Abstract
The introduction of a label which can be detected in living cells opens new possibilities for the direct analysis of dynamic processes in virus replication, such as the transport and assembly of structural proteins. Our aim was to generate a tool for the analysis of the trafficking of the main structural protein of human immunodeficiency virus type 1 (HIV-1), Gag, as well as for the analysis of virus-host cell interactions in an authentic setting. We describe here the construction and characterization of infectious HIV derivatives carrying a label within the Gag polyprotein. Based on our initial finding that a short epitope tag could be inserted near the C terminus of the matrix domain of Gag without affecting viral replication, we constructed HIV derivatives carrying the egfp gene at the analogous position, resulting in the expression of a Gag-EGFP fusion protein in the authentic viral context. Particles displaying normal viral protein compositions were released from transfected cells, and Gag-EGFP was efficiently processed by the viral protease, yielding the expected products. Furthermore, particles with mature morphology were observed by thin-section electron microscopy. The modified virus was even found to be infectious, albeit with reduced relative infectivity. By preparing mixed particles containing equimolar amounts of Gag-EGFP and Gag, we were able to obtain highly fluorescently labeled virion preparations which displayed normal morphology and full wild-type infectivity, demonstrating that the process of HIV particle assembly displays a remarkable flexibility. The fluorescent virus derivative is a useful tool for investigating the interaction of HIV with live cells.
Human immunodeficiency virus (HIV) virions are enveloped particles with a diameter of approximately 140 nm, the main constituent being the structural protein Gag. Gag is synthesized as a myristoylated polyprotein which traffics to the plasma membrane, where it assembles into immature spherical particles. Besides Gag, these particles contain the viral RNA genome and a number of other viral and cellular proteins, most prominently the Gag-Pol polyprotein comprising the viral enzymes and the envelope protein (Env). Concomitant with virus release, the Gag polyprotein is cleaved by the viral protease (PR) into its functional domains, the matrix (MA), capsid (CA), nucleocapsid (NC), and p6 domains, as well as two small spacer peptides. This cleavage leads to a morphological rearrangement of the virus core, which is crucial for virion infectivity. It has been shown that Gag alone is sufficient for the formation and release of virus-like particles (VLPs) (14). However, a number of other viral and cellular factors—most importantly the Env protein, genomic RNA, and the lipid membrane enveloping the virus particle—contribute to the assembly of HIV virions in an infected cell. How the various virion components come together at the site of assembly and interact to form a spherical particle is currently only partly understood. Although Gag expressed alone is able to traffic to the plasma membrane, studies of polarized cells and neurons indicate that the presence of Env can influence the trafficking pathway of retroviral Gag (26, 44). For murine leukemia virus (MLV) assembly, a model has been proposed in which the viral genomic RNA is recruited from the cytoplasm by a complex of Gag and Env formed at endosomal or lysosomal membranes (3).
Biochemical and structural studies have yielded detailed information about the architecture of immature and mature virions, as well as subviral complexes involved in viral replication steps. However, the dynamics of essential processes in replication escapes these analyses. In contrast, modern real-time imaging techniques allow direct observation of intracellular transport processes and protein interactions. A prerequisite for the application of these techniques is the introduction of a fluorescent label suitable for the monitoring of the protein of interest inside the living cell. Recently, the chemical labeling of nonenveloped viruses with fluorescent dyes has allowed the application of real-time imaging techniques to study virus-cell interactions. The microtubule-mediated intracellular transport of the nonenveloped adenoviruses could be visualized when the surfaces of the virions were chemically labeled with a fluorescent dye (25, 40). Similarly, the entry of adeno-associated virus into a host cell could be monitored after fluorescent labeling. In the case of adeno-associated virus, the method of single-virus tracing allowed for the first time the visualization of the entry pathway of single virions with millisecond time resolution (37). In the case of the enveloped HIV, the viral capsid is surrounded by a lipid membrane which fuses with the cellular membrane during the entry process. Therefore, virion-associated Gag is not accessible to chemical modification and any label inserted into the membrane of the particle would get lost during virus entry. Furthermore, chemical modification in vitro is not applicable for the labeling of progeny viral proteins to study transport processes during assembly and release. In recent years, fusion of the protein of interest to the green fluorescent protein (GFP) of Aequorea victoria or its derivatives has become an important tool for monitoring transport processes inside living cells. Viral proteins fused to GFP have been used to investigate the entry of the enveloped alphaherpesviruses (4, 39) as well as for studying vaccinia virus particle formation (13, 36, 43). McDonald et al. (28) have recently reported the labeling of HIV particles by the coexpression of GFP fused to the accessory protein Vpr, which can be incorporated into virions via its interaction with Gag. Their studies indicated that the intracellular transport of incoming virus involves, as with other viruses, retrograde transport along microtubuli. The fusion of GFP to the C terminus of HIV type 1 (HIV-1) Gag has also been reported (18, 32). In these studies, Gag trafficking could be analyzed after the expression of Gag-GFP alone or upon the coexpression of Env. The gag-gfp fusion gene used cannot be transferred to the viral context, since in the HIV-1 genome, the 3′ region of the gag open reading frame (ORF) overlaps with the viral pol gene (Fig. 1A) and a translational frame shift is required for synthesis of the Gag-Pol precursor protein. Thus, any fusion of foreign sequences to the 3′ end of gag in the viral genome will result in the alteration or destruction of pol and interfere with the balanced gene expression of HIV. Gag-GFP expressed by itself directs the budding of VLPs from cells. However, these particles do not contain any other viral components, nor do they display a regular spherical distribution of electron-dense material as observed in immature HIV (33) or undergo morphological maturation.
FIG. 1.
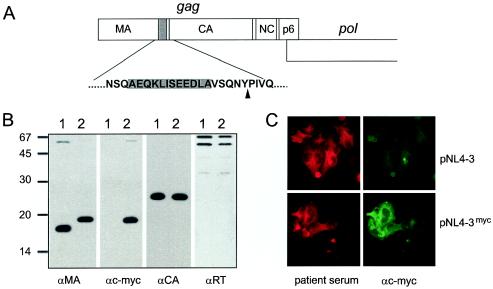
Insertion of a c-myc epitope tag into the HIV-1 Gag protein. (A) The coding sequence for the c-myc epitope tag sequence, flanked by an alanine residue at either side, was inserted into the gag ORF as indicated, resulting in the introduction of 12 additional amino acids between Gln127 and Val128 of MA. (B) Expression of the myc-tagged MA protein in the viral context. VLPs were prepared from 293T cells transfected with pKHIV (lanes 1) or pKHIVmyc (lanes 2). Viral proteins were separated by SDS-polyacrylamide gel electrophoresis (17.5% polyacrylamide; 200:1), transferred to nitrocellulose, and reacted with the indicated antisera. Molecular mass markers (in kilodaltons) are indicated on the left. (C) Detection of c-myc-tagged MA protein in transfected cells. HeLaP4 cells were transfected with pNL4-3 or pNL4-3myc. Forty-eight hours after transfection, cells were fixed and stained with patient serum or a monoclonal antibody against c-myc, respectively.
With the aim of generating a tool for the analysis of Gag trafficking and virus-cell interactions in the viral context, we decided to insert a label at a position within the gag ORF. We describe here the construction and detailed characterization of an HIV-1NL4-3 provirus derivative carrying the egfp gene within the gag ORF and show that it is possible to generate a fluorescent HIV derivative displaying wild-type morphology and infectivity. Fluorescent HIV derivatives represent a useful tool for analyzing the interaction with live host cells in real time as well as for studying the dynamics of Gag trafficking and assembly in an authentic context. Furthermore, they reveal the remarkable plasticity of the viral assembly process.
MATERIALS AND METHODS
Antisera.
Rabbit polyclonal antisera against HIV-1 CA, MA, and reverse transcriptase (RT) as well as against enhanced GFP (EGFP) were raised against purified recombinant proteins. Polyclonal rabbit antiserum against HIV-1 gp120 was kindly provided by Valerie Bosch (DKFZ, Heidelberg, Germany). Monoclonal antibody against c-myc was obtained from Calbiochem.
Plasmids.
Plasmid constructs were based on the infectious proviral plasmid pNL4-3 (1). The infectious proviral plasmid pNLC4-3 expresses the authentic genomic RNA from pNL4-3 under the control of the cytomegalovirus (CMV) promoter (4a). The noninfectious subviral construct pKHIV contains the SstI-XhoI fragment from pNL4-3, comprising the major splice site, the RNA packaging signal, and the complete coding sequence except for a portion of the nef gene, under the control of the CMV promoter. For the introduction of foreign sequences, the subviral construct pBSMA containing the BsshII-ApaI fragment from pNL4-3 in Bluescript KS(−) was used. A 597-bp-long NsiI-EcoRI fragment from this plasmid was exchanged for the modified fragment containing the codons for the c-myc epitope tag flanked by alanine residues immediately 3′ of codon Gln127 of MA, which was generated by overlap PCR. Following confirmation by sequence analysis, the mutated BssHII-ApaI fragment was recloned into pNL4-3 and pKHIV. For the construction of egfp-carrying plasmids, a unique ClaI site was inserted into pBSMA at position 1171 of the NL4-3 sequence (3′ of the codon for Gln127 of MA) by overlap PCR. By using this newly generated restriction site, the complete coding sequence of EGFP amplified by PCR from plasmid pEGFPc1 (Invitrogen) flanked by ClaI sites was introduced. A BssHII-SphI fragment comprising the modified MA sequence was cloned into pNLC4-3 and pKHIV, resulting in plasmids pNLC4-3EGFP and pKHIVEGFP, respectively. Construction of a late-domain-defective variant was carried out by cloning an ApaI-SdaI fragment from a pNL4-3 derivative coding for a PTAP-to-LIRL exchange within p6 (20) into the context of pKHIVEGFP. Detailed cloning procedures and primer sequences are available upon request.
Tissue culture and transfections.
293T, HeLaP4, and TZM cells (JC53BL cells which express β-galactosidase and luciferase under the control of the HIV-1 long terminal repeat [45]) were cultivated in Dulbecco's modified Eagle medium (Invitrogen) supplemented with 10% fetal calf serum. PM-1 cells were grown in RPMI 1640 medium (Invitrogen) supplemented with 10% fetal calf serum. The transfection of cells was carried out either by calcium phosphate precipitation according to standard procedures or by lipofection with FuGENE 6 reagent (Roche) according to the manufacturers' instructions.
Particle preparation.
HeLaP4 or 293T cells were transfected with the plasmids indicated in the figure legends by using calcium phosphate precipitation. At 48 h after transfection, the tissue culture supernatant was harvested, cleared by low-speed centrifugation (10 min; 400 × g), and passed through a 0.45-μm-pore-size filter. Particles were pelleted from the filtrate by ultracentrifugation through a 20% (wt/wt) sucrose cushion in phosphate-buffered saline (PBS) (90 min; 130,000 × g at 4°C) and resuspended in PBS. Cells were scraped from the plates, washed with PBS, and lysed by boiling in sodium dodecyl sulfate (SDS) sample buffer. Proteins from cell lysates and purified particles were separated by SDS-polyacrylamide gel electrophoresis and analyzed by immunoblotting by using the antisera indicated in the figure legends.
PCR analysis of viral cDNA.
At 48 h after transfection of HeLaP4 cells with pNL4-3 and pNL4-3myc, virus containing supernatant was harvested, passed through a 0.45-μm-pore-size filter, and used to infect C8166 cells in quadruplicate. The tissue culture supernatant from infected cells was serially passaged on C8166 cells. After five passages, cells were harvested and total DNA was prepared by using DNAzol reagent (Invitrogen) according to the manufacturer's instructions. DNA was subjected to PCR, with a primer pair spanning a region from nucleotides 1090 to 1300 in the pNL4-3 sequence. The resulting PCR fragments were separated by agarose gel electrophoresis.
Infectivity assay.
293T cells (2 × 106) were transfected with either 5 μg of pNLC4-3, 5 μg of pNLC4-3EGFP, or 5 μg of a mixture of both plasmids. Forty-eight hours after transfection, the medium was harvested, passed through a 0.45-μm-pore-size filter, and stored frozen in aliquots at −80°C. The particle concentrations in these virus stocks were determined by a quantitative enzyme-linked immunosorbent assay (ELISA) detecting the viral CA protein (23). Serial fivefold dilutions of virus stocks were used to infect TZM indicator cells in 96-well plates. At 48 h postinfection, cells were fixed with ice-cold methanol-acetone (50:50) and stained for β-galactosidase. Blue cells from suitable dilutions were counted, and the numbers of infected cells were normalized to the amount of CA used for infection.
Electron microscopy.
HeLaP4 cells were transfected with plasmid pNLC4-3 or pNLC4-3EGFP or an equimolar mixture of both plasmids by using FuGENE 6 lipofection reagent (Roche) according to the manufacturer's instructions. At 44 h after transfection, the medium was removed, and the cells were briefly rinsed with PBS, fixed, and prepared for thin-section microscopy as previously described (16). Micrographs were taken with a Zeiss EM-10 electron microscope at 80 kV. The magnification indicator was routinely controlled by the use of a grating replica.
RESULTS
Insertion of an epitope tag within HIV-1 Gag is compatible with viral replication.
Our aim was to label the main structural protein of HIV-1, Gag, in the context of the infectious virus. The introduction of traceable markers within Gag should permit the monitoring of trafficking of HIV structural proteins in an authentic setting, as well as the direct observation of virus particles during the interaction of HIV with its host cell. Based on the results of structural studies of Gag domains as well as immature HI virions, we considered the C-terminal region of the MA domain best suited for the insertion of foreign sequences into Gag. Cryoelectron microscopic analysis of immature HIV particles had previously revealed a perpendicular radial arrangement of Gag molecules apposed to the lipid envelope. The functional domains of Gag are aligned as separately folded domains along the elongated Gag molecule, and an approximately 40-Å-wide region of low protein density separates the membrane-associated globular domain of MA from the N-terminal domain of CA (12, 46). Consistent with this fact, the model of HIV-1 MA derived from crystallographic analyses shows that the amino terminus of MA forms a compact globular structure, whereas amino acids 105 to 121 are arranged in an extended α-helix projecting away from the body of the protein (19, 27). The structure of the C terminus of mature MA (amino acids 123 to 132) is not resolved in structural analyses, indicating high flexibility of these residues. In the precursor protein, the region connecting the folded MA and CA domains (amino acids 123 to 143) is highly mobile and exists in a random coil conformation (41). Thus, we reasoned that insertions into the extended region between the globular MA and CA domains might be possible without interfering with the overall arrangement of Gag functional domains and particle formation.
In initial experiments, we introduced a nucleotide sequence coding for a 12-amino-acid-long epitope tag derived from the c-myc protein close to the 3′ end of the MA coding region of the infectious proviral plasmid pNL4-3. An analogous construct was made based on the noninfectious subgenomic plasmid pKHIV, which lacks the viral long terminal repeat sequences and expresses all HIV-1 proteins except Nef under the control of a CMV promoter. As illustrated in Fig. 1A, the minimal recognition site for the viral PR between the MA and CA domains (SQNYPIV) was retained. Transfection of the variant plasmids into HeLaP4 cells revealed no differences from the wild-type virus with respect to Gag expression and particle production (data not shown). Particles were pelleted from the supernatant of transfected cells, and their protein composition was analyzed by immunoblotting by using antisera against the c-myc epitope, as well as against various viral proteins. As shown in Fig. 1B, the insertion of the c-myc epitope did not interfere with the expression of Gag or its processing into the functional domains. Due to the addition of 12 amino acids, the modified MA protein (Fig. 1B, lanes 2) showed slightly decreased mobility compared to that in the wild type (lanes 1). The attachment of the c-myc epitope to the MA protein of the variant virus (Fig. 1B, lanes 2) was demonstrated by the reactivity of the MA-derived protein with a c-myc-directed monoclonal antibody. Except for a weak band migrating at 55 kDa, the size of the Gag precursor, only a single band reactive with anti-MA antiserum or anti-c-myc antibody, respectively, was observed, demonstrating that the fusion protein was stable. No differences in expression levels and amounts of particles released were observed, and immunoblots against viral proteins other than MA revealed no differences from the wild-type control (Fig. 1B and data not shown). Thus, the presence of the epitope tag was compatible with particle formation and Gag processing. Immunofluorescence staining of cells transfected with pNL4-3myc demonstrated that all cells expressing HIV proteins also reacted with anti-c-myc antibody (Fig. 1C).
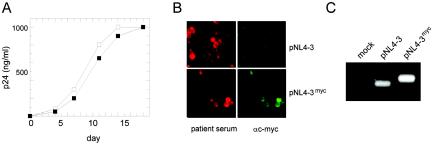
To investigate the effect of the introduced epitope tag on viral replication, the supernatants from transfected HeLaP4 cells were used to infect PM-1 cells and viral replication was monitored by quantitation of CA release. As shown in Fig. 2A, the variant virions were fully infectious and replicated with kinetics similar to those of the wild-type virus in PM-1 cells. To analyze the stability of the introduced sequence upon continuous viral replication, serial passages of virus on MT-4 and C8166 cells were carried out. Following multiple rounds of infection, we tested for the presence of the epitope tag by immunofluorescence (Fig. 2B) and PCR of viral cDNA sequences (Fig. 2C). More than 90% of the cells expressing viral proteins were clearly positive for c-myc immunofluorescence after three passages (Fig. 2B). To test for potential loss of the epitope coding sequence, we performed a PCR analysis of viral cDNA isolated after five passages with primers covering the tag insertion site. As seen in Fig. 2C, a strong band of the expected size for the tagged variant (346 bp), but no shorter fragment corresponding to the wild-type sequence (310 bp; compare with the NL4-3 control), was detected in the case of NL4-3myc-infected cells, demonstrating the stability of the introduced sequence. In summary, the introduction of a small foreign sequence into the C-terminal region of MA did not interfere with viral replication in tissue culture, and the inserted sequence could be propagated stably over multiple rounds of infection.
FIG. 2.
Infectivity of NL4-3myc and stability of the epitope tag. (A) Supernatants from HeLaP4 cells transfected with pNL4-3 (open squares) or pNL4-3myc (filled squares) were used to infect PM-1 cells (10 ng of p24 CA per 106 cells). The release of p24 CA into the supernatant was monitored by ELISA. (B) Supernatants from transfected HeLa cells were used to infect MT-4 cells. Following three passages on MT-4 cells, cells were fixed with patient antiserum and stained with a monoclonal antibody against c-myc. (C) After five rounds of replication in C8166 cells, DNA was prepared from infected cultures, and gag-derived sequences were amplified by PCR with primers flanking the epitope tag insertion site as described in Materials and Methods. Samples were separated by electrophoresis on an agarose gel. Amplification of the wild-type sequence yielded a 210-bp fragment; in the case of the myc-tagged variant, the expected size of the PCR product is 246 bp.
Labeling of the HIV-1 Gag protein in the viral context by the insertion of an EGFP domain.
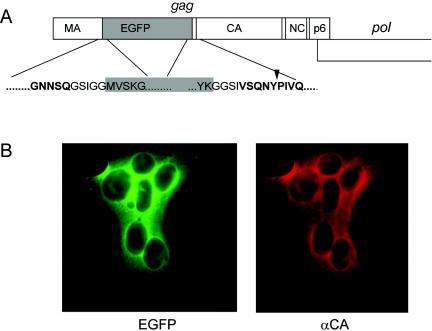
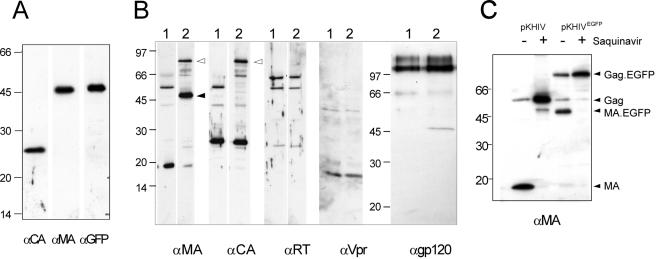
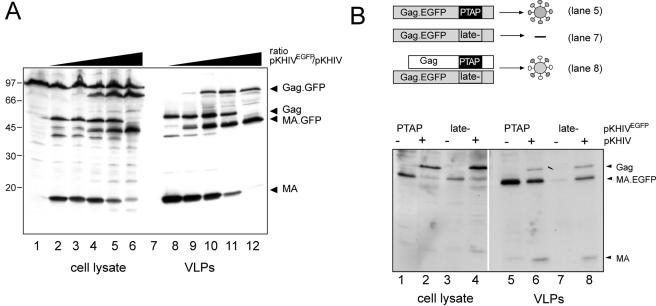
The c-myc epitope can thus be used as a marker to detect the variant Gag or MA protein inside an infected cell after fixation and antibody staining. Since we also wanted to monitor the viral structural protein within live cells, it was of interest to test whether the insertion of the gfp gene into the analogous position within gag was possible. The gene for enhanced GFP (egfp) was inserted close to the 3′ end of the MA coding region of pNLC4-3 and of pKHIV, yielding the coding sequence for the fusion protein shown in Fig. 3A. The EGFP domain was separated from HIV-derived sequences by short glycine-rich linkers, and the PR recognition site between MA and CA was again retained to allow processing of the modified Gag precursor by PR. Initial characterization of the labeled virus was carried out by using the subviral derivative pKHIVEGFP. Following transfection of this plasmid into HeLaP4 cells, cytoplasmic EGFP fluorescence was observed. All EGFP-positive cells also expressed HIV-1 CA, as determined by immunofluorescence (Fig. 3B). In addition, 48 h after transfection, nearly all fluorescent cells were found within syncytia, indicating the presence of functional Env protein on their plasma membrane. We concluded that EGFP-positive cells expressed both major HIV structural proteins, Gag and Env. To test whether the expected Gag fusion protein was stable and could assemble into VLPs released from cells, particles were pelleted from the supernatant of transfected 293T cells and analyzed by immunoblotting. As shown in Fig. 4A, antiserum against HIV-1 MA as well as serum against GFP reacted with a protein with an apparent molecular mass of 45 kDa, which corresponds well with the calculated molecular mass for the MA-EGFP fusion protein. Neither free MA (17 kDa) nor free EGFP (27 kDa) was observed in the preparation, indicating that the resulting fusion protein was stable. Antiserum against HIV-1 CA detected a band migrating at a position corresponding to the molecular mass of unmodified CA (24 kDa) (Fig. 4A). We concluded that the Gag-EGFP fusion protein was expressed in the viral context, efficiently released from cells, and processed to yield the expected subunits. For a more detailed characterization, we probed EGFP-labeled VLPs (Fig. 4B, lanes 2) with antisera raised against different HIV-1 proteins and compared the patterns to those of unmodified VLPs (Fig. 4B, lanes 1). As shown in Fig. 4B, the only differences detected were the expected 27-kDa increases in apparent molecular masses of the Gag precursor and MA in the case of KHIVEGFP. Antiserum against CA detected processed CA protein in both cases and also detected residual Gag precursor, migrating at 55 kDa in the case of the wild type and at 82 kDa (55 + 27 kDa) in the case of pKHIVEGFP. No differences were observed by using antisera against RT, Vpr, and Env, and the relative amounts of Gag (MA or CA) to Pol (RT), to Env (gp120), and to Vpr, which is incorporated into virions via interaction with the p6 domain of Gag (22, 31), appeared to be unaltered. To confirm that the processing of Gag-EGFP was carried out at the authentic site by the viral PR, rather than being due to unspecific cleavage between the EGFP and CA domains, particles were purified from transfected cells grown in the presence of the specific PR inhibitor saquinavir and the apparent molecular masses of MA containing proteins were determined by immunoblotting (Fig. 4C). In the untreated samples, mostly mature MA or MA-EGFP and only small amounts of the respective precursor protein were detected. In contrast, anti-MA-reactive proteins were detected only as uncleaved precursors (55 or 82 kDa) in VLPs released from saquinavir-treated cells, demonstrating that processing in untreated cells was the result of cleavage by the viral PR. In summary, the insertion of the egfp gene did not interfere with particle release, incorporation of other virion proteins, or PR-mediated Gag processing.
FIG. 3.
(A) Construction of an HIV-1 variant carrying the egfp gene. The schematic drawing shows the gag ORF with the egfp gene inserted between the MA and CA coding regions. The expanded regions show the derived amino acid sequence at the domain borders, with HIV sequences displayed in boldface type and EGFP sequences shown in black type on a gray background. An arrowhead indicates the PR cleavage site between MA and CA. (B) Expression of EGFP and Gag in HeLaP4 cells. Cells were transfected with pKHIVEGFP, and immunofluorescence using anti-CA antiserum was carried out 44 h posttransfection. The images show a syncytium displaying EGFP fluorescence and red immunofluorescence.
FIG. 4.
Detection of HIV proteins in preparations of wild-type and EGFP-labeled particles. 293T cells were transfected with pKHIV or pKHIVEGFP, and particles were purified from the supernatant 48 h posttransfection by centrifugation through a sucrose cushion. HIV proteins and EGFP were detected in the preparations by enhanced chemiluminescence immunoblotting by using the indicated antisera. Molecular mass standards (in kilodaltons) are indicated at the left of each blot. (A) Immunoblot of a KHIVEGFP preparation, showing that the protein of approximately 45 kDa is recognized by antisera against MA as well as against GFP. (B) Comparison of viral protein patterns of parallel preparations of KHIV (lanes 1) and KHIVEGFP (lanes 2). (C) Inhibition of Gag-GFP processing by the HIV PR inhibitor saquinavir. Particles were prepared from 293T cells grown in the absence or presence of 5 μM saquinavir and analyzed by immunoblotting by using antiserum against MA.
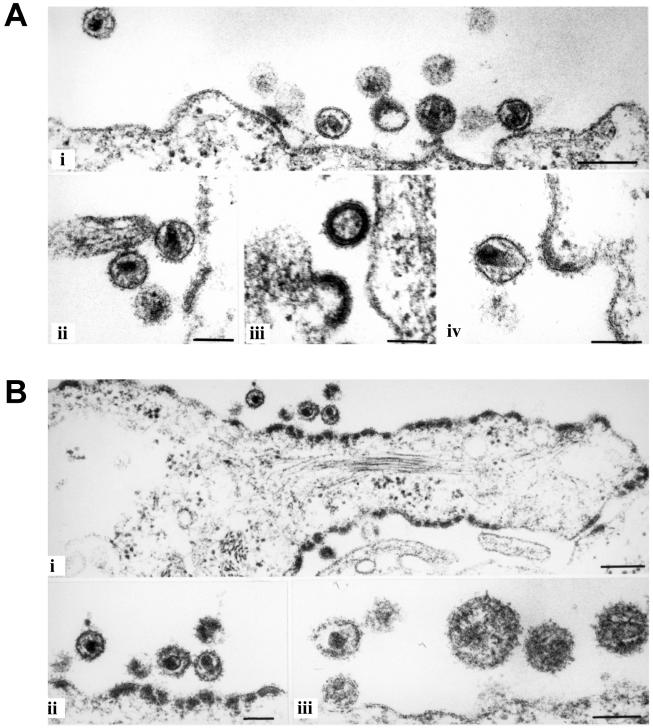
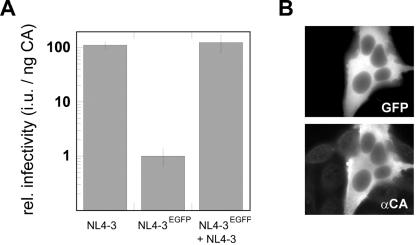
Since the insertion of the large EGFP domain represents a substantial change of the overall Gag molecule architecture, we wanted to determine whether this modification affected particle morphology. HeLaP4 cells transfected with pNLC4-3 or with the fluorescent derivative pNLC4-3EGFP were analyzed by thin-section electron microscopy (Fig. 5A and B). In both cases, the accumulation of protein at the plasma membrane, viral budding structures, and released particles were seen. However, electron micrographs of cells transfected with pNLC4-3EGFP showed significantly more protein accumulations or early budding structures at the plasma membrane (Fig. 5B, panel i) than the wild-type control; at the same time, the numbers of free virions detected in the images were reduced (Fig. 5B and data not shown). In contrast, Gag release from cells into tissue culture supernatants as determined by ELISA or estimated from anti-CA immunoblots of pelleted particles (Fig. 4 and data not shown) was not strongly reduced in the case of NL4-3EGFP compared to NL4-3. The quantification of NL4-3EGFP release by CA ELISA yielded Gag concentrations in the medium of between 40 and 95% of that of the wild type (average result from six independent experiments, 68%). Thus, we hypothesize that the difference in the steady-state Gag distribution displayed in electron micrographs reflects a change in budding kinetics, resulting in temporal accumulation of virions at an early stage of budding rather than a block of particle release. Whereas no obvious differences in particle size were observed, electron density within the MA layer underneath the membrane appeared unevenly distributed in some of the NL4-3EGFP particles (Fig. 5B, panel iii). Importantly, however, most free EGFP-labeled virions observed in these images displayed the typical core morphology of mature HIV particles (Fig. 5B, panel ii), indicating undisturbed morphological rearrangements of Gag-derived proteins following PR processing. We also tested the infectivity of EGFP-labeled virions. Virus stocks were prepared by transfection of 293T cells with pNLC4-3 or pNLC4-3EGFP and used to infect TZM cells, which carry a β-galactosidase reporter gene under the control of the HIV long terminal repeat (45). Infectivity was determined by counting the infected cells and was normalized to the amount of p24 CA used for infection. As shown in Fig. 6A, EGFP-labeled virions were indeed found to be infectious. The relative infectivity of the labeled particles per nanogram of CA protein was approximately 2 orders of magnitude lower than that of the wild type, but remained at a titer of approximately 5 × 103 per ml. To exclude the possibility that any β-galactosidase-positive cells resulted from the uptake of residual DNA from the initial transfections, a control experiment was carried out in the presence of the RT inhibitor zidovudine during infection. Inhibition of reverse transcription completely abolished the infectivity of the labeled particles (data not shown), demonstrating that the positive cells were indeed products of infection. Cells infected with NL4-3EGFP expressed p24 CA, as determined by immunofluorescence, and displayed green fluorescence, indicating the presence of the egfp gene (Fig. 6B).
FIG. 5.
Morphology of NL4-3EGFP particles. HeLaP4 cells were transfected with the proviral plasmids pNLC4-3 (A) and pNLC4-3EGFP (B). Twenty-four hours posttransfection, cells were harvested, prepared, and analyzed by thin-section electron microscopy as described in Materials and Methods. Bars, 200 nm (panels i) or 100 nm (A, panels ii, iii, and iv, and B, panels ii and iii).
FIG. 6.
Infectivity of fluorescently labeled HIV derivatives. (A) 293T cells were transfected with pNLC4-3, pNLC4-3EGFP, or an equimolar mixture of both plasmids. Serial dilutions of supernatants were used to infect TZM cells, and relative infectivities were determined as described in Materials and Methods. Data points are mean values obtained from two independent transfections, with all samples titrated in duplicate. Several independent experiments yielded similar relative differences. (B) TZM cells infected with NL4-3EGFP displayed green fluorescence (top) and expressed HIV-1 Gag as detected by immunofluorescence using polyclonal serum against CA (bottom).
Generation of labeled HIV derivatives displaying wild-type morphology and infectivity.
The finding that the insertion of the large EGFP domain within Gag was compatible with virion infectivity was in itself astonishing. However, our aim was to obtain labeled particles with infectivity properties identical to those of the wild type. We reasoned that the infectivity of the labeled virions might be increased by reducing the numbers of EGFP molecules incorporated per particle. To accomplish this, we attempted to generate mixed particles by cotransfecting cells with different molar ratios of pNLC4-3 and pNLC4-3EGFP DNA while keeping the total amount of DNA constant. As shown in Fig. 7A, this experiment resulted in the coexpression of Gag and Gag-EGFP at ratios corresponding to the relative amounts of the respective DNAs used for transfection. When an equimolar ratio of both DNAs was used (Fig. 7A, lanes 5 and 11), approximately equal amounts of Gag plus MA compared to Gag-EGFP plus MA-EGFP were detected in the VLP preparation (lane 11). The ratios of Gag to Gag-EGFP detected in pelleted particles matched those detected within the respective producing cells. However, this result by itself did not allow us to conclude that Gag and Gag-EGFP coassembled into mixed particles rather than forming two separate virion populations. Thus, a late-domain mutant rescue experiment was carried out to confirm mixed particle formation. The efficient release of Gag depends on the presence of the late-domain motif P(T/S)AP within the p6 domain. The mutation or deletion of this motif results in budding arrest and the accumulation of particles at the plasma membrane. Particles which are released display an immature morphology, and Gag processing is inhibited (15, 20). It has been shown that the release of late-domain-deficient retroviral Gag can be rescued by the coexpression of wild-type Gag, presumably through the formation of mixed particles (47). Thus, we generated a defective pKHIVEGFP variant by exchanging the PTAP motif for LIRL, which has been shown to block particle release (20) and tested Gag-EGFPlate− release from transfected cells in the absence or presence of wild-type Gag (Fig. 6 B). Lanes 1, 2, 5, and 6 show the control experiment, demonstrating that Gag-EGFP is found in the virion samples in the presence or absence of wild-type Gag. Gag-EGFPlate− was equally well expressed (lane 3), but not efficiently released into the tissue culture supernatant (compare lane 7 to lane 5), showing that Gag-EGFP release occurred in a late-domain-dependent manner. However, the coexpression of wild-type Gag rescued Gag-EGFPlate− release, as indicated by its presence in the virion sample (lane 8), confirming the formation of mixed particles.
FIG. 7.
Generation of mixed particles by cotransfection of the wild type and the EGFP-labeled variant. (A) Variation of the ratio between the wild-type and the fluorescence-labeled variant. 293T cells were transfected with a mixture of pKHIV and pKHIVEGFP, while the total amount of DNA was kept constant. The relative amounts of pKHIVEGFP used were 0% (lanes 2 and 8), 4% (lanes 3 and 9), 20% (lanes 4 and 10), 50% (lanes 5 and 11), and 100% (lanes 6 and 12). At 44 h posttransfection, cells were lysed and particles were purified from the tissue culture supernatant by centrifugation through a sucrose cushion. Gag-derived proteins were detected in the cell lysate and purified particles by enhanced chemiluminescence immunoblotting by using antiserum against MA. (B) Rescue of a release-deficient variant of pKHIVEGFP by cotransfection with wild-type pKHIV. PKHIVEGFP and pKHIVEGFP late− were either transfected alone (lanes 1, 3, 5, and 7) or cotransfected with an equimolar amount of wild-type pKHIV (lanes 2, 4, 6, and 8). At 44 h posttransfection, cells were lysed and particles were prepared from the tissue culture supernatant. Gag-derived proteins were detected in the samples by immunoblotting with polyclonal antiserum against MA.
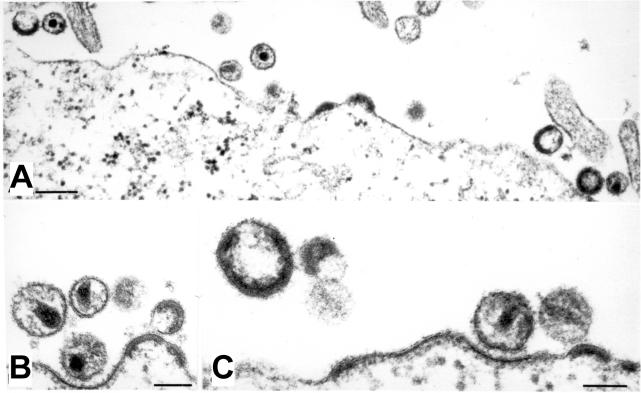
To determine the morphology of mixed particles, HeLaP4 cells were cotransfected with equimolar amounts of pNLC4-3 and pNLC4-3EGFP. Analysis by thin-section electron microscopy (Fig. 8) revealed that the amount and morphology of particles released from cotransfected cells was indistinguishable from that of the wild-type control (compare Fig. 8 to Fig. 5A). The accumulation of electron-dense material at the plasma membrane, which had been observed in the case of pNLC4-3EGFP alone (Fig. 5B), was not detected upon the cotransfection of this variant with the wild type. Budding structures and immature as well as mature particles were of the same size and their morphology was apparently identical to that of unmodified HIV particles. This restoration of wild-type properties was also reflected in the relative infectivity of the virions: particles generated by cotransfection of pNLC4-3 and its egfp-carrying derivative displayed the same relative infectivity as the pNLC4-3 control (Fig. 6A). A careful analysis of the stability of the introduced egfp ORF through multiple rounds of viral replication was prevented by the low titers of infectious virus stocks obtained following the transfection of pNLC4-3EGFP alone. The cotransfection of pNLC4-3EGFP with wild-type NLC4-3 yielded fluorescent particle preparations with high titers of infectious virus stocks, which were serially passaged. In this case, the egfp-carrying virus genome was rapidly lost (data not shown), presumably by competition with the shorter wild-type RNA present in the same cells for packaging into virus particles.
FIG. 8.
Morphology of partly fluorescently labeled particles. HeLaP4 cells were transfected with a mixture of equimolar amounts of pNLC4-3 and pNLC4-3EGFP. At 44 h posttransfection, cells were harvested, prepared, and analyzed by thin-section electron microscopy as described in Materials and Methods. Bars, 200 nm (A) or 100 nm (B and C).
DISCUSSION
In this study, we have shown that long foreign sequences can be introduced between functional domains of the HIV-1 Gag polyprotein without affecting virus assembly or virion infectivity. The results presented here demonstrate that HIV Gag assembly displays a remarkable tolerance towards structural modification. The insertion of 12 amino acids within Gag did not affect particle formation and virion infectivity in tissue culture, and the inserted sequence was stably propagated through multiple rounds of infection. The myc-tagged virus variant is useful for analyzing the intracellular localization of Gag and MA and for the purification of replication intermediates containing these proteins. The presence of a detectable tag which does not interfere with viral replication can also be exploited in studies which require the presence of two equally well replicating but distinguishable variants of HIV within the same cell. We are currently using the c-myc epitope-tagged variant in combination with labels introduced near the 3′ end of the genome in studies investigating viral recombination. The finding that the C terminus of HIV MA can be modified without affecting the infectivity of the virus is consistent with results reported by Nelle and Wills, who showed by deletion analysis that the C-terminal half of MA is not essential for Rous sarcoma virus budding and infectivity (30), as well as with the findings of Reil et al., who reported that under certain conditions up to 90% of the HIV-1 MA protein can be deleted (35). The observed tolerance of the C terminus of HIV-1 MA towards the insertion of a small epitope tag is also in agreement with recent results reported by Auerbach and colleagues (2), who scanned a large portion of the MLV gag gene, including the 31 C-terminal codons of MA, by genetic footprinting for insertions compatible with replication competence. They reported that most of the C-terminal region of MA and the adjacent p12 protein tolerated 12-amino-acid-long insertions. Their finding that the region analogous to the insertion site used there was covered by an insertion footprint in the case of MLV may be explained by the fact that the borders of this footprint were not well defined and extended into the processing site between MA and p12.
Even in light of the results obtained with the c-myc-tagged HIV variant, it appears surprising that the insertion of a whole additional protein domain with a higher molecular mass than any of the functional domains of Gag itself (27 kDa, compared to 17, 24, 7, and 6 kDa for MA, CA, NC, and p6, respectively) within the structural polyprotein is compatible with virion assembly and infectivity. This finding demonstrates the high structural flexibility of the immature retroviral capsid. Such flexibility has already been inferred from cryoelectron microscopy analyses of immature virions and VLPs (12, 46). These studies revealed that HIV particles released from infected cells are heterogeneous in size and shape and that the molecule arrangement within the Gag shell displays no overall symmetry, but rather consists of arrays of local order. It appears that the compactly folded 27-kDa EGFP domain can be fitted into this flexible structure without enforcing fundamental changes in Gag arrangement, as evident from the unaltered size and mature morphology of particles viewed in thin-section electron micrographs. However, some influence of the EGFP domain on the assembly process is implicated from the images of pNLC4-3EGFP transfected cells. Significantly more electron-dense material than observed for the wild type accumulated at the plasma membrane in the steady state (Fig. 5B, panel i), suggesting delayed kinetics of spherical capsid formation. The presence of wild-type Gag in equimolar amounts was able to overcome this apparent assembly or budding delay, since the pronounced electron-dense accumulations at the plasma membrane were not observed in coexpression experiments (Fig. 8). The particles released in this case appeared indistinguishable from those of the wild type (Fig. 5A), although the possibility of subtle disturbances of Gag or MA packaging due to the attached EGFP domain, which are undetectable by thin-section electron microscopy, cannot be absolutely excluded. However, even if such disturbances exist, they did not have any detectable functional consequences, as indicated by the wild-type infectivity of the mixed particles. Sherer and colleagues have recently reported that MLV particles containing up to 25% of a Gag-GFP fusion protein display about half the infectious titer of wild-type MLV (38). In comparison to the findings presented here, this result appears to indicate a higher tolerance of the HIV particle structure towards a large modification; however, since those authors used a different approach (separate overexpression of a C-terminal Gag-GFP fusion together with the wild-type virus), this conclusion cannot be certain.
In the case of the fully labeled particles, potential irregularities in the matrix shell represent one possible cause for the observed reduction in relative infectivity. Decreased particle stability due to irregular structure may explain the discrepancy between a nearly wild-type amount of CA released from the cells and the reduced amount of infectious virions in the supernatant, as determined by titration on TZM cells. Since the arrangement of Gag molecules within the virion cannot be resolved by thin-section electron microscopy, cryoelectron microscopy analyses aimed at determining the fine architecture of immature and mature labeled particles in comparison to that of wild-type HIV are under way. The reduced relative infectivity of the EGFP-labeled particles might also be caused by deficiencies in packaging their longer and modified genomic RNA. Passage of virus containing the supernatants from cells cotransfected with egfp-carrying and wild-type HIV proviral DNAs resulted in rapid loss of the modified variant, possibly by preferred packaging of the shorter wild-type RNA present in the same cells. Careful competition analyses will be required to determine the relative packaging efficiencies of wild-type and egfp-containing RNAs. Finally, the possibility that the presence of the EGFP domain interferes with a function of the mature MA protein in virus replication cannot be excluded. While most of the virion-associated MA appears to stay tightly associated with the viral envelope and is thus not associated with the viral core after entry, it has been reported that at least some MA can be detected in the viral preintegration complex (8, 29). The role of these MA molecules in early stages of viral replication is presently unclear. A function of MA in the nuclear import of the preintegration complex in nondividing cells had been suggested (7, 17, 42), but this idea was not supported by later studies (10, 11, 35). It has been shown that under certain conditions, at least, MA is dispensable for viral replication (35). On the other hand, mutagenesis studies have indicated a role for MA in early postentry steps of HIV replication prior to reverse transcription (21, 34, 48). In particular, it has been reported that a short deletion introduced at the C terminus of MA (amino acids 116 to 128), covering the site used here for the insertion of EGFP, resulted in delayed replication kinetics in Sup-T1 cells and significantly decreased the amount of viral DNA synthesis (48). Since the majority of MA molecules are lost at the membrane during virus entry, any function of MA in early replication may be carried out by wild-type MA in the case of the mixed virus preparations. Experiments aimed at determining the cause for the reduced infectivity of fully fluorescently labeled particles are under way. Preliminary results suggest that the observed 30- to 100-fold reduction in infectious titer compared to that of the wild type results from synergy among minor defects at several stages of viral replication.
The EGFP-labeled HIV variant described here is highly suitable for studying Gag trafficking and interactions with viral and host factors in live cells by confocal microscopy as well as by biophysical techniques detecting intracellular protein interactions and protein movements (fluorescence energy resonance transfer and fluorescence recovery after photobleaching). Fluorescence energy resonance transfer has recently been applied for the study of Gag assembly after the expression of Gag-GFP alone (9, 24). By using the construct characterized in the present study, such analyses can be performed in the context of a virus-producing cell, and Gag interactions with viral and cellular factors leading to the release of infectious virus can be monitored.
By cotransfection of pNLC4-3EGFP and pNLC4-3, we have generated a fluorescently labeled HIV-1 derivative displaying wild-type protein composition, morphology, and infectivity. This labeling can be exploited for analyses of virus-cell interaction during the early steps of HIV replication, in particular virus binding and fusion with the host cell. Based on an average number of 5,000 molecules of Gag per HIV particle (6), mixed particles purified from cotransfection supernatants would carry approximately 2,500 molecules of EGFP, making the preparations highly suitable for investigating virus-cell interactions by time-lapse confocal microscopy and single virus tracing (5) analyses. Ongoing studies using these techniques are aimed towards a kinetic analysis of fast steps in the HIV entry process.
Acknowledgments
We thank Birgit Hub for excellent technical assistance. We gratefully acknowledge the gifts of a late-domain-defective HIV derivative by Eric Freed and of rabbit αgp120 antiserum by Valerie Bosch.
This work was supported in part by a grant from the Deutsche Forschungsgemeinschaft to H.G.K. and B.M. (KR 906/4-1) and by SFB544.
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Auerbach, M. R., C. Shu, A. Kaplan, and I. R. Singh. 2003. Functional characterization of a portion of the Moloney murine leukemia virus gag gene by genetic footprinting. Proc. Natl. Acad. Sci. USA 100:11678-11683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basyuk, E., T. Galli, M. Mougel, J. M. Blanchard, M. Sitbon, and E. Bertrand. 2003. Retroviral genomic RNAs are transported to the plasma membrane by endosomal vesicles. Dev. Cell 5:161-174. [DOI] [PubMed] [Google Scholar]
- 4.Bearer, E. L., X. O. Breakefield, D. Schuback, T. S. Reese, and J. H. LaVail. 2000. Retrograde axonal transport of herpes simplex virus: evidence for a single mechanism and a role for tegument. Proc. Natl. Acad. Sci. USA 97:8146-8150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4a.Bohne, J., and H.-G. Kräusslich. 2004. Mutation of the major 5′ splice site renders a CMV-driven HIV-1 proviral clone Tat-dependent: connections between transcription and splicing. FEBS Lett. 563:113-118. [DOI] [PubMed] [Google Scholar]
- 5.Bräuchle, C., G. Seisenberger, T. Endress, M. U. Ried, H. Buning, and M. Hallek. 2002. Single virus tracing: visualization of the infection pathway of a virus into a living cell. ChemPhysChem 3:299-303. [DOI] [PubMed] [Google Scholar]
- 6.Briggs, J. A. G., M. N. Simon, I. Gross, H.-G. Kräusslich, S. D. Fuller, V. M. Vogt, and M. C. Johnson. 2004. The stoichiometry of Gag protein in HIV-1. Nat. Struct. Mol. Biol. 11:672-675. [DOI] [PubMed] [Google Scholar]
- 7.Bukrinsky, M. I., S. Haggerty, M. P. Dempsey, N. Sharova, A. Adzhubel, L. Spitz, P. Lewis, D. Goldfarb, M. Emerman, and M. Stevenson. 1993. A nuclear localization signal within HIV-1 matrix protein that governs infection of non-dividing cells. Nature 365:666-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bukrinsky, M. I., N. Sharova, T. L. McDonald, T. Pushkarskaya, W. G. Tarpley, and M. Stevenson. 1993. Association of integrase, matrix, and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc. Natl. Acad. Sci. USA 90:6125-6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Derdowski, A., L. Ding, and P. Spearman. 2004. A novel fluorescence resonance energy transfer assay demonstrates that the human immunodeficiency virus type 1 Pr55Gag I domain mediates Gag-Gag interactions. J. Virol. 78:1230-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fouchier, R. A., B. E. Meyer, J. H. Simon, U. Fischer, and M. H. Malim. 1997. HIV-1 infection of non-dividing cells: evidence that the amino-terminal basic region of the viral matrix protein is important for Gag processing but not for post-entry nuclear import. EMBO J. 16:4531-4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freed, E. O., G. Englund, and M. A. Martin. 1995. Role of the basic domain of human immunodeficiency virus type 1 matrix in macrophage infection. J. Virol. 69:3949-3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuller, S. D., T. Wilk, B. E. Gowen, H.-G. Kräusslich, and V. M. Vogt. 1997. Cryo-electron microscopy reveals ordered domains in the immature HIV-1 particle. Curr. Biol. 7:729-738. [DOI] [PubMed] [Google Scholar]
- 13.Geada, M. M., I. Galindo, M. M. Lorenzo, B. Perdiguero, and R. Blasco. 2001. Movements of vaccinia virus intracellular enveloped virions with GFP tagged to the F13L envelope protein. J. Gen. Virol. 82:2747-2760. [DOI] [PubMed] [Google Scholar]
- 14.Gheysen, D., E. Jacobs, F. de Foresta, C. Thiriart, M. Francotte, D. Thines, and M. De Wilde. 1989. Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell 59:103-112. [DOI] [PubMed] [Google Scholar]
- 15.Göttlinger, H. G., T. Dorfman, J. G. Sodroski, and W. A. Haseltine. 1991. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc. Natl. Acad. Sci. USA 88:3195-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gottwein, E., J. Bodem, B. Müller, A. Schmechel, H. Zentgraf, and H.-G. Kräusslich. 2003. The Mason-Pfizer monkey virus PPPY and PSAP motifs both contribute to virus release. J. Virol. 77:9474-9485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heinzinger, N. K., M. I. Bukinsky, S. A. Haggerty, A. M. Ragland, V. Kewalramani, M. A. Lee, H. E. Gendelman, L. Ratner, M. Stevenson, and M. Emerman. 1994. The Vpr protein of human immunodeficiency virus type 1 influences nuclear localization of viral nucleic acids in nondividing host cells. Proc. Natl. Acad. Sci. USA 91:7311-7315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hermida-Matsumoto, L., and M. D. Resh. 2000. Localization of human immunodeficiency virus type 1 Gag and Env at the plasma membrane by confocal imaging. J. Virol. 74:8670-8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hill, C. P., D. Worthylake, D. P. Bancroft, A. M. Christensen, and W. I. Sundquist. 1996. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly. Proc. Natl. Acad. Sci. USA 93:3099-3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang, M., J. M. Orenstein, M. A. Martin, and E. O. Freed. 1995. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J. Virol. 69:6810-6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kiernan, R. E., A. Ono, G. Englund, and E. O. Freed. 1998. Role of matrix in an early postentry step in the human immunodeficiency virus type 1 life cycle. J. Virol. 72:4116-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kondo, E., F. Mammano, E. A. Cohen, and H. G. Gottlinger. 1995. The p6gag domain of human immunodeficiency virus type 1 is sufficient for the incorporation of Vpr into heterologous viral particles. J. Virol. 69:2759-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Konvalinka, J., M. A. Litterst, R. Welker, H. Kottler, F. Rippmann, A.-M. Heuser, and H.-G. Kräusslich. 1995. An active-site mutation in the human immunodeficiency virus type 1 proteinase (PR) causes reduced PR activity and loss of PR-mediated cytotoxicity without apparent effect on virus maturation and infectivity. J. Virol. 69:7180-7186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larson, D. R., Y. M. Ma, V. M. Vogt, and W. W. Webb. 2003. Direct measurement of Gag-Gag interaction during retrovirus assembly with FRET and fluorescence correlation spectroscopy. J. Cell Biol. 162:1233-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leopold, P. L., B. Ferris, I. Grinberg, S. Worgall, N. R. Hackett, and R. G. Crystal. 1998. Fluorescent virions: dynamic tracking of the pathway of adenoviral gene transfer vectors in living cells. Hum. Gene Ther. 9:367-378. [DOI] [PubMed] [Google Scholar]
- 26.Lodge, R., H. Göttlinger, D. Gabuzda, E. A. Cohen, and G. Lemay. 1994. The intracytoplasmic domain of gp41 mediates polarized budding of human immunodeficiency virus type 1 in MDCK cells. J. Virol. 68:4857-4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Massiah, M. A., M. R. Starich, C. Paschall, M. F. Summers, A. M. Christensen, and W. I. Sundquist. 1994. Three-dimensional structure of the human immunodeficiency virus type 1 matrix protein. J. Mol. Biol. 244:198-223. [DOI] [PubMed] [Google Scholar]
- 28.McDonald, D., M. A. Vodicka, G. Lucero, T. M. Svitkina, G. G. Borisy, M. Emerman, and T. J. Hope. 2002. Visualization of the intracellular behavior of HIV in living cells. J. Cell Biol. 159:441-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller, M. D., C. M. Farnet, and F. D. Bushman. 1997. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J. Virol. 71:5382-5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nelle, T. D., and J. W. Wills. 1996. A large region within the Rous sarcoma virus matrix protein is dispensable for budding and infectivity. J. Virol. 70:2269-2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paxton, W., R. I. Connor, and N. R. Landau. 1993. Incorporation of Vpr into human immunodeficiency virus type 1 virions: requirement for the p6 region of gag and mutational analysis. J. Virol. 67:7229-7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perrin-Tricaud, C., J. Davoust, and I. M. Jones. 1999. Tagging the human immunodeficiency virus gag protein with green fluorescent protein. Minimal evidence for colocalisation with actin. Virology 255:20-25. [DOI] [PubMed] [Google Scholar]
- 33.Pornillos, O., D. S. Higginson, K. M. Stray, R. D. Fisher, J. E. Garrus, M. Payne, G. P. He, H. E. Wang, S. G. Morham, and W. I. Sundquist. 2003. HIV Gag mimics the Tsg101-recruiting activity of the human Hrs protein. J. Cell Biol. 162:425-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reicin, A. S., A. Ohagen, L. Yin, S. Hoglund, and S. P. Goff. 1996. The role of Gag in human immunodeficiency virus type 1 virion morphogenesis and early steps of the viral life cycle. J. Virol. 70:8645-8652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reil, H., A. A. Bukovsky, H. R. Gelderblom, and H. G. Göttlinger. 1998. Efficient HIV-1 replication can occur in the absence of the viral matrix protein. EMBO J. 17:2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rietdorf, J., A. Ploubidou, I. Reckmann, A. Holmstrom, F. Frischknecht, M. Zettl, T. Zimmermann, and M. Way. 2001. Kinesin-dependent movement on microtubules precedes actin-based motility of vaccinia virus. Nat. Cell Biol. 3:992-1000. [DOI] [PubMed] [Google Scholar]
- 37.Seisenberger, G., M. U. Ried, T. Endress, H. Buning, M. Hallek, and C. Bräuchle. 2001. Real-time single-molecule imaging of the infection pathway of an adeno-associated virus. Science 294:1929-1932. [DOI] [PubMed] [Google Scholar]
- 38.Sherer, N. M., M. J. Lehmann, L. F. Jimenez-Soto, A. Ingmundson, S. M. Horner, G. Cicchetti, P. G. Allen, M. Pypaert, J. M. Cunningham, and W. Mothes. 2003. Visualization of retroviral replication in living cells reveals budding into multivesicular bodies. Traffic 4:785-801. [DOI] [PubMed] [Google Scholar]
- 39.Smith, G. A., S. P. Gross, and L. W. Enquist. 2001. Herpesviruses use bidirectional fast-axonal transport to spread in sensory neurons. Proc. Natl. Acad. Sci. USA 98:3466-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suomalainen, M., M. Y. Nakano, S. Keller, K. Boucke, R. P. Stidwill, and U. F. Greber. 1999. Microtubule-dependent plus- and minus end-directed motilities are competing processes for nuclear targeting of adenovirus. J. Cell Biol. 144:657-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang, C., Y. Ndassa, and M. F. Summers. 2002. Structure of the N-terminal 283-residue fragment of the immature HIV-1 Gag polyprotein. Nat. Struct. Biol. 9:537-543. [DOI] [PubMed] [Google Scholar]
- 42.von Schwedler, U. K., T. L. Stemmler, V. Y. Klishko, S. Li, K. H. Albertine, D. R. Davis, and W. I. Sundquist. 1998. Proteolytic refolding of the HIV-1 capsid protein amino-terminus facilitates viral core assembly. EMBO J. 17:1555-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ward, B. M., and B. Moss. 2001. Visualization of intracellular movement of vaccinia virus virions containing a green fluorescent protein-B5R membrane protein chimera. J. Virol. 75:4802-4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weclewicz, K., M. Ekström, K. Kristensson, and H. Garoff. 1998. Specific interactions between retrovirus Env and Gag proteins in rat neurons. J. Virol. 72:2832-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wei, X., J. M. Decker, H. Liu, Z. Zhang, R. B. Arani, J. M. Kilby, M. S. Saag, X. Wu, G. M. Shaw, and J. C. Kappes. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 46:1896-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilk, T., I. Gross, B. E. Gowen, T. Rutten, F. de Haas, R. Welker, H.-G. Kräusslich, P. Boulanger, and S. D. Fuller. 2001. Organization of immature human immunodeficiency virus type 1. J. Virol. 75:759-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiang, Y., C. E. Cameron, J. W. Wills, and J. Leis. 1996. Fine mapping and characterization of the Rous sarcoma virus Pr76gag late assembly domain. J. Virol. 70:5695-5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu, X., Q.-C. Yu, T.-H. Lee, and M. Essex. 1992. The C terminus of human immunodeficiency virus type 1 matrix protein is involved in early steps of the virus life cycle. J. Virol. 66:5667-5670. [DOI] [PMC free article] [PubMed] [Google Scholar]