Abstract
RNA interference (RNAi) is an antiviral mechanism that is activated when double-stranded RNA is cleaved into fragments, called short interfering RNA (siRNA), that prime an inducible gene silencing enzyme complex. We applied RNAi against a herpes simplex virus type 1 (HSV-1) gene, glycoprotein E, which mediates cell-to-cell spread and immune evasion. In an in vitro model of infection, human keratinocytes were transfected with siRNA specific for glycoprotein E and then infected with wild-type HSV-1. RNAi-mediated gene silencing reproduced the small plaque phenotype of a gE-deletion mutant virus. The specificity of gene targeting was demonstrated by flow cytometry and Northern blot analyses. Exogenous siRNA can suppress HSV-1 glycoprotein E expression and function during active infection in vitro through RNAi. This work establishes RNAi as a genetic tool for the study of HSV and provides a foundation for development of RNAi as a novel antiviral therapy.
RNA-mediated gene suppression was first described for plants as a defense mechanism against viruses and transposons. Subsequently, this mechanism was also described for ciliates, fungi, Caenorhabditis elegans, mammals, and human cell lines (11, 12, 32, 42, 44, 46). RNA interference (RNAi) is now the term used to describe the mechanism of RNA-inducible gene silencing. Double-stranded RNA is cleaved into 21- to 25-bp short interfering RNAs (siRNAs) by an enzyme called Dicer with RNase III activity (4, 11, 19, 30, 43). These double-stranded siRNAs prime an RNA-inducible silencing complex. The activated RNA-inducible silencing complex is thought to bind mRNA with a sequence complementary to the siRNA and bisect the area of homology (35). This renders the mRNA transcripts inert and more degradable, resulting in a specific reduction (knockdown) in expression of the targeted gene. In mammals, RNA-mediated gene silencing has not yet been demonstrated to play an active endogenous role in antiviral defense. However, the components of this ancient system have been retained and contribute to multiple aspects of genome regulation (18).
Herpes simplex virus type 1 (HSV-1) is a double-stranded DNA virus that infects epithelial and neuronal cells. Glycoprotein E (gE) from HSV-1 strain NS is a 552-amino-acid type I transmembrane protein that is essential for cell-to-cell spread (8) as well as viral evasion from complement and antibody (3, 10, 26). The deletion of gE from the NS strain of HSV-1 (gEnull) produces a defect in cell-to-cell spread. This is observed as smaller viral plaques in vitro and less cutaneous ulceration in vivo (2, 37, 45). We chose the in vitro plaque model of HSV-1 gE deletion to study the effects of siRNA because it provides a reproducible and visibly measurable phenotype. The purpose of this study was to determine whether exogenously delivered siRNA could suppress the expression of an HSV-1 gene during acute infection.
MATERIALS AND METHODS
Antibodies.
The following were used as primary antibodies: 1BA10, mouse anti-gE monoclonal antibody (MAb); ID3, mouse anti-gD MAb; M26, mouse anti-p24 (of human immunodeficiency virus) MAb used as a negative control (13). Nonimmune polyclonal mouse immunoglobulin G (Sigma, St. Louis, Mo.) was also used as a negative control in some trials. Secondary antibody was fluorescein isothiocyanate-labeled F(ab′)2 sheep anti-mouse immunoglobulin G (Sigma).
Cells and viruses.
HaCaT cells (human keratinocytes) were propagated in Dulbecco's modified Eagle medium supplemented with 10% sterile, heat-inactivated, fetal bovine serum, 15 mM HEPES buffer, 2 mM l-glutamine, 10 μg of gentamicin/ml, and 1 μg of Fungizone/ml at 37°C in a 5% CO2 incubator. The HSV-1 strain NS is a low-passage, clinical isolate that was used for all wild-type (WT) infections (14). The gE-deletion mutant (gEnull) was derived from strain NS HSV-1 and has been described previously (37).
siRNA generation.
A double-stranded DNA template containing a T7 RNA polymerase promoter and the target gene sequence were constructed, and sense and antisense RNA strands were made according to the manufacturer's instructions (Silencer siRNA construction kit; Ambion, Austin, Tex.) (31). The RNA strands were annealed overnight in a cabinet incubator at 37°C and then digested with both single-stranded RNase and DNase for 1 h at 37°C. The NS strain gE coding sequence targeted by the siRNA is identical to the published GenBank strain 17 HSV-1 sequence and was HSV-specific by GenBank BLAST analysis. The target sequences were 5′-AATATACGAATCGTGTCTGTA-3′ (sigE) of NS strain US8, corresponding to positions 142067 to 142087 of the GenBank HSV-1 complete genome, and 5′-AATCGGGCAGTTGTTTGAGAT-3′ (siCRK), corresponding to positions 1023 to 1043 of Leishmania CRK1, a protein kinase chosen as a negative control (26, 29, 33).
Plaque assays.
HaCaT cells were grown in 12-well plates to 80 to 90% confluence and then transfected with specific or control siRNA (10 to 75 nM) by Lipofectin complexing (24). After 24 h, the cells were infected with 25 PFU of WT or gEnull HSV-1 strain NS. Cells were overlaid with 2 ml of a 1:1 mixture of low-melting-temperature agarose (SeaPlaque agarose; Biowhittaker Molecular Applications, Rockland, Maine) and 2× Dulbecco's modified Eagle medium to allow only cell-to-cell spread of virus. At 48 to 72 h postinfection, plates were stained with neutral red for 6 h to measure plaque areas or with crystal violet for 10 min without the agarose overlay to photograph the monolayer. Plaque sizes were measured along perpendicular diameters by using an inverted light microscope (Leitz, Wetzlar, Germany) with an eyepiece micrometer at an objective of ×40. The area was calculated as πr2. Overlapping plaques and plaques at the edge of the well were counted as a single plaque for the purpose of deriving the number of plaques per well. However, only clearly demarcated plaques were included for the measurement of plaque areas. More confluent plaques occurred in the WT and control siRNA samples.
Flow cytometry.
HaCaT cells were grown in 12-well plates and processed as described above for in vitro plaque assays. However, instead of staining with neutral red, the overlay was removed and the cells were dissociated from the plate by washing with phosphate-buffered saline (PBS) without Ca2+ or Mg2+ (PBS−/−) (Invitrogen, Grand Island, N.Y.) followed by 500 μl of 1 mM EDTA-PBS−/−/well for 1 h at 37°C. The dissociated cells were washed in PBS−/−, spun at 500 × g for 5 min at room temperature (Beckman centrifuge, GH 3.8 rotor), and stained with 50 μl of primary antibody on ice for 1 h. The cells were then washed, and 50 μl of secondary antibody was added and incubated for 1 h on ice. After a final wash, the cells were fixed with 2% paraformaldehyde (Fisher Scientific, Fairlawn, N.J.) in PBS−/− and analyzed on a FACScan flow cytometer (Becton Dickinson, Mountain View, Calif.). For total cellular gE and gD measurements, the dissociated cells were prepared as described above, except that all washes and antibody staining (30 min on ice) were performed with 1% fetal bovine serum supplemented with 0.1% saponin (Sigma)-PBS−/−.
Northern blot analysis.
Northern blot analyses were performed with total RNA isolated from HaCaT cells with guanidinium thiocyanate (Master Blaster; Bio-Rad, Hercules, Calif.) 48 h after infection. To enhance the RNA yield, 70 μg of glycogen (Boehringer Mannheim, Indianapolis, Ind.) was added as carrier, and the precipitation was performed in siliconized tubes at −20°C overnight. The RNA pellet was reconstituted in 25 μl of nuclease-free water (Promega) and stored at −20°C. RNA samples (5 μl) containing ∼1 μg of RNA were denatured and then separated in a 1.4% denaturing agarose, 0.22 M formaldehyde gel that was submerged into morpholinepropanesulfonic acid (MOPS)-EDTA-sodium acetate buffer (Sigma) supplemented with formaldehyde (0.22 M). RNA was transferred to NYTRAN SuperCharge filters (Schleicher and Schuell, Keene, N.H.) and UV cross-linked. The filters were prehybridized at 68°C for 1 h in MiracleHyb (Stratagene, La Jolla, Calif.). To probe the Northern blots, 50 ng of DNA was labeled with Redivue [α-32P]dCTP (Amersham, Arlington Heights, Ill.) with a random prime-labeling kit (Boehringer Mannheim). The filters were hybridized at 68°C for 20 h with MiracleHyb containing the labeled and denatured probe. The filters were washed and exposed to Kodak film with an intensifier screen at −70°C for 2 to 72 h.
Probes.
Templates for gE and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probes were excised and gel purified, respectively, from plasmids pCMV3gE (9) and pHcGAP (obtained from the American Type Culture Collection).
Enzyme-linked immunosorbent assay for IFN-β.
The detection of human beta interferon (IFN-β) from HaCaT cells was done by using an IFN-β enzyme-linked immunosorbent assay kit (Fujirebio, Inc., Tokyo, Japan) according to the manufacturer's instructions.
Statistical analysis.
The measurements for the groups were evaluated by using the Wilcoxon rank sum test (Stati, version 7.0, software). The Wilcoxon rank sum (nonparametric) test was chosen for group median comparison because the plaque area measurements were not normally distributed.
RESULTS AND DISCUSSION
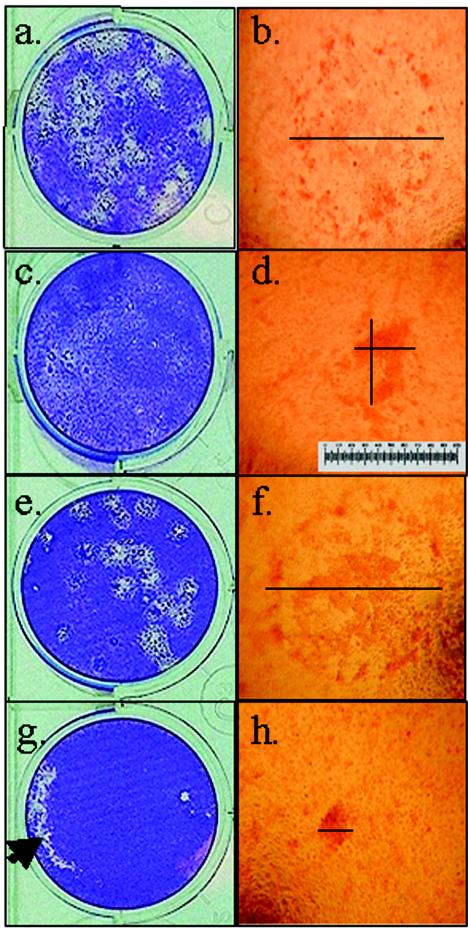
WT HSV-1 produces visible plaques on HaCaT cells at 48 h postinoculation that develop central clearing as the virus spreads outward (Fig. 1a and b). gEnull virus produces plaques that are characteristically smaller than the WT because of reduced cell-to-cell spread (Fig. 1c and d) (37, 45). HaCaT cells were transfected with gE-specific siRNA (sigE) or negative control siRNA (siCRK) and then infected with 25 PFU of WT HSV-1. Transfection with control siRNA (Fig. 1e and f) had no effect upon viral plaque size following infection. By contrast, WT HSV-1 infection of HaCaT cells that were transfected with sigE produced small plaques, similar to those of gEnull virus (Fig. 1g and h versus 1c and d, respectively).
FIG. 1.
gE-specific siRNA treatment of HaCaT cells demonstrates a reduced plaque size with WT HSV-1 infection, similar to that observed with gEnull virus. HaCaT cell monolayers were stained with crystal violet (a, c, e, and g) or neutral red (b, d, f, and h). Individual plaques are shown at a magnification of ×40. Transfection with siRNA was done 24 h prior to inoculation with 25 PFU of HSV-1. (a and b) WT HSV-1 alone; (c and d) gEnull HSV-1 phenotype control; (e and f) negative control siRNA (siCRK)-transfected, WT-infected cells; (g and h) sigE-transfected, WT-infected cells. The diameter (line) was measured twice for each plaque. Both measurements are shown for panel d to illustrate that the largest diameter was chosen for asymmetric plaques. A micrometer scale is inserted in panel d. The large unstained area of the cell monolayer in panel g represents an artifact from lifting the agarose overlay to apply the crystal violet (arrow). For plaque area measurements, the agarose overlay was not removed and neutral red was used to prevent disruption of the cell monolayer. This experiment was repeated five times with similar results.
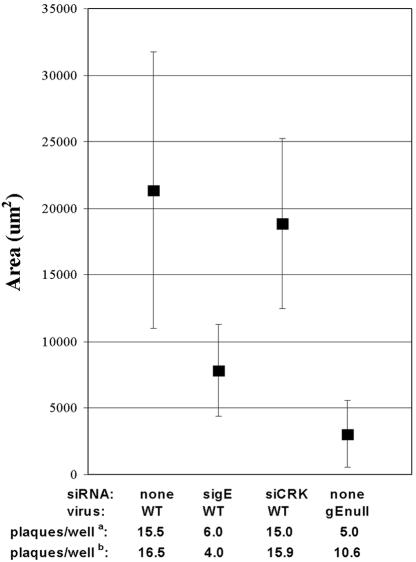
Figure 2 shows the median plaque areas from a single experiment, representative of the results from five repetitions. The median plaque area for the sigE group was significantly different from that of untransfected (P = 0.003) and control siRNA-transfected (P = 0.016) cells. By contrast, the control siRNA group did not differ from untreated WT infection (P = 0.41). Thus, transfection with exogenous siRNA, specific for gE, reproduced the phenotype of the gEnull virus in plaque assays. The small plaque phenotype, characteristic of gE deletion, was observed without sequence-independent suppression at both 70 and 10 nM siRNA concentrations.
FIG. 2.
Median plaque areas for a single experiment show that the gE-specific siRNA reproduces the small plaque phenotype of a gEnull HSV-1. The median plaque area (square) is shown ± SD (error bars) for WT HSV-1 alone, sigE-transfected and WT-infected cells, siCRK-transfected and WT-infected cells, and cells infected with the gEnull HSV-1. The numbers of plaques for this experiment (a) and for the average of the results from five experiments (b) are given. The inoculum of 25 PFU was determined by measuring the virus titer on Vero cells. These data are representative of the results from five separate experiments.
A single transfection of siRNA was administered 24 h prior to infection, and this proved sufficient. If infections were allowed to continue, the plaques in the non-gE siRNA-treated, WT-infected wells coalesced and an increased cytopathic effect became evident in the sigE-transfected wells but not in the gEnull-infected wells, suggesting that the effect of the siRNA diminished after 5 days (data not shown). This is consistent with the duration of induced RNAi in other dividing mammalian cells (6). RNAi is not propagated or perpetuated in mammals; therefore, each cell division dilutes the amount of siRNA per cell and, thus, the suppressive effect. This loss of effect may also explain why the median plaque area of the sigE group was slightly larger than that of the gEnull group.
The number of formed plaques was less than expected in all groups. A likely explanation of this general finding is that PFU were determined with Vero cells, whereas plaque area measurements were conducted with HaCaT cells. The reduction in expected plaque number was more notable for the sigE-transfected and gEnull virus-infected conditions. This is shown at the bottom of Fig. 2 as the average of the results from five experiments: 4 plaques/well versus 15.9 and 16.5 plaques/well for control siRNA and WT, respectively, despite an inoculum of 25 PFU per well. The gEnull virus-infected cells also showed fewer plaques than expected on HaCaT cells: 25 PFU gave 10.6 plaques/well on HaCaT cells. These observations likely represent that both gE deletion and knockdown produce smaller plaques or no plaques, which would be consistent with the gE-mediated reduction of viral plaque size that has been reported for other gE-deficient HSV-1 strains (2).
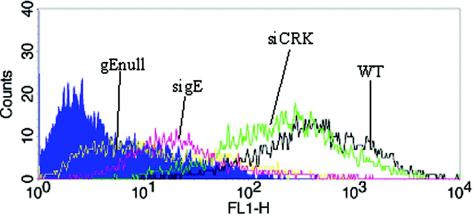
Plaque assay experiments demonstrated that sigE induced a small plaque size in WT-infected cells, which was similar to that of gEnull virus. The effect of sigE treatment upon WT HSV-1 protein expression was determined next by flow cytometry. The sigE-treated, WT-infected HaCaT cells demonstrated reduced cell surface expression of gE, resembling gEnull-infected cells (Fig. 3). Treatment with the nonspecific siRNA control did not significantly affect WT gE expression. This result shows that sigE specifically and efficiently lowers gE protein expression in HSV-1-infected cells.
FIG. 3.
Flow cytometry analysis demonstrates that sigE reduces cell surface gE protein expression. HaCaT cells were transfected with 70 nM siRNA and then infected with 25 PFU of WT or gEnull HSV-1 (where indicated). On day 2 postinfection, the cells were harvested for flow cytometry. Color key: blue, isotype control; yellow, gEnull virus; red, sigE; black, WT without siRNA; green, siCRK. This result is representative of the results from four repetitions.
A late time point was chosen for Fig. 3 to correspond with the plaque assays; however, to exclude effects of diminished viral spread upon protein expression, an earlier time point, during a single cycle of viral replication, was studied. The expression of gD, an unrelated HSV-1 protein important for viral entry, was measured at 17 h postinfection and compared with gE expression (40). It was necessary to increase the viral inoculum to quantify glycoprotein expression at this earlier time point. The HaCaT monolayer pretreated with the gE-specific siRNA (10 nM) and then infected with 25,000 PFU of HSV-1 showed a decreased level of total cellular gE (Fig. 4a) but no effect upon cell surface (Fig. 4b) and total cellular (Fig. 4c) gD protein expression.
FIG. 4.
gD expression is not affected by the gE-specific siRNA. HaCaT cells were transfected with 10 nM siRNA and then infected with 25,000 PFU of WT HSV-1. At 17 h postinfection, gD and gE expression on the cell surface or after permeabilization were measured by flow cytometry. (a) Total gE; (b) cell surface gD; (c) total gD. Color key: blue, isotype control; red, sigE; black, WT without siRNA; green (shown in panel b), siCRK. This result is representative of the results from three repetitions.
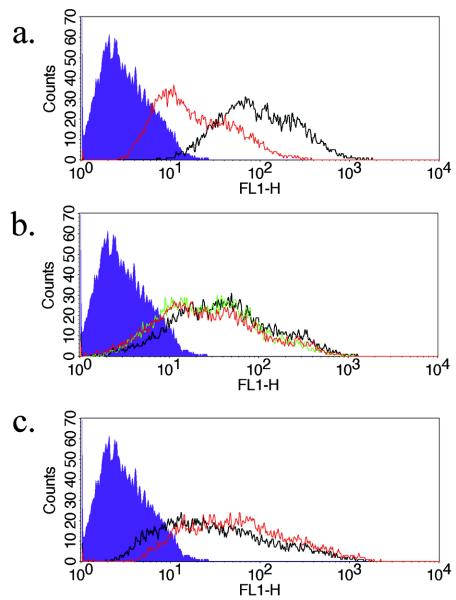
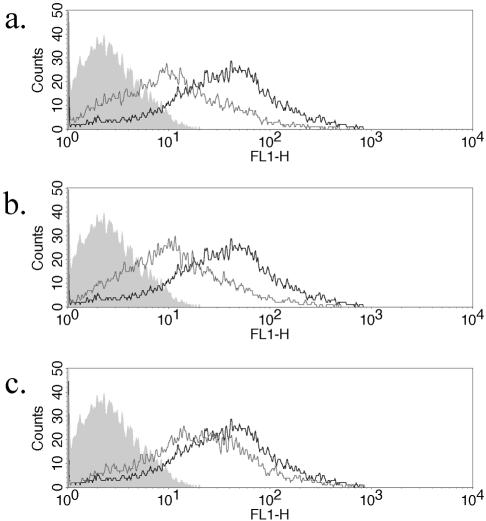
To further characterize the potency of the gE-specific siRNA, a titration of cell surface gE knockdown was done. The knockdown of cell surface gE was demonstrated at 70, 10, and 5 nM (Fig. 5a, b, and c, respectively). When compared with Fig. 3, some gE expression was noted after siRNA knockdown. This is likely due to the higher viral challenge used (25,000 PFU) and was more notable at the 5 nM concentration, representing the approach of the lower limit of effect. This shows that the siRNA effect is dependent upon both the concentration of siRNA used and upon the magnitude of viral challenge given. Therefore, validation of siRNAs should include a measurement of both parameters. The ability to titrate the concentrations of added siRNA remains a distinct advantage of exogenous delivery over intracellular, plasmid-generated siRNA in the initial steps of characterizing specificity.
FIG. 5.
Titration of gE-specific siRNA shows inhibition at 10 nM concentration. The HaCaT monolayer was transfected with 70 nM (a), 10 nM (b), and 5 nM (c) sigE and then infected with 25,000 PFU of WT HSV-1. gE cell surface expression was measured at 17 h postinfection. Color key: grey (filled), isotype control; grey (line), sigE; black, WT without siRNA.
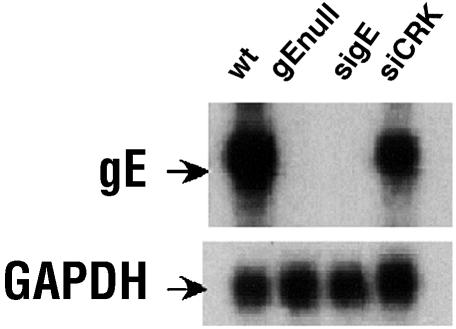
RNAi functions by identifying and degrading mRNA that shares sequence complementarity with the siRNA. Therefore, Northern blot analysis was used to analyze the effect of sigE on gE mRNA expression (Fig. 6). The level of gE mRNA in the sigE-treated, WT-infected cells was almost completely reduced (increased exposure time demonstrated a small gE band in the sigE-treated cells) compared with that of control siRNA-transfected or untransfected, WT-infected cells. GAPDH was used as a cellular mRNA control and showed no effect of the siRNA treatment.
FIG. 6.
mRNA expression from infected HaCaT cells demonstrates specific suppression of the gE transcript by sigE. siRNA-treated and untreated cells were harvested from 12-well plates 48 h after inoculation with 25 PFU of HSV-1. Total RNA was isolated, separated by polyacrylamide gel electrophoresis, and transferred to a nitrocellulose membrane for Northern blot analysis. WT, WT HSV-1 infection; gEnull, gE-deletion virus; sigE, gE-specific siRNA followed by WT infection; siCRK, negative control siRNA followed by WT infection. The membrane was probed for gE and GAPDH as indicated. This result is representative of the results from three trials.
RNAi was thought to be a gene suppression system that is independent of the IFN-α/β antiviral system. However, IFN-mediated sequence-independent gene suppression has been described that is induced by siRNA and mediated, in part, by Toll-like receptor 3 and double-stranded-RNA-dependent protein kinase (5, 23, 38). Therefore, siRNA appear to possess a mixture of gene-specific and non-gene-specific suppression activity, and it is important to characterize the relative contributions of each. Low concentrations of certain siRNAs can produce large levels of nonspecific effects, and high concentrations of others have little nonspecificity (P. Bhuyan and D. Weissman, unpublished data). Higher concentrations of siRNA exhibit more-measurable nonspecific gene suppression activity (23). Therefore, the ideal siRNA should have little sequence-independent properties at high concentrations and yet retain potency at low concentrations, which we demonstrate. The factors that predict nonspecific activity remain poorly defined but may be a combination of sequence-related and target-cell-dependent properties.
To determine the nonspecific contribution of the sigE, we also measured IFN-β production by HaCaT cells, induced by siRNA treatment. The amount of IFN-β was at the lower limit of detection (average ± standard deviation [SD], 0.173 ± 0.387 IU) 24 h after 70 nM sigE treatment, resembling untreated cells (0.007 ± 0.029 IU). As a reference point, the IFN-β response from HaCaT cells exposed to a siRNA with known, high-level, nonspecific gene suppression properties was 45.7 ± 10.9 IU (data not shown). Furthermore, the antiviral contribution of IFN is unlikely because HSV-1 has escape mechanisms mediated by viral genes such as virion host shutoff and γ134.5 (36, 41).
As an additional control for the nonspecificity of sigE, the effect of sigE upon gEnull infection was tested by plaque assay. Any impact upon plaque number or size should represent the proportion of gE-independent activity of sigE transfection. A comparison of sigE-transfected with untransfected conditions produced gEnull plaque numbers of 20 versus 36 (at 50 PFU) and median areas ± SDs of 208 ± 206 μm2 versus 307 ± 247 μm2, respectively, at 48 h. The sum of experimental evidence supports the notion that the contribution of the nonspecific effect of sigE transfection is minimal compared to the specific effect of sigE upon WT HSV-1.
Some viruses, particularly in plants, exhibit RNAi escape mechanisms. These mechanisms include virus-encoded RNAi suppressor proteins, some of which have been demonstrated to bind and sequester double-stranded RNA (25). HSV-1 contains many suppressors of host antiviral responses, including ICP47, virion host shutoff, γ134.5, gC, and gE (10, 15, 16, 20, 36). Our study did not demonstrate an effective escape mechanism from siRNA-mediated RNAi by HSV-1. If such an escape mechanism for RNAi existed, it would likely have been detected as we analyzed the effect of siRNA on infection phenotype and gE protein and mRNA expression. However, this study would not have detected an HSV mechanism to inhibit the generation of siRNA, such as an inhibitor of Dicer function, because we bypass the Dicer-mediated siRNA generation step by doing the transfection with premade siRNA. Furthermore, the virus could theoretically develop a mutation in the targeted gene sequence and thereby mediate escape, but mutational escape is infrequent in DNA viruses compared with RNA viruses. If HSV-specific siRNA were to be used therapeutically, either a multiple sequence cocktail approach or the targeting of a sequence that could not tolerate sequence changes could be used to minimize this potential problem.
RNAi will be an increasingly valuable tool for the study of viral pathogenesis. sigE closely approximates the gEnull phenotype as determined by plaque formation and the expression of mRNA and protein. One of the advantages of using RNAi instead of gene deletion is that siRNA-mediated gene knockdown allows the observation of partial phenotypes. This will be particularly important when studying genes that would produce a nonreplicating or lethal phenotype when completely deleted. Synergy has been reported for siRNA knockdowns, making it possible to improve the degree of gene suppression if necessary (6). Another powerful advantage of RNAi is that it allows the targeting of multiple genes simultaneously or at different time points during infection.
RNAi is also a novel antiviral mechanism that can be developed as a therapeutic modality. There are many precedents for RNAi being effective against human viral pathogens in vitro. The growing list includes human immunodeficiency virus, poliovirus, hepatitis C virus, human papillomavirus, dengue virus, hepatitis delta virus, and murine gammaherpesviruses (1, 6, 7, 17, 21, 22, 28, 34). HSV-1 is a human pathogen that infects up to 80% of individuals by adulthood worldwide and causes significant morbidity among immunocompromised hosts. It produces a wide spectrum of diseases and remains one of the major causes of infectious blindness. The severity of illness varies from asymptomatic to fatal disease and affects people of all age groups. The development of siRNA-mediated antiviral therapy may be of greatest benefit among populations infected with virus strains resistant to conventional antivirals and in cases of severe or recrudescent disease.
In vivo activity of RNAi has been demonstrated for both viral and host genes of mice (39). These models used hydrodynamic infusion in which a large volume of siRNA is rapidly injected though the tail vein. Intravenous injection (tail vein) with manual liver massage has also been used to successfully deliver naked plasmid DNA into mice (27). These data suggest that siRNA can be developed as a parenteral drug; however, better delivery strategies are needed. The in vitro system described will enable us to more easily pursue further studies of gE function and provides a foundation for the development of in vivo applications of siRNA.
Acknowledgments
This work was supported by grants T32 A107634 (NIH, NIAID postdoctoral training grant), R01 A133063 (NIH, NIAID), and EGPAF no. 27-PG-51257 (Pediatric AIDS Foundation).
We thank Vincent Lo Re for assistance with statistical analysis and Edward Bell for assistance with photography.
REFERENCES
- 1.Adelman, Z. N., I. Sanchez-Vargas, E. A. Travanty, J. O. Carlson, B. J. Beaty, C. D. Blair, and K. E. Olson. 2002. RNA silencing of dengue virus type 2 replication in transformed C6/36 mosquito cells transcribing an inverted-repeat RNA derived from the virus genome. J. Virol. 76:12925-12933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balan, P., N. Davis-Poynter, S. Bell, H. Atkinson, H. Browne, and T. Minson. 1994. An analysis of the in vitro and in vivo phenotypes of mutants of herpes simplex type 1 lacking glycoproteins gG, gE, gI, of the putative gJ. J. Gen. Virol. 75:1245-1258. [DOI] [PubMed] [Google Scholar]
- 3.Basu, S., G. Dubin, M. Basu, V. Nguyen, and H. M. Friedman. 1995. Characterization of regions of herpes simplex virus type 1 glycoprotein E involved in binding the Fc domain of monomeric IgG and in forming a complex with glycoprotein I. J. Immunol. 154:260-267. [PubMed] [Google Scholar]
- 4.Bernstein, E., A. A. Caudy, S. M. Hammond, and G. J. Hannon. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409:363-366. [DOI] [PubMed] [Google Scholar]
- 5.Bridge, A. J., S. Pebernard, A. Ducraux, A. L. Nicoulaz, and R. Iggo. 2003. Induction of an interferon response by RNAi vectors in mammalian cells. Nat. Genet. 34:263-264. [DOI] [PubMed] [Google Scholar]
- 6.Capodici, J., K. Kariko, and D. Weissman. 2002. Inhibition of HIV-1 infection by small interfering RNA-mediated RNA interference. J. Immunol. 169:5196-5201. [DOI] [PubMed] [Google Scholar]
- 7.Chang, J., and J. M. Taylor. 2003. Susceptibility of human hepatitis delta virus RNAs to small interfering RNA action. J. Virol. 77:9728-9731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dingwell, K. S., C. R. Brunetti, R. L. Hendricks, Q. Tang, M. Tang, A. J. Rainbow, and D. C. Johnson. 1994. Herpes simplex virus glycoproteins E and I facilitate cell-to-cell spread in vivo and across junctions of cultured cells. J. Virol. 68:834-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dubin, G., S. Basu, D. L. Mallory, M. Basu, R. Tal-Singer, and H. M. Friedman. 1994. Characterization of domains of herpes simplex virus type 1 glycoprotein E involved in Fc binding activity for immunoglobulin G aggregates. J. Virol. 68:2478-2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dubin, G., E. Socolof, I. Frank, and H. M. Friedman. 1991. Herpes simplex virus type 1 Fc receptor protects infected cells from antibody-dependent cellular cytotoxicity. J. Virol. 65:7046-7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elbashir, S. M., W. Lendeckel, and T. Tuschl. 2001. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 15:188-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fire, A., S. Xu, M. K. Montgomery, S. A. Kostas, S. E. Driver, and C. C. Mello. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391:806-811. [DOI] [PubMed] [Google Scholar]
- 13.Friedman, H. M., G. H. Cohen, R. J. Eisenberg, C. A. Seidel, and D. B. Cines. 1984. Glycoprotein C of herpes simplex virus 1 acts as a receptor for the C3b complement component on infected cells. Nature 309:633-635. [DOI] [PubMed] [Google Scholar]
- 14.Friedman, H. M., E. J. Macarak, R. R. MacGregor, J. Wolfe, and N. A. Kefalides. 1981. Virus infection of endothelial cells. J. Infect. Dis. 143:266-273. [DOI] [PubMed] [Google Scholar]
- 15.Fries, L. F., H. M. Friedman, G. H. Cohen, R. J. Eisenberg, C. H. Hammer, and M. M. Frank. 1986. Glycoprotein C of herpes simplex virus 1 is an inhibitor of the complement cascade. J. Immunol. 137:1636-1641. [PubMed] [Google Scholar]
- 16.Fruh, K., K. Ahn, H. Djaballah, P. Sempe, P. M. van Endert, R. Tampe, P. A. Peterson, and Y. Yang. 1995. A viral inhibitor of peptide transporters for antigen presentation. Nature 375:415-418. [DOI] [PubMed] [Google Scholar]
- 17.Gitlin, L., S. Karelsky, and R. Andino. 2002. Short interfering RNA confers intracellular antiviral immunity in human cells. Nature 418:430-434. [DOI] [PubMed] [Google Scholar]
- 18.Grosshans, H., and F. J. Slack. 2002. Micro-RNAs: small is plentiful. J. Cell Biol. 156:17-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hammond, S. M., E. Bernstein, D. Beach, and G. J. Hannon. 2000. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404:293-296. [DOI] [PubMed] [Google Scholar]
- 20.Hill, A., P. Jugovic, I. York, G. Russ, J. Bennink, J. Yewdell, H. Ploegh, and D. Johnson. 1995. Herpes simplex virus turns off the TAP to evade host immunity. Nature 375:411-415. [DOI] [PubMed] [Google Scholar]
- 21.Jia, Q., and R. Sun. 2003. Inhibition of gammaherpesvirus replication by RNA interference. J. Virol. 77:3301-3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang, M., and J. Milner. 2002. Selective silencing of viral gene expression in HPV-positive human cervical carcinoma cells treated with siRNA, a primer of RNA interference. Oncogene 21:6041-6048. [DOI] [PubMed] [Google Scholar]
- 23.Kariko, K., P. Bhuyan, J. Capodici, H. Ni, and D. Weissman. siRNA treatment of mammalian cell induces specific and non-specific suppression. J. Investig. Dermatol., in press.
- 24.Kariko, K., A. Kuo, E. S. Barnathan, and D. J. Langer. 1998. Phosphate-enhanced transfection of cationic lipid-complexed mRNA and plasmid DNA. Biochim. Biophys. Acta 1369:320-334. [DOI] [PubMed] [Google Scholar]
- 25.Lichner, Z., D. Silhavy, and J. Burgyan. 2003. Double-stranded RNA-binding proteins could suppress RNA interference-mediated antiviral defenses. J. Gen. Virol. 84:975-980. [DOI] [PubMed] [Google Scholar]
- 26.Lin, X., J. M. Lubinski, and H. M. Friedman. 2004. Immunization strategies to block the herpes simplex virus type 1 immunoglobulin G Fc receptor. J. Virol. 78:2562-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu, F., and L. Huang. 2002. Noninvasive gene delivery to the liver by mechanical massage. Hepatology 35:1315-1319. [DOI] [PubMed] [Google Scholar]
- 28.McCaffrey, A. P., L. Meuse, T. T. Pham, D. S. Conklin, G. J. Hannon, and M. A. Kay. 2002. RNA interference in adult mice. Nature 418:38-39. [DOI] [PubMed] [Google Scholar]
- 29.McGeoch, D. J., A. Dolan, S. Donald, and F. J. Rixon. 1985. Sequence determination and genetic content of the short unique region in the genome of herpes simplex virus type 1. J. Mol. Biol. 181:1-13. [DOI] [PubMed] [Google Scholar]
- 30.Mette, M. F., W. Aufsatz, J. van der Winden, M. A. Matzke, and A. J. Matzke. 2000. Transcriptional silencing and promoter methylation triggered by double-stranded RNA. EMBO J. 19:5194-5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Milligan, J. F., and O. C. Uhlenbeck. 1989. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 180:51-62. [DOI] [PubMed] [Google Scholar]
- 32.Morel, J. B., and H. Vaucheret. 2000. Post-transcriptional gene silencing mutants. Plant Mol. Biol. 43:275-284. [DOI] [PubMed] [Google Scholar]
- 33.Mottram, J. C., B. P. McCready, K. G. Brown, and K. M. Grant. 1996. Gene disruptions indicate an essential function for the LmmCRK1 cdc2-related kinase of Leishmania mexicana. Mol. Microbiol. 22:573-583. [DOI] [PubMed] [Google Scholar]
- 34.Novina, C. D., M. F. Murray, D. M. Dykxhoorn, P. J. Beresford, J. Riess, S. K. Lee, R. G. Collman, J. Lieberman, P. Shankar, and P. A. Sharp. 2002. siRNA-directed inhibition of HIV-1 infection. Nat. Med. 8:681-686. [DOI] [PubMed] [Google Scholar]
- 35.Nykanen, A., B. Haley, and P. D. Zamore. 2001. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell 107:309-321. [DOI] [PubMed] [Google Scholar]
- 36.Roizman, B., and A. Sears. 1996. Herpes simplex viruses and their replication, p. 2231-2278. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed., vol. 2. Lippincott-Raven, Philadelphia, Pa. [Google Scholar]
- 37.Saldanha, C. E., J. Lubinski, C. Martin, T. Nagashunmugam, L. Wang, H. van Der Keyl, R. Tal-Singer, and H. M. Friedman. 2000. Herpes simplex virus type 1 glycoprotein E domains involved in virus spread and disease. J. Virol. 74:6712-6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sledz, C. A., M. Holko, M. J. de Veer, R. H. Silverman, and B. R. Williams. 2003. Activation of the interferon system by short-interfering RNAs. Nat. Cell Biol. 5:834-839. [DOI] [PubMed] [Google Scholar]
- 39.Song, E., S. K. Lee, J. Wang, N. Ince, N. Ouyang, J. Min, J. Chen, P. Shankar, and J. Lieberman. 2003. RNA interference targeting Fas protects mice from fulminant hepatitis. Nat. Med. 9:347-351. [DOI] [PubMed] [Google Scholar]
- 40.Spear, P. G., and R. Longnecker. 2003. Herpesvirus entry: an update. J. Virol. 77:10179-10185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suzutani, T., M. Nagamine, T. Shibaki, M. Ogasawara, I. Yoshida, T. Daikoku, Y. Nishiyama, and M. Azuma. 2000. The role of the UL41 gene of herpes simplex virus type 1 in evasion of non-specific host defense mechanisms during primary infection. J. Gen. Virol. 81:1763-1771. [DOI] [PubMed] [Google Scholar]
- 42.Svoboda, P., P. Stein, H. Hayashi, and R. M. Schultz. 2000. Selective reduction of dormant maternal mRNAs in mouse oocytes by RNA interference. Development 127:4147-4156. [DOI] [PubMed] [Google Scholar]
- 43.Thomas, C. L., L. Jones, D. C. Baulcombe, and A. J. Maule. 2001. Size constraints for targeting post-transcriptional gene silencing and for RNA-directed methylation in Nictotiana benthamiana using a potato virus X vector. Plant J. 25:417-425. [DOI] [PubMed] [Google Scholar]
- 44.Vance, V., and H. Vaucheret. 2001. RNA silencing in plants—defense and counterdefense. Science 292:2277-2280. [DOI] [PubMed] [Google Scholar]
- 45.Weeks, B. S., P. Sundaresan, T. Nagashunmugam, E. Kang, and H. M. Friedman. 1997. The herpes simplex virus-1 glycoprotein E (gE) mediates IgG binding and cell-to-cell spread through distinct gE domains. Biochem. Biophys. Res. Commun. 235:31-35. [DOI] [PubMed] [Google Scholar]
- 46.Wianny, F., and M. Zernicka-Goetz. 2000. Specific interference with gene function by double-stranded RNA in early mouse development. Nat. Cell Biol. 2:70-75. [DOI] [PubMed] [Google Scholar]