb FK2 against cUb followed by anti-mouse IgG conjugated to Alexa 488 (α-cUb) (right column). Bars, 10 μm.
To determine whether the temperature-dependent phenotypes of T3DN μ2 map to Ser208, we performed the same experiment with CV-1 cells transfected with plasmids to express T3DN-S208P μ2 or T1L-P208S. Similar to what we observed for T3DN μ2, T1L-P208S μ2, which formed aggregates that colocalized with cUb in most cells at 37°C, was shifted to a filamentous distribution and associated with cUb in many fewer cells at 31°C (Table 4; see Fig. S4 in the supplemental material). Moreover, similar to what we observed for T1L μ2, the properties of T3DN-S208P μ2 (association with microtubules and less common colocalization with cUb) were similar at both temperatures (Table 4; see Fig. S4 in the supplemental material). These results support the role of residue Ser208 in determining the temperature-dependent phenotypes of T3DN μ2.
DISCUSSION
Ubiquitination is a pervasive modification that plays key roles in protein turnover and regulation of protein functions in eukaryotic cells (reviewed in reference 39). Viral proteins intersect ubiquitination pathways in a variety of ways, both as substrates and as regulators (reviewed in reference 17). Ubiquitination of reovirus proteins has not been shown in previous publications but has been discussed at conferences (J. L. Mbisa and E. G. Brown, personal communication). The findings for μ2 in this report are therefore among the first linking aspects of reovirus infection and cellular ubiquitination pathways, a topic that may affect diverse aspects of reovirus replication, assembly, effects on host cells, and pathogenesis. The location(s) in the μ2 sequence where Ub chains are added and which cellular factors (E3 Ub ligases, etc. [reviewed in reference 39]) are required remain to be identified. Having alanine at residue 2 makes μ2 a poor candidate for Ub-based degradation by the N-end rule pathway (reviewed in reference 49).
Proposal that misfolding of T3DN μ2 is a primary, temperature-dependent defect that leads to increased aggregation, higher levels of Ub-μ2, and decreased association with microtubules.
We have previously reported that a strain-dependent difference in microtubule association of viral factories between reovirus strains T1L and T3DN maps to μ2 residue 208 (38). But how does the presence of Ser208 versus Pro208 determine this difference? Since in this study we showed that both T3DN and T1L-P208S μ2 formed ubiquitinated aggregates in transfected cells in a temperature-dependent manner, we conclude that Ser208 makes μ2 more prone to misfolding as the primary defect. Increased aggregation, higher levels of Ub-μ2, and decreased association with microtubules are then secondary effects according to our model. Although other explanations are possible, we believe that the temperature-dependent nature of the covariant phenotypes particularly favors the misfolding hypothesis.
Notably, the P208S mutation does not inhibit μ2 from associating with viral protein μNS, which forms the matrix of the factories (8, 38). The capacity of Ser208-containing μ2 to continue to associate with μNS in factories or factory-like inclusions suggests that the μNS-binding region of μ2 either is not subject to misfolding or can otherwise continue to function in the face of the Ser208-based effects. One possibility is that the μNS-binding region of μ2 is distinct from the region that binds microtubules and that the folding defect that leads to aggregation and higher levels of Ub-μ2 directly affects only the microtubule-binding region.
Other sequence differences in T1L and T3DN μ2 and their roles in ubiquitination and degradation.
Although T3DN-S208P μ2 accumulated ubiquitinated forms to lower levels than did T3DN μ2, it did so to higher levels than did T1L μ2 (Fig. 5B, bottom panel; Table 4). This observation suggests that at least one residue unique to T3DN in addition to Ser208 contributes to the higher levels of ubiquitinated T3DN μ2 in transfected cells. Since none of the sequence differences between T3DN and T1L μ2 involves a lysine residue (38), the ubiquitination differences are unlikely to be explained by the number of potential Ub ligation sites. Higher levels of Ub-μ2 in transfected or infected cells could reflect more frequent ubiquitination and/or less efficient degradation once ubiquitination has occurred. Given the accumulation of T1L and T3DN-S208P μ2 after a 6-h treatment with MG132, we infer that these proteins were subject to substantial degradation over this period in the untreated cells. These results and others indicate that even when μ2 is free of the effects of Ser208, it is a substrate for the Ub-proteasome system. T3DN and T1L-P208S μ2, in contrast, were less subject to degradation over the 6-h period in untreated cells, possibly because they were aggregated and therefore more resistant to proteasomal degradation (3). Thus, although at least one of the other amino acid differences between T3DN and T1L μ2 determines the higher levels of ubiquitinated T3DN-S208P μ2 in transfected cells, it is specifically Ser208 in T3DN μ2 that results in a protein that is prominently aggregated and putatively misfolded.
Roles of μ2 in viral replication and pathogenesis.
As a component of viral cores (11, 34) and an enzyme capable of nucleoside and RNA triphosphatase activities (25, 37), μ2 is thought to play a key role in viral mRNA synthesis (50) and possibly in minus-strand synthesis of the viral genome as well (10). Despite the proposed folding defect in T3DN μ2 at 37°C, this strain can generally replicate to high yields in L929 cells at that temperature, and its virions assembled at 37°C have a relative infectivity similar to those of other strains (data not shown). Thus, a large enough fraction of the T3DN μ2 molecules expressed in infected L929 cells at 37°C must be available in functional form for the essential role(s) that μ2 plays in replication and assembly of infectious particles in those cells.
A number of other strain-dependent phenotypes have been genetically mapped to the μ2-encoding M1 genome segment. These include the different infectious yields of T3D (Fields laboratory derivatives) and T1L upon replication in murine heart cells (30), bovine aortic endothelial cells (29), or the livers of severe combined immunodeficiency mice (23). In each of these cases, the yields of T1L are higher than those of T3D. Although the mechanistic basis of these M1/μ2 effects on viral replication remains largely unknown, a new hypothesis based on evidence in this report is that levels of functional T3D μ2 may not be high enough for efficient replication in certain cell types. It also remains possible that microtubule association of factories by T1L μ2 confers a replication advantage in these cell types.
Aggresome model for reovirus factories.
A previous study has noted several similarities between reovirus factories and cellular aggresomes (38). Because many of the misfolded proteins in aggresomes have been tagged for destruction by the Ub-proteasome system, immunostaining for multiubiquitinated proteins can often be used to identify these structures (reviewed in references 19 and 26). In the present study, we found that ubiquitinated proteins regularly colocalized with the factories formed by reovirus strains T3DN and T3C12 but not with those formed by 14 other strains. Moreover, even with T3DN and T3C12, the extent to which ubiquitinated proteins colocalized with factories was temperature dependent, decreasing at reduced temperature. Based on the evidence for strain- and temperature-dependent effects on the levels of Ub-μ2 as a primary determinant of the colocalization of cUb with reovirus factories, we consider it improper to conclude that the factories are consistently “aggresome-like.” It nevertheless remains possible that some or all reoviruses co-opt elements of the cellular aggresome response to aid in forming the factories.
Roles of λ3 and λ2 in factory costaining for cUb.
Two other T3D genome segments, L1 (λ3 protein) and L2 (λ2 protein), increased the extent of factory costaining for cUb in the presence of T1L M1/μ2 in reassortant viruses. λ3 is the viral RNA-dependent RNA polymerase, and λ2 possesses both guanylyltransferase and methyltransferase activities that are involved in adding a 5′ cap to the viral plus-strand RNAs (18, 40, 46). We have not yet determined the mechanism by which these genome segments or their encoded proteins can affect the levels of ubiquitinated proteins in viral factories. Nonetheless, the results with these reassortants containing T1L M1 clearly show that there must be additional mechanisms by which ubiquitinated proteins can accumulate in viral factories in the absence of Ser208-containing μ2. By continuing to investigate these different mechanisms, we hope to learn more about the effects of the Ub-proteasome system on reovirus replication, assembly, and host cells.
Acknowledgments
We express our sincere thanks to Elaine Freimont for lab maintenance and technical assistance and to other members of our laboratories for helpful discussions.
This work was supported by NIH grants R01 AI47904 (to M.L.N.), K08 AI52209 (to J.S.L.P), and F32 AI56939 (to C.L.M.) and also by a junior faculty research grant from the Giovanni Armenise-Harvard Foundation (to M.L.N.). C.L.M. received additional support from NIH grant T32 AI07061 to the Combined Infectious Diseases Training Program at Harvard Medical School during earlier stages of this work. M.J.P. received additional support from NIH grant T32 AI07245 to the Viral Infectivity Training Program at Harvard Medical School. C.D.S.P. received additional support from the Harvard College Research Program.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Becker, M. M., M. I. Goral, P. R. Hazelton, G. S. Baer, S. E. Rodgers, E. G. Brown, K. M. Coombs, and T. S. Dermody. 2001. Reovirus σNS protein is required for nucleation of viral assembly complexes and formation of viral inclusions. J. Virol. 75:1459-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becker, M. M., T. R. Peters, and T. S. Dermody. 2003. Reovirus σNS and μNS proteins form cytoplasmic inclusion structures in the absence of viral infection. J. Virol. 77:5948-5963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bence, N. F., R. M. Sampat, and R. R. Kopito. 2001. Impairment of the ubiquitin-proteasome system by protein aggregation. Science 292:1552-1555. [DOI] [PubMed] [Google Scholar]
- 4.Bodkin, D. K., and B. N. Fields. 1989. Growth and survival of reovirus in intestinal tissue: role of the L2 and S1 genes. J. Virol. 63:1188-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brentano, L., D. L. Noah, E. G. Brown, and B. Sherry. 1998. The reovirus protein μ2, encoded by the M1 gene, is an RNA-binding protein. J. Virol. 72:8354-8357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Broering, T., A. McCutcheon, V. Centonze, and M. Nibert. 2000. Reovirus nonstructural protein μNS binds to reovirus cores but does not inhibit their transcription activity. J. Virol. 74:5516-5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Broering, T. J., J. Kim, C. L. Miller, C. D. S. Piggott, J. B. Dinoso, M. L. Nibert, and J. S. L. Parker. 2004. Reovirus nonstructural protein μNS recruits viral core surface proteins and entering core particles to factory-like inclusions. J. Virol. 78:1882-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Broering, T. J., J. S. L. Parker, P. L. Joyce, J. Kim, and M. L. Nibert. 2002. Mammalian reovirus nonstructural protein μNS forms large inclusions and colocalizes with reovirus microtubule-associated protein μ2 in transfected cells. J. Virol. 76:8285-8297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown, E. G., M. L. Nibert, and B. N. Fields. 1983. The L2 gene of reovirus serotype 3 controls the capacity to interfere, accumulate deletions, and establish persistent infection, p. 275-288. In R. W. Compans and D. H. L. Bishop (ed.), Double-stranded RNA viruses. Elsevier, New York, N.Y.
- 10.Coombs, K. M. 1996. Identification and characterization of a double-stranded RNA-reovirus temperature-sensitive mutant defective in minor core protein μ2. J. Virol. 70:4237-4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coombs, K. M. 1998. Stoichiometry of reovirus structural proteins in virus, ISVP, and core particles. Virology 243:218-228. [DOI] [PubMed] [Google Scholar]
- 12.Coombs, K. M., B. N. Fields, and S. C. Harrison. 1990. Crystallization of the reovirus type 3 Dearing core. Crystal packing is determined by the λ2 protein. J. Mol. Biol. 215:1-5. [DOI] [PubMed] [Google Scholar]
- 13.Dales, S., P. Gomatos, and K. C. Hsu. 1965. The uptake and development of reovirus in strain L cells followed with labelled viral ribonucleic acid and ferritin-antibody conjugates. Virology 25:193-211. [DOI] [PubMed] [Google Scholar]
- 14.Drayna, D., and B. N. Fields. 1982. Activation and characterization of the reovirus transcriptase: genetic analysis. J. Virol. 41:110-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujimuro, M., H. Sawada, and H. Yokosawa. 1994. Production and characterisation of monoclonal antibodies specific to multi-ubiquitin chains of polyubiquitinated proteins. FEBS Lett. 349:173-180. [DOI] [PubMed] [Google Scholar]
- 16.Furlong, D. B., M. L. Nibert, and B. N. Fields. 1988. Sigma 1 protein of mammalian reoviruses extends from the surfaces of viral particles. J. Virol. 62:246-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Furman, M. H., and H. L. Ploegh. 2002. Lessons from viral manipulation of protein disposal pathways. J. Clin. Investig. 110:875-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Furuichi, Y., S. Muthukrishnan, J. Tomasz, and A. J. Shatkin. 1976. Caps in eukaryotic mRNAs: mechanism of formation of reovirus mRNA 5′-terminal m7GpppGm-C. Prog. Nucleic Acid Res. Mol. Biol. 19:3-20. [DOI] [PubMed] [Google Scholar]
- 19.Garcia-Mata, R., Y. S. Gao, and E. Sztul. 2002. Hassles with taking out the garbage: aggravating aggresomes. Traffic 3:388-396. [DOI] [PubMed] [Google Scholar]
- 20.Goldberg, A. L. 2003. Protein degradation and protection against misfolded or damaged proteins. Nature 426:895-899. [DOI] [PubMed] [Google Scholar]
- 21.Gomatos, P. J., I. Tamm, S. Dales, and R. M. Franklin. 1962. Reovirus type 3: physical characteristics and interactions with L cells. Virology 17:441-454. [DOI] [PubMed] [Google Scholar]
- 22.Goral, M. I., M. Mochow-Grundy, and T. S. Dermody. 1996. Sequence diversity within the reovirus S3 gene: reoviruses evolve independently of host species, geographic locale, and date of isolation. Virology 216:265-271. [DOI] [PubMed] [Google Scholar]
- 23.Haller, B. L., M. L. Barkon, G. P. Vogler, and H. W. Virgin IV. 1995. Genetic mapping of reovirus virulence and organ tropism in severe combined immunodeficient mice: organ-specific virulence genes. J. Virol. 69:357-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hrdy, D. B., L. Rosen, and B. N. Fields. 1979. Polymorphism of the migration of double-stranded RNA genome segments of reovirus isolates from humans, cattle, and mice. J. Virol. 31:104-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim, J., J. S. L. Parker, K. E. Murray, and M. L. Nibert. 2004. Nucleoside and RNA triphosphatase activities of Orthoreovirus transcriptase cofactor μ2. J. Biol. Chem. 279:4394-4403. [DOI] [PubMed] [Google Scholar]
- 26.Kopito, R. R. 2000. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 10:524-530. [DOI] [PubMed] [Google Scholar]
- 27.Leary, T. P., J. C. Erker, M. L. Chalmers, A. T. Cruz, J. D. Wetzel, S. M. Desai, I. K. Mushahwar, and T. S. Dermody. 2002. Detection of mammalian reovirus RNA by using reverse transcription-PCR: sequence diversity within the λ3-encoding L1 gene. J. Clin. Microbiol. 40:1368-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee, D. H., and A. L. Goldberg. 1998. Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol. 8:397-403. [DOI] [PubMed] [Google Scholar]
- 29.Matoba, Y., W. S. Colucci, B. N. Fields, and T. W. Smith. 1993. The reovirus M1 gene determines the relative capacity of growth of reovirus in cultured bovine aortic endothelial cells. J. Clin. Investig. 92:2883-2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matoba, Y., B. Sherry, B. N. Fields, and T. W. Smith. 1991. Identification of the viral genes responsible for growth of strains of reovirus in cultured mouse heart cells. J. Clin. Investig. 87:1628-1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mbisa, J. L., M. M. Becker, S. Zou, T. S. Dermody, and E. G. Brown. 2000. Reovirus μ2 protein determines strain-specific differences in the rate of viral inclusion formation in L929 cells. Virology 272:16-26. [DOI] [PubMed] [Google Scholar]
- 32.Miller, C. L., T. J. Broering, J. S. L. Parker, M. M. Arnold, and M. L. Nibert. 2003. Reovirus σNS protein localizes to inclusions through an association requiring the μNS amino-terminus. J. Virol. 77:4566-4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller, C. L., and D. J. Pintel. 2001. The NS2 protein generated by the parvovirus minute virus of mice is degraded by the proteasome in a manner independent of ubiquitin chain elongation or activation. Virology 285:346-355. [DOI] [PubMed] [Google Scholar]
- 34.Mustoe, T. A., R. F. Ramig, A. H. Sharpe, and B. N. Fields. 1978. Genetics of reovirus: identification of the ds RNA segments encoding the polypeptides of the μ and σ size classes. Virology 89:594-604. [DOI] [PubMed] [Google Scholar]
- 35.Nibert, M. L., R. L. Margraf, and K. M. Coombs. 1996. Nonrandom segregation of parental alleles in reovirus reassortants. J. Virol. 70:7295-7300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nibert, M. L., and L. A. Schiff. 2001. Reoviruses and their replication, p. 1679-1728. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 37.Noble, S., and M. L. Nibert. 1997. Core protein μ2 is a second determinant of nucleoside triphosphatase activities by reovirus cores. J. Virol. 71:7728-7735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parker, J. S., T. J. Broering, J. Kim, D. E. Higgins, and M. L. Nibert. 2002. Reovirus core protein μ2 determines the filamentous morphology of viral inclusion bodies by interacting with and stabilizing microtubules. J. Virol. 76:4483-4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pickart, C. M. 2004. Back to the future with ubiquitin. Cell 116:181-190. [DOI] [PubMed] [Google Scholar]
- 40.Reinisch, K. M., M. L. Nibert, and S. C. Harrison. 2000. Structure of the reovirus core at 3.6 Å resolution. Nature 404:960-967. [DOI] [PubMed] [Google Scholar]
- 41.Rhim, J. S., L. E. Jordan, and H. D. Mayor. 1962. Cytochemical, fluorescent-antibody and electron microscopic studies on the growth of reovirus (ECHO 10) in tissue culture. Virology 17:342-355. [DOI] [PubMed] [Google Scholar]
- 42.Sharpe, A. H., L. B. Chen, and B. N. Fields. 1982. The interaction of mammalian reoviruses with the cytoskeleton of monkey kidney CV-1 cells. Virology 120:399-411. [DOI] [PubMed] [Google Scholar]
- 43.Sherry, B., and B. N. Fields. 1989. The reovirus M1 gene, encoding a viral core protein, is associated with the myocarditic phenotype of a reovirus variant. J. Virol. 63:4850-4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Silverstein, S. C., and P. H. Schur. 1970. Immunofluorescent localization of double-stranded RNA in reovirus-infected cells. Virology 41:564-566. [DOI] [PubMed] [Google Scholar]
- 45.Sturzenbecker, L. J., M. Nibert, D. Furlong, and B. N. Fields. 1987. Intracellular digestion of reovirus particles requires a low pH and is an essential step in the viral infectious cycle. J. Virol. 61:2351-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tao, Y., D. L. Farsetta, M. L. Nibert, and S. C. Harrison. 2002. RNA synthesis in a cage—structural studies of the reovirus polymerase λ3. Cell 111:733-745. [DOI] [PubMed] [Google Scholar]
- 47.Touris-Otero, F., J. Martinez-Costas, V. N. Vakharia, and J. Benavente. 2004. Avian reovirus nonstructural protein μNS forms viroplasm-like inclusions and recruits protein σNS to these structures. Virology 319:94-106. [DOI] [PubMed] [Google Scholar]
- 48.Tyler, K. L. 2001. Mammalian reoviruses, p. 1729-1748. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 49.Varshavsky, A. 2003. The N-end rule and regulation of apoptosis. Nat. Cell Biol. 5:373-376. [DOI] [PubMed] [Google Scholar]
- 50.Yin, P., M. Cheang, and K. M. Coombs. 1996. The M1 gene is associated with differences in the temperature optimum of the transcriptase activity in reovirus core particles. J. Virol. 70:1223-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yin, P., N. D. Keirstead, T. J. Broering, M. M. Arnold, J. S. L. Parker, M. L. Nibert, and K. M. Coombs. Comparisons of the M1 genome segments and encoded μ2 proteins of different reovirus isolates. Virol. J. 1:6. [DOI] [PMC free article] [PubMed]
- 52.Zar, J. H. 1984. Biostatistical analysis, 2nd ed. Prentice Hall, Englewood Cliffs, N.J.