Abstract
Since 1997 two distinct geminivirus species, Tomato yellow leaf curl Sardinia virus (TYLCSV) and Tomato yellow leaf curl virus (TYLCV), have caused a similar yellow leaf curl disease in tomato, coexisted in the fields of southern Spain, and very frequently doubly infected single plants. Tomatoes as well as experimental test plants (e.g., Nicotiana benthamiana) showed enhanced symptoms upon mixed infections under greenhouse conditions. Viral DNA accumulated to a similar extent in singly and doubly infected plants. In situ tissue hybridization showed TYLCSV and TYLCV DNAs to be confined to the phloem in both hosts, irrespective of whether they were inoculated individually or in combination. The number of infected nuclei in singly or doubly infected plants was determined by in situ hybridization of purified nuclei. The percentage of nuclei containing viral DNA (i.e., 1.4% in tomato or 6% in N. benthamiana) was the same in plants infected with either TYLCSV, TYLCV, or both. In situ hybridization of doubly infected plants, with probes that discriminate between both DNAs, revealed that at least one-fifth of infected nuclei harbored DNAs from both virus species. Such a high number of coinfected nuclei may explain why recombination between different geminivirus DNAs occurs frequently. The impact of these findings for epidemiology and for resistance breeding concerning tomato yellow leaf curl diseases is discussed.
Yellow leaf curl diseases are one of the most important pests affecting tomatoes (Lycopersicon esculentum L.) and have induced severe damages in crops from tropical and subtropical regions (6, 7, 30, 36, 39). They are caused by several distinct single-stranded-DNA-containing geminivirus species of the genus Begomovirus that are transmitted by the whitefly Bemisia tabaci Genn. and most of which possess a monopartite genome (9, 44). Geminiviruses replicate in the nuclei of host cells by a rolling-circle mechanism (14, 24) and by a recombination-dependent mechanism (19, 40). In the Mediterranean basin, two virus species cause diseases: Tomato yellow leaf curl Sardinia virus (TYLCSV), previously known as TYLCV-Sar (21), and the mild strain of Tomato yellow leaf curl virus (TYLCV), previously known as TYLCV-Is, originating in Israel (1). Their DNA sequences are 74% identical with the same genome structure and six partially overlapping open reading frames (Fig. 1). TYLCSV was first described as causing considerable losses in tomato crops of southeast Spain in 1992 (34). The second viral species, TYLCV, was detected in 1997 (31, 33). The two species differ in host range, but both are able to infect tomato and Nicotiana benthamiana (32, 47, 48). Tomato plants coinfected with the two species have been frequently detected in the fields (47).
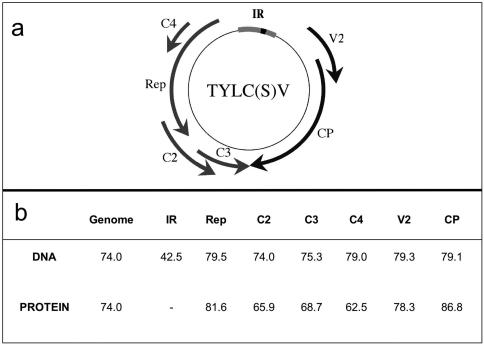
FIG. 1.
(a) Genome map of TYLCV and TYLCSV showing the positions of the intergenic region (IR) and the open reading frames (arrows) numbered in virion (V) and complementary sense (C). Rep, replication-associated protein; CP, coat protein. (b) Percentages of nucleotide and amino acid identity of IR and open reading frames between TYLCSV-ES[2] and TYLCV-[ES7297] (GenBank accession numbers L27708 and AF071228).
Although TYLCV is generally regarded as phloem limited (5, 42), one in situ localization study suggests that it may be present in other tissues within the inoculated leaf (27). The tissue tropism of TYLCSV, in single or mixed infection with TYLCV, is shown here for the first time.
Recombination contributes to the genetic diversification of geminiviruses giving rise to new epidemics (10, 15, 28, 35, 38, 41, 49, 50, 57-59). In Spain, a recombination event between TYLCV and TYLCSV has recently been reported (29). Homologous or heterologous recombinations are usually judged rare events for plant viruses; but in the case of begomoviruses, recombination might be directly involved in replication to repair viral single-stranded DNA defects (19, 40) and is therefore widespread (35).
A prerequisite for such a recombination is the encounter of different viruses within the same cell and nucleus. To determine the relevance of this possibility, we have investigated to which extent TYLCV and TYLCSV enter the same nucleus upon mixed infections.
MATERIALS AND METHODS
Microorganisms and general methods.
Manipulations of nucleic acid and bacteria were carried out by standard methods (45).
Plants and viruses.
N. benthamiana Domin and Lycopersicon esculentum L. cv. Turín, a commercial Spanish hybrid cultivar (Semillas Fitó, Barcelona, Spain), were grown in an insect-free greenhouse with supplementary lighting. Agroinoculation of seedlings was carried out with Agrobacterium tumefaciens LBA4404 (16) as previously described (22). Seedlings were agroinoculated with plasmid pGTYA2, a binary vector containing a dimer of the TYLCSV-ES[2] genome (GenBank accession no. L27708) for TYLCSV, and plasmid pGTYILES, a binary vector containing a dimer of TYLCV-[ES7297] (GenBank accession no. AF071228), for TYLCV. For mixed agroinoculations, equal amounts of both inocula were combined. For control, plants were mock inoculated either with water or with A. tumefaciens clone AbB (11), harboring a dimer of DNA B of Abutilon mosaic begomovirus (AbMV) in pBIN19 and incapable of replication by itself. Prior to localization experiments, systemically infected leaves of every single plant were tested for infection status by virus-discriminating blot hybridizations (see below).
Preparation of plant tissue sections.
Tissue sections of leaves, stems, axillary buds, and flowers were prepared as described previously (55, 56).
Purification of nuclei.
Nuclei were purified with Percoll gradient centrifugation as described previously (37), fixed with formaldehyde (freshly prepared from paraformaldehyde) at a final concentration of 3% in microtubule stabilizing buffer (MTSB) {50 mM PIPES [piperazine-N,N'-bis(2-ethanesulfonic acid)] [pH 6.9] with KOH, 5 mM EGTA, 5 mM MgSO4} for 1 h at 4°C, spread in droplets onto poly-l-lysine (Sigma)-coated slides, air dried, and hybridized.
Hybridization probes.
PCR-amplified DNAs comprising the intergenic regions of TYLCSV-ES [2] or TYLCV-[ES7297] were used to generate virus-discriminating probes. Primers oGM1 (5′-ACCAATTGACCTCAGATTCA-3′) and oGM2 (5′-CCATCAATATGTGGGA-3′) (for TYLCSV) and primers oTYA15 (5′-TATGGTCAATGAGTACC-3′) and oTYA16 (5′-GCAATATGTGGGATCCAC-3′) (for TYLCV) were used on templates pTYA58, a dimer clone of TYLCSV in pBluescript II SK(+) (M. I. Franco and E. R. Bejarano, unpublished data) or on pSP72/97 (32), respectively. For in situ hybridization, PCR fragments were labeled by nick translation (catalogue no. 1745824, Biotin-Nick translation mixture; Roche Diagnostics, Mannheim, Germany) to incorporate biotin-16-dUTP and were combined to detect both viral DNAs simultaneously (see below). To discriminate between the viruses, TYLCSV-specific probes were labeled with biotin as described above, and TYLCV-specific ones were labeled with digoxigenin (DIG) (catalogue no. 1745816, DIG-Nick translation mixture; Roche Diagnostics). Unincorporated nucleotides were removed by anion-exchange chromatography (Tip-5 column; QIAGEN, Hilden, Germany). Probes were used separately or combined and then adjusted to equal sensitivity by spot blot hybridization on nylon membranes.
For Southern or tissue blot hybridizations, the same virus-discriminating PCR-generated fragments were labeled with a DIG-High Prime labeling kit (catalogue no. 1585606, Roche Diagnostics).
Southern and tissue blot hybridization.
To determine the infection status of individual plants, Southern and tissue blot hybridizations were performed. Total nucleic acids were isolated from single systemically infected leaves, as described previously (51). Tissue blots were made by gently printing fresh cross-sections of leaves or petioles onto dry nylon membranes (Hybond-NX; Amersham) and UV cross-linking the attached nucleic acids (UV cross-linker UV setting, 700 μJ/cm2; Amersham).
In situ hybridization and probe detection. Tissue sections and isolated nuclei were hybridized as described previously (55). Biotinylated probes were detected with either streptavidin-alkaline phosphatase (AP) conjugate (catalogue no. 1093266, Roche Diagnostics) at a concentration of 2.3 U/ml or DIG-labeled probes with anti-DIG-horseradish peroxidase conjugate (catalogue no. 1207733; Roche Diagnostics) at a concentration of 2.3 U/ml. Slides were preincubated in phosphate-buffered saline (pH 7.2) for 10 min, blocked with 5% bovine serum albumin (BSA) in PBS, incubated with one conjugate or a mixture of both conjugates (diluted in 100 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% [wt/vol] bovine serum albumin) for 90 min at room temperature, and washed three times in PBS. For AP-mediated detection, the slides were equilibrated with AP buffer 3 (100 mM Tris-HCl [pH 9.5], 100 mM NaCl, 50 mM MgCl2) at room temperature and stained with nitroblue tetrazolium (NBT)-5-bromo-4-chloro-3-indolylphosphate (BCIP) (NBT, 500 μg/ml, Roche catalogue no. 1383213; BCIP, 190 μg/ml, Roche catalogue no. 1383221) in AP buffer 3, generating a brown stain. For double detection, the Vector Red alkaline phosphatase substrate kit I (catalogue no. SK-5100; Vector Laboratories, Burlingame, Calif.,), generating a red stain, was used in combination with the 3,3′,5,5′-tetramethylbenzidine (TMB) substrate kit for horseradish peroxidase (catalogue no. SK-4400; Vector Laboratories), generating a blue stain. All kit procedures were carried out according to manufacturers' recommendations. Staining was stopped after 5 to 30 min by several rinses in water and air drying.
Microscopy.
Rehydrated specimens were analyzed with epifluorescence microscopes (Axioskop, Carl Zeiss, Jena, Germany; and Nikon Eclipse E-800). To enhance contrast of plant tissues, differential interference contrast (DIC) microscopy was used. To visualize DNA, sections were stained with fluorescent DAPI (4′, 6-diamidino-2-phenylindole; 1 μg/ml in water) for 5 min. Photographs were taken on Kodak Ektachrome 64T film with Zeiss MC100 spot photograph equipment or with a Nikon DXM 1200 digital camera.
RESULTS
Symptoms of tomato and N. benthamiana plants infected with TYLCSV and TYLCV.
In four independent sets of experiments, at least 10 tomato plants and 25 N. benthamiana plants per treatment were agroinfected with infectious clones of either TYLCSV-ES[2], TYLCV-ES[7297], or both. As controls, 5 tomato plants and 25 N. benthamiana plants were mock infected in each experiment.
TYLCSV or TYLCV caused severe diseases in single infections of either host species, typically from 14 days postinfection (dpi) onwards. In general, symptoms were similar for both viruses: curling of leaflet margins and leaf blade reduction. In N. benthamiana, but not in tomato plants, some differences between the symptoms caused by TYLCSV and TLYCV were noticed. Leaflets cupped upwards upon TYLCSV infection (Fig. 2, panel d2) but downwards for TYLCV (Fig. 2, panel d3). On the contrary, tomato plants showed only slight downward- or upward-curling upon either viral infection (Fig. 2, b2 and b3). In accordance with previous reports (5, 36), infected plants exhibited reduced size due to a decrease in internode length. Stunting was more intense with TYLCV than with TYLCSV infection in both host species (Fig. 2, panels a3 and c3 versus a2 and c2).
FIG. 2.
Representative systemic symptoms in L. esculentum (a and b) and N. benthamiana (c and d) plants 4 wpi, either mock inoculated (1) or inoculated with TYLCSV (2), TYLCV (3), or both (4).
Coinfected plants developed the first symptoms 14 dpi as did singly infected plants, but they were smaller and showed reduced internode lengths (Fig. 2, panels a4 and c4 versus a2, a3, c2, and c3). Tomato plants developed striking upward curling of leaves (Fig. 2, panel b4), frequently combined with an increased number of vestigial lateral shoots in the apical region (data not shown). In N. benthamiana (Fig. 2, panel d4), leaf curling resembled that caused by TYLCV alone (Fig. 2, panel d3).
Tissue tropism of TYLCSV and TYLCV.
All plants were analyzed by tissue blot hybridization to check for the presence of the inoculated viruses at 14 dpi. In addition, total DNA was extracted from systemically infected leaves to quantify viral DNA. Southern blot analysis of singly and doubly infected plants did not reveal any significant differences in the amount of viral DNA (data not shown), confirming previous results (47). Both host plants were analyzed by in situ hybridization to investigate whether differences in tissue tropism might account for the observed symptom variations. Tissue samples from stems, leaves, petioles, and flowers were taken from infected and control plants at 4 to 7 weeks postinfection (wpi). They were fixed, paraffin embedded, sectioned, and hybridized against biotin-labeled probes specific for TYLCSV or TYLCV. For each infection type and host plant species, about 280 sections from 14 specimens from 8 tomato plants and 17 N. benthamiana plants with typical phenotypes were examined in six independent in situ hybridization experiments. Four repeated embedding procedures were carried out from 4 to 7 wpi.
In summary, both viruses, in either single or double infection, were confined to the phloem in all tested tissues of both host plants (Fig. 3 to 5), irrespective of whether plants were analyzed at 4 or 7 wpi.
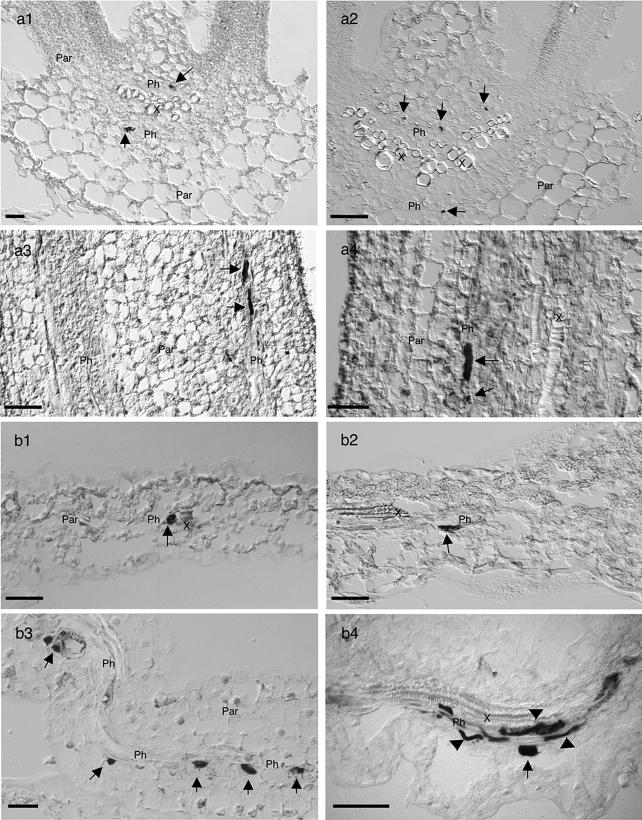
FIG. 3.
In situ hybridization of TYLCV and TYLCSV DNA in infected L. esculentum (a) and N. benthamiana (b) tissues, from leaves (a1, a2, b1, and b2), stems (a3, and a4), and petals (b3 and b4) infected with TYLCSV (left panels) or TYLCV (right panels), collected 21 dpi, and hybridized with biotin-labeled probes. Infected nuclei are indicated by dark NBT-BCIP staining (arrows). Accumulation of TYLCSV or TYLCV is mainly restricted to the nuclei of phloem parenchymal and companion cells (arrows); the companion cells may be very elongated or spindle shaped. For comparison, see the DAPI staining in Fig. 5, panel a2 or c2. In addition, some signals were detected in sieve tubes (b4, arrowheads). Sections were examined by DIC microscopy. Bars, 50 μm.
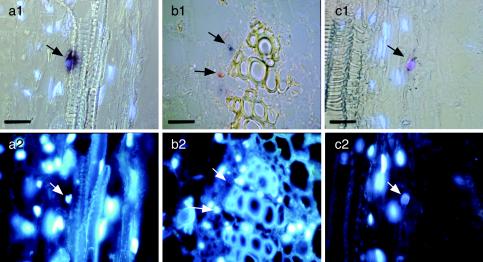
FIG. 5.
In situ detection discriminating between TYLCSV and TYLCV in N. benthamiana tissues from stems of doubly infected N. benthamiana, hybridized with a biotin-labeled TYLCSV probe staining red (b) or a DIG-labeled TYLCV probe staining blue (a and b). Violet color (c) indicates the presence of both viruses. Samples were examined by either combined DIC and epifluorescence microscopy (a1 and c1), DIC microscopy (b1), or epifluorescence microscopy alone to visualize DAPI-stained nuclei (a2, b2, and c2). Bar, 25 μm. Arrows point at the same sites in the corresponding pictures. Note the predominantly TYLCV- or TYLCSV-infected nuclei in neighboring cells (b1) and the presence of both viruses within one nucleus (c1).
Shape and localization of the signals suggested that viral DNA accumulated predominantly in nuclei, which was confirmed by DAPI counterstaining (Fig. 5), although accumulations in sieve tubes were also observed (Fig. 3, panel b4).
The specificity of DNA detection was confirmed by several tests; sections of uninfected tissues of comparable ages were included in all experiments. In addition, sections of TYLCSV- or TYLCV-infected plants were mock incubated (omitting the probe from the hybridization solution) or hybridized with a probe specific for the noninoculated virus, e.g., TYLCSV-infected plants probed with TYLCV probe and vice versa (data not shown). No hybridization signals were detected in control tests.
A high proportion of mixed-infected nuclei.
To trace both viruses simultaneously in doubly infected plants and to assess how often the two viruses met each other within the cells, tissue sections at different infection status were hybridized with a mixture of virus-discriminating probes labeled with distinct haptens, staining TYLCSV DNA red and TYLCV DNA blue. As a control, singly infected specimens were treated by the complete double-hybridization and detection procedure, which did not develop any mixed stain. In contrast, nuclei and sieve elements of doubly infected leaf tissues exhibited a wide range of hues from red via purple and violet to blue, which was obvious even at first glance. In tissue sectors where several infected nuclei occurred in close vicinity, the signal hue frequently differed from nucleus to nucleus. A mixture of apparently singly (completely red or blue) and doubly infected (purple to violet) nuclei with variable TYLCSV:TYLCV ratios, sometimes adjacent to each other, was the typical distribution pattern in virus-infected tissues.
Since viral DNA was only present in a small portion of the veinal nuclei and since sectioning to survey sufficiently large areas of infected leaves is extremely laborious, quantitative data about viral coinvasion of single nuclei were acquired from purified nuclei by the double-detection procedure. As a first step, the overall amount of infected nuclei within the plants with a different infection status was determined. Nuclei were purified from leaves of four tomato plants or five N. benthamiana plants, each systemically infected with either TYLCSV, TYLCV, or both at 3 to 4 wpi. The nuclei were counted, fixed, mounted on slides, and hybridized with probes specific for TYLCSV or TYLCV. Table 1 summarizes the results of five independent experiments, where the percentage of infected nuclei was calculated to be from 450 to 1,850 nuclei per infection. As a control, nuclei from mock-inoculated plants were hybridized with both viral probes giving no significant signal.
TABLE 1.
Percentage of infected nucleia
| Test plant | n | Yield (106) | m | % of infected nucleib
|
||
|---|---|---|---|---|---|---|
| TYLCSV | TYLCV | Both | ||||
| L. esculentum | 2 | 1.8 ± 1.0 | 630-1,340 | 1.8† | 1.3† | 1.3† |
| N. benthamiana | 3 | 1.0 ± 0.3 | 480-1,880 | 7.4* | 5.5* | 6.4* |
Yield, the number of infected nuclei per gram (fresh weight) isolated from two host species after infection with TYLCSV, TYLCV, or both; values are means ± standard deviations. n, number of independent experiments; m, number of nuclei analyzed per treatment in five independent in situ experiments.
χ2 test: for α < 0.05, no significant differences for values labeled with the same characters (* or †), and significant differences for values labeled with different characters.
The percentage of infected nuclei from N. benthamiana plants (6%) was about four times higher than that from tomato plants (1.5%). However, no significant difference was observed between plants infected by one viral species or both, irrespective of the host species. Moreover, both viruses exhibited an unexpectedly high rate of infected nuclei compared to predictions gained from tissue sectioning.
To estimate the probability of viral recombination within an individual doubly infected plant, the percentage of nuclei infected with both viruses was determined. To this aim, purified nuclei from both coinfected host plants were simultaneously hybridized with TYLCSV- and TYLCV-discriminating probes of equal sensitivities, as described above, staining TYLCSV DNA red, TYLCV DNA blue, and staining doubly infected nuclei violet to purple. For comparison and control, nuclei from singly infected plants were hybridized under the same conditions, in parallel. Figure 6 shows representative results, and Table 2 summarizes the evaluation obtained after analyzing at least 200 randomly chosen infected nuclei in about 15,000 tomato and 4,200 N. benthamiana nuclei. Whereas 35 to 50% of the nuclei contained only one type of either viral DNA, approximately 20% of the infected nuclei were unequivocally classified as doubly infected, which was similar in both hosts. The percentage of double infection was probably rather underestimated, because nuclei were classified as such only if they contained well-detectable amounts of both DNAs and produced a clear purple-to-violet appearance.
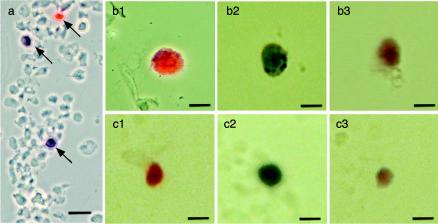
FIG. 6.
In situ hybridization of purified nuclei from N. benthamiana and L. esculentum plants agroinoculated with TYLCSV (b1 and c1), TYLCV (b2 and c2), or both (a, b3, and c3). Nuclei were fixed and hybridized simultaneously with a biotin-labeled TYLCSV-specific probe and a DIG-labeled TYLCV-specific probe. Red coloring represents TYLCSV, blue coloring represents TYLCV, and purple coloring represents both viruses. (a) General view of isolated nuclei in N. benthamiana by DIC microscopy. Arrows indicate nuclei infected with TYLCSV (red) or TYLCV (blue). (b and c) Close-up images of isolated nuclei from N. benthamiana (b1, b2, and b3) or L. esculentum (c1, c2, and c3) plants by light microscopy are shown. Bars, 25 μm (a) or 10 μm (b1, b2, b3, c1, c2, and c3).
TABLE 2.
Percentage of nuclei infected by both virusesa
| Test plant | n | m | % of nuclei infected with:
|
Calculated frequency of nuclei infected with
|
||||
|---|---|---|---|---|---|---|---|---|
| TYLCSV | TYLCV | Both | TYLCSV | TYLCV | Both | |||
| L. esculentum | 15,000 | 200 | 37 ± 4 | 42 ± 7 | 22 ± 7 | 0.48 | 0.52 | 0.25 |
| N. benthamiana | 4,200 | 272 | 48 ± 6 | 34 ± 7 | 18 ± 6 | 0.56 | 0.44 | 0.25 |
Percentage (mean ± standard deviation) of infected nuclei which contain DNA from TYLCSV, TYLCV, or both, as judged by color evaluation after coinoculation of TYLSCV and TYLCV. n, total nuclei analyzed; m, infected nuclei analyzed (three experiments with L. esculentum and four experiments with N. benthamiana, respectively). To obtain the calculated frequencies, the numbers of nuclei infected by a particular virus individually or in combination with the other virus were added, resulting in new values for total TYLCSV- and TYLCV-infected nuclei. The probability that both TYLCSV and TYLCV would enter the same nucleus was calculated by using the product of the frequencies for the individual viruses. Within the range of the standard deviations, the calculated numbers for infection by both viruses fit well to the observed percentages.
DISCUSSION
Under field conditions, mixed viral infections are quite common. They may result in additive, synergistic, or mutually exclusive effects on symptoms and provide the possibility to evolve new more pathogenic or cross-protecting viruses by genetic recombination. The knowledge of these effects is important for epidemiology and resistance breeding, either by classical or gene technological methods (18). Concerning the safety assessment of gene technologically modified transgenic plants, synergistic effects or recombination between transgenes and infecting viruses has especially drawn attention from the public and researchers and has been reviewed in great detail by Hammond et al. (13). Obviously, a risk determined in this area must be compared to naturally occurring coinfection and to the sustainability of classically bred cultivars (18).
The case study of tomato yellow leaf curl disease is of particular, basic, and agronomic importance in this context. TYLCV has spread over continents and to the Carribean Islands, Mexico, and the United States during the last 10 years, founding new populations (30, 39). Coinfections of different TYLC virus species have expanded (47); a new virus isolate has evolved in the field by recombination between DNA from both species (29, 33), exhibiting new biological characteristics with respect to host range and symptom phenotype. The recombinant virus became prevalent in the area where it had been isolated (29). To repress agronomic losses due to TYLCV, tolerant tomato cultivars were introduced into the market (30), which reduce symptoms but propagate the virus. Safety assessment should consider all these circumstances.
In this report, we present first evidence for synergistic effects between two TYLC virus species and exclude effects of cross-protection between them. The possibility for recombination between these viruses under natural conditions is expected to be high, since both viruses frequently enter the same nucleus. We confirm that TYLCSV as well as TYLCV are phloem limited in systemically infected leaves, irrespective of whether they are inoculated separately or in combination.
Although both viruses alone produced symptoms of similar type and severity in both hosts, enhanced symptoms were observed in plants systemically infected with both viruses. The increase in symptom severity did not correlate with an elevated virus titer in doubly infected plants. A similar level of virus accumulation was found by Sanchez-Campos et al. (47) for single and mixed infections of TYLCV and TYLCSV in L. esculentum cv. ′Moneymaker.' In contrast to our studies, no symptom increase due to mixed infection was found in the field in Spain, where the cultivars were not specified (32). Under these field conditions, mixed-infected plants exhibited a severe phenotype similar to that induced by TYLCV alone. Under our greenhouse conditions, TYLCV or TYLCSV did not induce the maximum of symptoms in single infections, and therefore the synergistic effects were monitored from a lower basal level.
In situ hybridization showed that both viruses share the same tissues in all plants throughout extended periods of time. Both were restricted to the phloem, irrespective of the host. These findings are consistent with many tissue localization reports using manifold techniques for systemically infected leaves (3, 4, 42, 43). Michelson et al. (26) have observed TYLCV DNA in tissues other than phloem in the inoculated leaves of tomatoes, independently of whether the plants carried the Ty-1 TYLCV tolerance gene or not.
TYLCV DNA and, as shown here for the first time, TYLCSV DNA were localized in the nuclei of companion and phloem parenchyma cells, as well as in the lumen of sieve tubes, which has rarely been detected before. The accumulation in sieve tubes may correspond to virions or other viral transport complexes undergoing long-distance movement in the plants (for a review, see reference 12).
Similar patterns of infection have been observed for the majority of geminiviruses that infect dicotyledonous hosts: AbMV in Abutilon spp., N. benthamiana, and Malva parviflora plants (17, 55); Squash leaf curl virus in pumpkin plants (54); and Beet curly top virus in N. benthamiana (23). Escape from the phloem to other leaf tissues has been reported for some dicotyledonous host-infecting begomoviruses, such as African cassava mosaic virus and Tomato golden mosaic virus in N. benthamiana (55), Bean dwarf mosaic virus in N. benthamiana and bean plants (52, 53), Bean golden mosaic virus if coinfected with Tobacco mosaic virus (2), and mastreviruses which infect monocotyledonous hosts (25).
This limited tissue tropism may have various causes, depending on both the particular virus and the host plant. Either non-phloem cells cannot support replication, transcription or cell-to-cell movement of the particular virus, viral movement proteins lack the capability to facilitate transport out of the phloem, or defense mechanisms of the host may suppress invasion into and/or efficient multiplication in further cells.
In situ hybridization of tissue sections detected most viral DNA within nuclei because they are sites of accumulation. Viral DNA, as it is transported through cytoplasm and plasmodesmata or through sieve elements, is hardly detectable, probably because virus spread is a transient process during plant and infection development (reference 56 and references therein). Here, we show that although the transport through sieve elements is a rare event, it is accompanied by high concentrations of viral DNA if it occurs (Fig. 3).
Since both viruses are phloem limited, all purified infected nuclei should stem from this tissue. The percentage of infected nuclei in singly as well as in doubly infected plants was higher than expected from rough estimations of tissue-thin sectioning. Moreover, the percentages were quite constant in independent preparations from singly and doubly infected plants. However, the number of nuclei in tissues and in purified samples cannot be compared directly, since <10% of the total nuclei are usually obtained under the best purification conditions. It remains possible that vascular or infected nuclei are enriched during preparation. Nevertheless, this effect should be similar for independent experiments, a conclusion that is substantiated by the reproducibility concerning different experiments and different hosts (Table 1).
With respect to the low number of infected nuclei detected in tissue sections, one might be tempted to speculate that coinfection of nuclei is an extremely rare event and therefore that recombination between two geminiviruses is improbable under natural conditions. However, Tables 1 and 2 provide evidence to support the opposite conclusion. Most importantly, the percentages of infected nuclei for all sets of experiments are nearly the same for each host and for each infection status. If infecting nuclei were just an additive process driven by chance, an increase of infected nuclei would result from mixed infection. Therefore, we interpret from these results that only a limited and fixed number of phloem nuclei of a particular host are attainable by and/or susceptible to the virus. This notion is substantiated by the good fit of calculated frequencies (Table 2) and observed percentages of double infection. In terms of population genetics, the two viruses have different levels of fitness (Table 2; frequency p = 0.48 or 0.56 for TYLCSV; frequency q = 0.52 or 0.44 for TYLCV), and the coinfection can simply be described to be the product of the fitness data (Table 2; p×q = 0.25 for both viruses). The fact that the calculated values for coinfection are slightly higher than the observed ones can be attributed to underestimation of doubly colored nuclei as discussed above.
The presented data infer hit statistics to explain virus dynamics without any interference, such as cross-protection. This phenomenon contrasts with experiences with RNA viruses. For example, Dietrich and Maiss (8) found that upon coinfection, closely related potyviruses separate from each other in different tissue sectors.
If the probability that TYLCSV and TYLCV will meet in a single nucleus is high, what will this represent for recombination and creation of new viruses in the field? Recently, it has been shown that begomoviruses, including TYLCV, may use recombination to repair incomplete viral DNA during normal replication (19, 40). Therefore, molecular recombination should occur very frequently if two viruses meet each other. Nevertheless, a recombined molecule must reach a certain level of fitness to be maintained in the population. If its parents are well adapted in a certain host, this is improbable. In the case of TYLCSV, the population of an individual begomovirus strain can be very stable over an extended period of time (46). However, as observed in studies carried out in southern Spain, the incorporation of a second begomovirus species to the epidemy might result in a dramatic alteration of the population structure (47) and the appearance of new strains due to recombination. In a comparable analysis of Sida-infecting begomoviruses kept under isolation in greenhouses over a period of 25 years, a recombinant creating a begomovirus in statu nascendi was observed and stably maintained, although the parental viruses persisted in a population now composed of three distinct geminiviruses (20). Therefore, recombination among geminiviruses could be a frequent event, able to provide new viruses that may be harmful under new selection pressures, e.g., if a new host, cultivar, or a vector insect is introduced, as proposed earlier (35).
Against this background, the dissemination of tolerant cultivars has more safety impact than the release of genetically modified plants that, as the best safety measures indicate, normally harbor only parts of viral genomes, which could be dysfunctional for viruses.
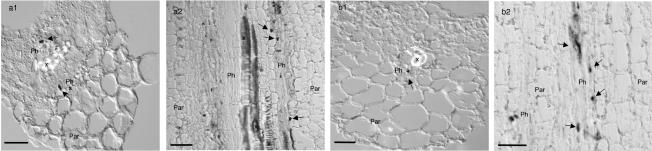
FIG. 4.
In situ hybridization codetecting TYLCSV and TYLCV in doubly infected L. esculentum (a1 and a2) and N. benthamiana (b1 and b2) tissues, from leaves (a1 and b1) and stems (a2 and b2) 21 to 28 dpi. Infected nuclei are indicated by dark staining precipitated from NBT-BCIP (indicated by arrows). All sections were hybridized with a biotin-labeled mixed probe containing TYLCSV plus TYLCV. Accumulation of both viruses is restricted to nuclei from phloem parenchymal cells and companion cells. X, xylem; Ph, phloem; Par, nonvascular parenchyma. Sections were examined by DIC microscopy. Bars, 50 μm.
Acknowledgments
This research was partially supported by grants from the Spanish Ministerio de Ciencia y Tecnología (AGF98-0439-C05-05 and AGL2001-2857-CO4-01). G.M. was awarded a Predoctoral Fellowship from the University of Málaga.
We thank Enrique Moriones for providing us with the TYLCV-[ES7297] clone. We are grateful to Jens O. Pohl, who established the two-color detection system. Many thanks to Cornelia Kocher, Nieves Santiago, Raul Heredia, Lucía Cruzado, Alexandra Schwierzok, and Sigrid Kober for their excellent technical assistance and to our gardeners, Diether Gotthardt and Irene Petschi. We thank Carmen Beuzón for critically reading the manuscript and making many helpful suggestions.
REFERENCES
- 1.Antignus, Y., and S. Cohen. 1994. Cloning of tomato yellow leaf curl virus (TYLCV) and the complete nucleotide sequence of a mild infectious clone. Phytopathology 84:707-712. [Google Scholar]
- 2.Carr, R. J., and K. S. Kim. 1983. Evidence that bean golden mosaic virus invades non-phloem tissue in double infections with tobacco mosaic virus. J. Gen. Virol. 64:2489-2492. [Google Scholar]
- 3.Channarayappa, V. Muniyappa, D. Schwegler-Berry, and G. Shivashankar. 1992. Ultrastructural changes in tomato infected with tomato leaf curl virus, a whitefly-transmitted geminivirus. Can. J. Bot. 70:1747-1753. [Google Scholar]
- 4.Cherif, C., and M. Russo. 1983. Cytological evidence of the association of a geminivirus with the tomato yellow leaf curl disease in Tunisia. Phytopathol. Z. 108:221-225. [Google Scholar]
- 5.Cohen, S., and Y. Antignus. 1994. Tomato yellow leaf curl virus (TYLCV), a whitefly-borne geminivirus of tomatoes. Adv. Dis. Vector Res. 10:259-288. [Google Scholar]
- 6.Czosnek, H., M. Ghanim, S. Morin, G. Rubinstein, V. Fridman, and M. Zeidan. 2001. Whiteflies: vectors, and victims (?), of geminiviruses. Adv. Virus Res. 57:291-322. [DOI] [PubMed] [Google Scholar]
- 7.Czosnek, H., and H. Laterrot. 1997. A worldwide survey of tomato yellow leaf curl viruses. Arch. Virol. 142:1391-1406. [DOI] [PubMed] [Google Scholar]
- 8.Dietrich, C., and E. Maiss. 2003. Fluorescent labelling reveals spatial separation of potyvirus populations in mixed infected Nicotiana benthamiana plants. J. Gen. Virol. 84:2871-2876. [DOI] [PubMed] [Google Scholar]
- 9.Fauquet, C. M., D. M. Bisaro, R. W. Briddon, J. K. Brown, B. D. Harrison, E. P. Rybicki, D. C. Stenger, and J. Stanley. 2003. Revision of taxonomic criteria for species demarcation in the family Geminiviridae, and an updated list of begomovirus species. Arch. Virol. 148:405-421. [DOI] [PubMed] [Google Scholar]
- 10.Fondong, V. N., J. S. Pita, M. E. C. Rey, A. de Kockko, R. N. Beachy, and C. M. Fauquet. 2000. Evidence of synergism between African cassava mosaic virus and a new double-recombinant geminivirus infecting cassava in Cameroon. J. Gen. Virol. 81:287-297. [DOI] [PubMed] [Google Scholar]
- 11.Frischmuth, T., S. Roberts, A. von Arnim, and J. Stanley. 1993. Specificity of bipartite geminivirus movement proteins. Virology 196:666-673. [DOI] [PubMed] [Google Scholar]
- 12.Gafni, Y. 2003. Tomato yellow leaf curl virus, the intracellular dynamics of a plant DNA virus. Mol. Plant Pathol. 4:9-15. [DOI] [PubMed] [Google Scholar]
- 13.Hammond, J., H. Lecoq, and B. Raccah. 1999. Epidemiological risks from mixed virus infections and transgenic plants expressing viral genes. Adv. Virus Res. 54:189-314. [DOI] [PubMed] [Google Scholar]
- 14.Hanley-Bowdoin, L., S. B. Settlage, B. M. Orozco, S. Nagar, and D. Robertson. 1999. Geminiviruses: models for plant DNA replication, transcription, and cell cycle regulation. Crit. Rev. Plant Sci. 18:71-106. [PubMed] [Google Scholar]
- 15.Harrison, B. D., and D. J. Robinson. 1999. Natural genomic and antigenic variation in whitefly-transmitted geminiviruses (Begomoviruses). Annu. Rev. Phytopathol. 37:369-398. [DOI] [PubMed] [Google Scholar]
- 16.Hoekema, A., P. R. Hirsch, P. J. J. Hooykaas, and R. A. Schilperoort. 1983. A binary plant vector strategy based on separation of vir- and T-region of the Agrobacterium tumefaciens Ti-plasmid. Nature 303:179-180. [Google Scholar]
- 17.Horns, T., and H. Jeske. 1991. Localization of Abutilon mosaic virus DNA within leaf tissue by in-situ hybridization. Virology 181:580-588. [DOI] [PubMed] [Google Scholar]
- 18.Jeske, H. 2002. Transgenic plants with increased resistance and tolerance against viral pathogens, p. 517-548. In K. M. Oksman-Caldentey and W. Barz (ed.), Plant biotechnology and transgenic plants. Marcel Dekker, New York, N.Y.
- 19.Jeske, H., M. Lütgemeier, and W. Preiss. 2001. Distinct DNA forms indicate rolling circle and recombination-dependent replication of Abutilon mosaic geminivirus. EMBO J. 20:6158-6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jovel, J., G. Reski, D. Rothenstein, M. Ringel, T. Frischmuth, and H. Jeske. 2004. Sida micrantha mosaic is associated with a complex infection of begomoviruses different from Abutilon mosaic virus. Arch. Virol. 149:829-841. [DOI] [PubMed] [Google Scholar]
- 21.Kheyr-Pour, A., M. Bendahmane, H. J. Matzeit, G. P. Accotto, S. Crespi, and B. Gronenborn. 1991. Tomato yellow leaf curl virus from Sardinia is a whitefly-transmitted geminivirus. Nucleic Acids Res. 19:6763-6769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klinkenberg, F. A., S. Ellwood, and J. Stanley. 1989. Fate of African cassava mosaic virus coat protein deletion mutants after agroinoculation. J. Gen. Virol. 70:1837-1844. [DOI] [PubMed] [Google Scholar]
- 23.Latham, J. R., K. Saunders, M. S. Pinner, and J. Stanley. 1997. Induction of plant cell division by beet curly top virus gene C4. Plant J. 11:1273-1283. [Google Scholar]
- 24.Laufs, J., W. Traut, F. Heyraud, V. Matzeit, S. G. Rogers, J. Schell, and B. Gronenborn. 1995. In vitro cleavage and joining at the viral origin of replication by the replication initiator protein of tomato yellow leaf curl virus. Proc. Natl. Acad. Sci. USA 92:3879-3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lucy, A. P., M. I. Boulton, J. W. Davies, and A. J. Maule. 1996. Tissue specificity of Zea mays infection by maize streak virus. Mol. Plant-Microbe Interact. 9:22-31. [Google Scholar]
- 26.Michelson, I., D. Zamir, and H. Czosnek. 1994. Accumulation and translocation of tomato yellow leaf curl virus (TYLCV) in a Lycopersicon esculentum breeding line containing the L. chilense TYLCV tolerance gene Ty-1. Phytopathology 84:928-933. [Google Scholar]
- 27.Michelson, I., M. Zeidan, D. Zamir, and H. Czosnek. 1997. Localization of tomato yellow leaf curl virus (TYLCV) in susceptible and tolerant nearly isogenic tomato lines. Acta Hortic. 447:407-414. [Google Scholar]
- 28.Moffat, A. 1999. Geminiviruses emerge as serious crop threat. Science 286:1835. [Google Scholar]
- 29.Monci, F., S. Sanchez-Campos, J. Navas-Castillo, and E. Moriones. 2002. A natural recombinant between the geminiviruses tomato yellow leaf curl Sardinia virus and tomato yellow leaf curl virus exhibits a novel pathogenic phenotype and is becoming prevalent in Spanish populations. Virology 303:317-326. [DOI] [PubMed] [Google Scholar]
- 30.Moriones, E., and J. Navas-Castillo. 2000. Tomato yellow leaf curl virus, an emerging virus complex causing epidemics worldwide. Virus Res. 71:123-134. [DOI] [PubMed] [Google Scholar]
- 31.Navas-Castillo, J., S. Sanchez-Campos, J. A. Diaz, E. Saez-Alonso, and E. Moriones. 1997. First report of tomato yellow leaf curl virus-Is in Spain: coexistence of two different geminiviruses in the same epidemic outbreak. Plant Dis. 81:1461. [DOI] [PubMed] [Google Scholar]
- 32.Navas-Castillo, J., S. Sanchez-Campos, J. A. Diaz, E. Saez-Alonso, and E. Moriones. 1999. Tomato yellow leaf curl virus causes a novel disease of common bean and severe epidemics in tomato in spain. Plant Dis. 83:29-32. [DOI] [PubMed] [Google Scholar]
- 33.Navas-Castillo, J., S. Sanchez-Campos, E. Noris, D. Louro, G. P. Accotto, and E. Moriones. 2000. Natural recombination between tomato yellow leaf curl virus-Is and tomato leaf curl virus. J. Gen. Virol. 81:2797-2801. [DOI] [PubMed] [Google Scholar]
- 34.Noris, E., E. Hidalgo, G. P. Accotto, and E. Moriones. 1994. High similarity among the tomato yellow leaf curl virus isolates from the west Mediterranean basin: the nucleotide sequence of an infectious clone from Spain. Arch. Virol. 135:165-170. [DOI] [PubMed] [Google Scholar]
- 35.Padidam, M., S. Sawyer, and C. M. Fauquet. 1999. Possible emergence of new geminiviruses by frequent recombination. Virology 285:218-225. [DOI] [PubMed] [Google Scholar]
- 36.Picó, B., M. J. Diez, and F. Nuez. 1996. Viral diseases causing the greatest economic losses to the tomato crop. II. The tomato yellow leaf curl virus-—a review. Sci. Hortic. 67:151-196. [Google Scholar]
- 37.Pilartz, M., and H. Jeske. 1992. Abutilon mosaic geminivirus double-stranded DNA is packed into minichromosomes. Virology 189:800-802. [DOI] [PubMed] [Google Scholar]
- 38.Pita, J. S., V. N. Fondong, A. Sangare, O. S. Otim-Nape, and C. M. Fauquet. 2001. Recombination, pseudorecombination and synergism of geminiviruses are determinant keys to the epidemic of severe cassava mosaic disease in Uganda. J. Gen. Virol. 82:655-665. [DOI] [PubMed] [Google Scholar]
- 39.Polston, J. E., and P. K. Anderson. 1997. The emergence of whitefly-transmitted geminiviruses in tomato in the western hemisphere. Plant Dis. 81:1358-1369. [DOI] [PubMed] [Google Scholar]
- 40.Preiss, W., and H. Jeske. 2003. Multitasking in replication is common among geminiviruses. J. Virol. 77:2972-2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ribeiro, S. G., L. P. Ambrozevicius, A. C. Avila, I. C. Bezerra, R. F. Calegario, J. J. Fernandes, M. F. Lima, R. N. de Mello, H. Rocha, and F. M. Zerbini. 2003. Distribution and genetic diversity of tomato-infecting begomoviruses in Brazil. Arch. Virol. 148:281-295. [DOI] [PubMed] [Google Scholar]
- 42.Rojas, M. R., H. Jiang, R. Salati, B. Xoconostle-Cazares, M. R. Sudarshana, W. J. Lucas, and R. L. Gilbertson. 2001. Functional analysis of proteins involved in movement of the monopartite begomovirus, Tomato yellow leaf curl virus. Virology 291:110-125. [DOI] [PubMed] [Google Scholar]
- 43.Russo, M., S. Cohen, and G. P. Martelli. 1980. Virus-like particles in tomato plants affected by the yellow leaf curl disease. J. Gen. Virol. 49:209-213. [Google Scholar]
- 44.Rybicki, E. P., R. W. Briddon, J. K. Brown, C. M. Fauquet, D. P. Maxwell, B. D. Harrison, P. G. Markham, D. M. Bisaro, D. Robinson, and J. Stanley. 2000. Family Geminiviridae, p. 285-297. In M. H. V. van Regenmortel, C. M. Fauquet, and D. H. L. Bishop (ed.), Virus taxonomy-—classification and nomenclature of viruses. Academic Press, San Diego, Calif.
- 45.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 46.Sanchez-Campos, S., J. A. Diaz, F. Monci, E. R. Bejarano, J. Reina, J. Navas-Castillo, M. A. Aranda, and E. Moriones. 2002. High genetic stability of the begomovirus tomato yellow leaf curl Sardinia virus in southern Spain over an 8-year period. Phytopathology 92:842-849. [DOI] [PubMed] [Google Scholar]
- 47.Sanchez-Campos, S., J. Navas-Castillo, R. Camero, C. Soria, J. A. Diaz, and E. Moriones. 1999. Displacement of tomato yellow leaf curl virus (TYLCV)-Sr by TYLCV-Is in tomato epidemics in Spain. Phytopathology 89:1038-1043. [DOI] [PubMed] [Google Scholar]
- 48.Sanchez-Campos, S., J. Navas-Castillo, F. Monci, J. A. Diaz, and E. Moriones. 2000. Mercurialis ambigua and Solanum luteum: two newly discovered natural hosts of tomato yellow leaf curl geminiviruses. Eur. J. Plant Pathol. 106:391-394. [Google Scholar]
- 49.Sanz, A. I., A. Fraile, F. Garcia-Arenal, X. Zhou, D. J. Robinson, S. Khalid, T. Butt, and B. D. Harrison. 2000. Multiple infection, recombination and genome relationships among begomovirus isolates found in cotton and other plants in Pakistan. J. Gen. Virol. 81:1839-1849. [DOI] [PubMed] [Google Scholar]
- 50.Schnippenkoetter, W. H., D. P. Martin, J. A. Willment, and E. P. Rybicki. 2001. Forced recombination between distinct strains of Maize streak virus. J. Gen. Virol. 82:3081-3090. [DOI] [PubMed] [Google Scholar]
- 51.Song, J. Y., and H. Jeske. 1994. The level of Abutilon mosaic geminivirus in leaf discs and wound callus. J. Phytopathol. 140:45-52. [Google Scholar]
- 52.Sudarshana, M. R., H. L. Wang, W. J. Lucas, and R. L. Gilbertson. 1998. Dynamics of bean dwarf mosaic geminivirus cell-to-cell and long-distance movement in Phaseolus vulgaris revealed, using the green fluorescent protein. Mol. Plant-Microbe Interact. 11(4):277-291. [Google Scholar]
- 53.Wang, H. L., R. L. Gilbertson, and W. J. Lucas. 1996. Spatial and temporal distribution of bean dwarf mosaic geminivirus in Phaseolus vulgaris and Nicotiana benthamiana. Phytopathology 86:1204-1214. [Google Scholar]
- 54.Ward, B. M., R. Medville, S. G. Lazarowitz, and R. Turgeon. 1997. The geminivirus BL1 movement protein is associated with endoplasmic reticulum-derived tubules in developing phloem cells. J. Virol. 71:3726-3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wege, C., K. Saunders, J. Stanley, and H. Jeske. 2001. Comparative analysis of tissue tropism of bipartite geminiviruses. J. Phytopathol. 149:359-368. [Google Scholar]
- 56.Zhang, S. C., C. Wege, and H. Jeske. 2001. Movement proteins (BC1 and BV1) of Abutilon mosaic geminivirus are cotransported in and between cells of sink but not of source leaves as detected by green fluorescent protein tagging. Virology 290:249-260. [DOI] [PubMed] [Google Scholar]
- 57.Zhou, X., Y. Liu, L. Calvert, C. Munoz, G. W. Otim-Nape, D. J. Robinson, and B. D. Harrison. 1997. Evidence that DNA-A of a geminivirus associated with severe cassava mosaic disease in Uganda has arisen by interspecific recombination. J. Gen. Virol. 78:2101-2111. [DOI] [PubMed] [Google Scholar]
- 58.Zhou, X., Y. Xie, X. Tao, Z. Zhang, Z. Li, and C. M. Fauquet. 2003. Characterization of DNAβ associated with begomoviruses in China and evidence for coevolution with their cognate viral DNA-A. J. Gen. Virol. 84:237-247. [DOI] [PubMed] [Google Scholar]
- 59.Zhou, X. P., Y. L. Liu, D. J. Robinson, and B. D. Harrison. 1998. Four DNA-A variants among Pakistani isolates of cotton leaf curl virus and their affinities to DNA-A of geminivirus isolates from okra. J. Gen. Virol. 79:915-923. [DOI] [PubMed] [Google Scholar]