Abstract
In positive-strand RNA viruses, 5′ untranslated regions (5′ UTRs) mediate many essential viral processes, including genome replication. Previously, we proposed that the 5′-terminal portion of the genomic leader sequence of Tomato bushy stunt virus (TBSV) forms an RNA structure containing a 3-helix junction, termed the T-shaped domain (TSD). In the present study, we have carried out structure-function analysis of the proposed TSD and have confirmed an important role for this domain in mediating efficient viral RNA amplification. Using a model TBSV defective interfering RNA replicon and a protoplast system, we demonstrated that various TSD subelements contribute to the efficiency of viral RNA replication. In particular, the stabilities of all three stems (S1, S2, and S4) forming the 3-helix junction are important, while stem-loop 3—a terminal extension of S2—is largely dispensable. Additionally, some of the sequences forming the 3-helix junction are required in an identity-dependent manner. Thus, both secondary structure and nucleotide identity are important for TSD-mediated viral RNA replication. Importantly, these results are fully consistent with the dual functions we defined previously for the sequences corresponding to loops 3 and 4, respectively, in facilitating 5′ cap- and 3′ poly(A) tail-independent translation of the genome by forming a loop-loop interaction with the 3′-proximal translational enhancer and in mediating viral RNA replication through formation of a pseudoknot with the adjacent downstream RNA domain. Also, since comparable TSDs and associated interactions are predicted in the 5′ UTRs of all sequenced Aureusvirus genomes, members of at least one other genus in the family Tombusviridae appear to utilize this type of multifunctional RNA domain.
Genome replication is an essential feature of virus reproduction. For positive-strand RNA viruses, efficient genome replication proceeds through a negative-strand RNA intermediate and is regulated by viral RNA elements, viral proteins, and host factors (2). The 5′ untranslated regions (5′ UTRs) in viral genomes contain primary, secondary, and tertiary RNA structures that are involved in genome replication. Accordingly, structural and functional assessments of these 5′ UTRs have led to important advances in our understanding of viral genome replication and related regulatory mechanisms (2).
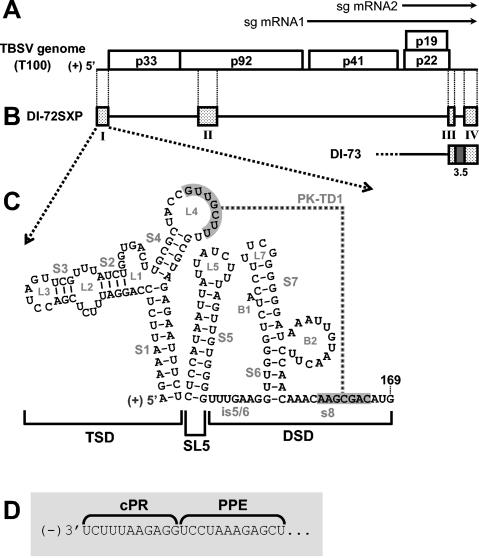
Tomato bushy stunt virus (TBSV), the prototype member of the genus Tombusvirus in the family Tombusviridae, is a monopartite positive-strand RNA virus (25). Its 4.8-kb genome encodes five viral proteins with known functions (10) (Fig. 1A). Viral RNA replication requires only p33 and its translational readthrough product p92 (the RNA-dependent RNA polymerase, RdRp) (17). These two proteins are also able to replicate defective interfering (DI) RNAs, which are small deleted forms of the genome that maintain cis-acting viral RNA replication elements (e.g., promoters and enhancers) (28). As such, tombusvirus DI RNAs have been very useful for defining RNA sequences and structures involved in the genome replication (3, 5, 6, 8, 9, 20, 21, 22). A prototypical TBSV DI RNA comprises four noncontiguous regions (RI through RIV) derived entirely from the viral genome (28) (Fig. 1B). Most relevant to this study is RI, which corresponds to the TBSV 5′ UTR and also includes the start codon for p33/p92 (Fig. 1B and C).
FIG. 1.
TBSV genome and relevant viral RNAs. (A) TBSV RNA genome. The genome is represented by a thick black line with coding regions depicted as boxes, and approximate molecular masses (in thousands) of the encoded proteins are shown, prefixed with “p.” (10). Two subgenomic mRNAs (sg mRNA1 and sg mRNA2) produced during infections are shown as arrows above the genome. (B) Prototypical TBSV DI RNAs DI-72SXP and DI-73. Shaded boxes represent TBSV genomic segments present in DI RNA, whereas black lines represent genomic segments that are absent. DI-73 is similar to DI-72SXP in that it contains RI through RIV (RI and RII are not shown). However, the former has an extra 3′-proximal sequence, region 3.5 (dark grey box), that is absent in the latter. (C) RNA secondary structure model for the TBSV 5′ UTR. The TBSV TSD, SL5, and the DSD are indicated by brackets below the structure (23, 32). Sequences forming a TSD-DSD pseudoknot (PK-TD1) are shaded in grey. RNA stem and loop structures are labeled, and TBSV genome coordinates are provided. is5/6, intervening sequence 5/6. (D) Sequences of the TBSV promoter (cPR) and enhancer (PPE) for positive-strand RNA synthesis located at the 3′ terminus of the DI RNA negative strand.
Previous studies have defined RI as an important sequence for efficient DI RNA replication in vivo (30). A surprising finding revealed by these analyses was that DI RNAs lacking all of RI are still capable of low-level replication (i.e., ∼10% that of wild-type [wt] DI RNAs), indicating the dispensability of the 5′ UTR (30). However, when RI is present, certain mutations introduced into RI abolish replication of DI RNAs (23, 32). Thus, while RI is dispensable for low levels of DI RNA replication, it can act in a cis-dominant-negative manner when present in a mutated form.
The secondary and tertiary structures of the TBSV 5′ UTR have been investigated previously within the context of a prototypical DI RNA (23, 32). The results support the existence of two major RNA domains, the T-shaped domain (TSD) and the downstream domain (DSD), both of which contribute to efficient DI RNA replication (23, 32) (Fig. 1C). These two RNA domains are separated by an intervening hairpin, termed stem-loop 5 (SL5), the formation of which is also necessary for robust DI RNA replication (23) (Fig. 1C). The TSD and DSD have been shown to communicate through the formation of a pseudoknot, PK-TD1, that also promotes efficient DI RNA replication (23). PK-TD1 is formed by base pairing between a sequence at the 3′ end of the DSD, designated s8, and the loop of the predicted SL4 in the TSD (Fig. 1C).
Stem 1 (S1), the largest predicted helix in the TSD, is the only domain subelement characterized in any detail thus far. Its formation in the positive strand has been shown to be critical for efficient DI RNA replication (32). The sequence complementary to the 5′ half of S1, i.e., the 3′-terminal nucleotides in the negative strand, also possesses activities important to viral RNA replication. This negative-strand sequence corresponds to the core promoter for positive-strand synthesis, termed the cPR, that was defined by in vitro studies using a partially purified tombusvirus RdRp preparation (16, 19) (Fig. 1C and D). Directly adjacent to the cPR in the negative strand is the promoter-proximal enhancer (PPE) (Fig. 1D). The PPE increases the efficiency of initiation of positive-strand synthesis from the 3′-terminal cPR and maps to the 5′ half of SL2 in the TSD (18) (Fig. 1C and D). The presence of important replication activities in corresponding sequences of opposite polarities necessitates that modifications in these regions be considered in both contexts.
The results of previous chemical and enzymatic solution probing studies are consistent with the structural model presented in Fig. 1C (32). However, with the exception of S1, the functional relevance of the other predicted subelements (i.e., SL2, SL3, and SL4)—as they relate to viral RNA replication in vivo—is unknown (32). In this study, we have used a TBSV DI RNA to analyze previously uncharacterized portions of the TSD and to define structural features that facilitate viral RNA replication in vivo. Our results are consistent with the RNA secondary structure model proposed previously (32) (Fig. 1C) and suggest a fundamental role for the TSD in viral RNA replication.
MATERIALS AND METHODS
Viral constructs.
Construction of a full-length TBSV genome plasmid, pT100 (10), and DI RNA pDI-72SXP (21) and DI-73 (29) has been described previously. Mutations were introduced into pDI-72SXP templates by using oligonucleotide-mediated PCR mutagenesis (Table 1). Mutations corresponding to mutants ΔSL2, ΔSL3, ΔSL2/SL3, S2-1a, S2-1b, ΔL1-1, ΔL1-2, S1-1a, S1-2a, S1-3a, sub5, and sub6 were generated using a common P9 primer and PR129, PR130, PR131, PR132, PR133, PR127, PR128, PR162, PR164, PR166, PRB1, and PRB2, respectively, from a pDI-72SXP template. PCR fragments were digested with SacI and XbaI, isolated, and ligated into a pDI-72SXP vector digested with SacI and XbaI. S2-1c was constructed by using PR132-P9 to generate a mutant PCR product from an S2-1b template, digesting with SacI and XbaI, and inserting the isolated fragment into a similarly digested pDI-72SXP vector. Many additional constructs were made using three-part ligations. For mutants ΔL4-1, SL4-1a, SL4-1b, SL4-1c, SL4-2a, SL4-2b, SL4-2c, SL4-3a, SL4-3b, SL4-3c, S4-1U1, and S4-1A1, a wt fragment was generated by PCR using phosphorylated oligonucleotides PR83 and PF7. Respective mutant PCR fragments were generated using phosphorylated oligonucleotides PR90, PR84, PR85, PR86, PR87, PR88, PR89, PR138, PR139, PR140, PR108,and PR109 and P9 from a pDI-72SXP template. For mutants L1-42, L1-46G, and L1-46A, a common wt PCR fragment was generated using phosphorylated PR170 and P9, whereas mutant PCR products were produced using PF7 and phosphorylated PR159, PR157, and PR158, respectively. For mutants ΔSL4, S1-1b, S1-2b, S1-3b, HN54, HN55, HN56, and HN57, mutant PCR products were generated using primers PF7 and phosphorylated PR65, PR163, PR165, PR167, PHN54, PHN55, PHN56, and PHN57, respectively, and common wt PCR products were made using phosphorylated PR171 and P9. Identical common wt PCR fragments were used for the construction of mutants S1-1c, S1-2c, and S1-3c; however, mutant PCR fragments were made using phosphorylated primers PR162-PR163, PR164-PR165, and PR166-PR167, respectively. All mutant and wt fragments were digested with SacI and XbaI, respectively, isolated, and ligated into a pDI-72SXP vector digested with identical enzymes. For mutants HN64, HN65, HN66, and HN67, PCR products were generated from HN54, HN55, HN56, and HN57 templates, respectively, using primers PF7 and PR152. These fragments were digested with SacI and XbaI, isolated, and ligated into a similarly treated pDI-72SXP vector. All PCR-derived regions in constructs were sequenced to ensure that only the intended modification was present.
TABLE 1.
Oligonucleotides used in the study
| Oligo- nucleotide | Positiona | REb site | Sequencec | Sensed |
|---|---|---|---|---|
| P9 | 4757-4776 | SphI/SmaI | GGCGGCCCGCATCCCGGGCTGCATTTCTGCAATGTTCC | − |
| P50 | 4754-4776 | GGAACATTGCAGAAATGCAGCCC | + | |
| P51 | 1420-1443 | BamHI | GCGCCGGATCCAGCGGTGCGAAACTCCGTACTTAC | + |
| PF6 | 4398-4417 | PstI | GCGCGCTGCAGAGCGAGTAAGACAGACTCTT | + |
| PF7 | 1-20 | SacI | GGCGGAGCTCTAATACGACTCACTATAGGAAATTCTCCAGGATTTCTC | + |
| PR21 | 4645-4668 | NsiI | CCGCGCATGCATATTCCTGTTTACGAAAGTTAGG | + |
| PR29 | 4568-4595 | CCGAAGGGTGAGATCAACCGTGTCTGGG | − | |
| PR35 | 4437-4467 | GGAGATGAGTGTAAATCTGGCATAGCATACAGGTTACTGCA | + | |
| PR37 | 4686-4708 | GACCCAGACACGGTTGATCTCAC | + | |
| PR38 | 4727-4739 | GATCGCTGGAAGCACTACCGGAC | + | |
| PR47 | 4730-4749 | GTCCGGTAGTGCTTCCAGCG | − | |
| PR63 | 1426-1445 | GCGAAACTCCGTACTTACAC | + | |
| PRB1 | 1-66 | SacI | GGCGGAGCTCTAATACGACTCACTATAGGAAATTCTCCAGGATTTCTCGACCTAGTTCGTTTATCTGGTGACTTACGCTACCGTTGCTTTGCG | + |
| PRB2 | 1-82 | SacI | GGCGGAGCTCTAATACGACTCACTATAGGAAATTCTCCAGGATTTCTCGACCTAGTTCGTTTATCTGGTGACTTGCGCTACCGTTGCTTTGCCTAGAGAATTTCTCTCC | + |
| PR65 | 21-78 | AGAAATTCTCAGTCACCAGATAAACGAACTAGGTC | − | |
| PR83 | 24-46 | AGTCACCAGATAAACGAACTAGG | − | |
| PR84 | 47-82 | TGCCGTACCGTTGCTTTGCGTAGAGAATTTCTCTCC | + | |
| PR85 | 47-82 | TGCGCTACCGTTGCTTTCGGTAGAGAATTTCTCTCC | + | |
| PR86 | 47-82 | TGCCGTACCGTTGCTTTCGGTAGAGAATTTCTCTCC | + | |
| PR87 | 47-82 | TGGCCTACCGTTGCTTTGCGTAGAGAATTTCTCTCC | + | |
| PR88 | 47-82 | TGCGCTACCGTTGCTTTGGCTAGAGAATTTCTCTCC | + | |
| PR89 | 47-82 | TGGCCTACCGTTGCTTTGGCTAGAGAATTTCTCTCC | + | |
| PR90 | 47-82 | TGCGCTTGCTTTGCGTAGAGAATTTCTCTCC | + | |
| PR108 | 47-94 | TGCGCTACCGTTGCTTTGTGTAGAGAATTTCTCTCCATAATTATTATC | + | |
| PR109 | 47-71 | TGCACTACCGTTGCTTTGCGTAGAG | + | |
| PR127 | 1-65 | SacI | GGCGGAGCTCTAATACGACTCACTATAGGAAATTCTCCAGGATTTCTCGACCTAGTTCGTTTATCTGGTTGCGCTACCGTTGCTTTGCG | + |
| PR128 | 1-65 | SacI | CGCGGAGCTCTAATACGACTCACTATAGGAAATTCTCCAGGATTTCTCGACCTAGTTCGTTTATCTGACTTGCGCTACCGTTGCTTTGCG | + |
| PR129 | 1-60 | SacI | GCCGGAGCTCTAATACGACTCACTATAGGAAATTCTCCACGACCTAGTTCGGGTGACTTGCGCTACCGTTGC | + |
| PR130 | 1-56 | SacI | GGCCGAGCTCTAATACGACTCACTATAGGAAATTCTCCAGGATTTCTTTTATCTGGTGACTTGCGCTACCG | + |
| PR131 | 1-60 | SacI | CGCGGAGCTCTAATACGACTCACTATAGGAAATTCTCCAGGTGACTTGCGCTACCGTTGC | + |
| PR132 | 1-41 | SacI | GCCGGAGCTCTAATACGACTCACTATAGGAAATTCTCCAGCTCTTCTCGACCTAGTTCGTTTATCTGG | + |
| PR133 | 1-56 | SacI | GGCGGAGCTCTAATACGACTCACTATATAGGAAATTCTCCAGGATTTCTCGACCTAGTTCGTTTGAGTGGTGACTTGCGCTACCG | + |
| PR138 | 47-66 | AGCGCTACCGTTGCTTTGCG | + | |
| PR139 | 47-82 | TGCGCTACCGTTGCTTTGCGTTGAGAATTTCTCTCC | + | |
| PR140 | 47-82 | AGCGCTACCGTTGCTTTGCGTTGAGAATTTCTCTCC | + | |
| PR152 | 130-168 | XbaI | CCGCGTCTAGACATGTGGAATGTTTGTTGGAAGTTACAATTTATCCCCCCG | − |
| PR157 | 24-46 | CGTCACCAGATAAACGAACTAGG | − | |
| PR158 | 24-46 | TGTCACCAGATAAACGAACTAGG | − | |
| PR159 | 24-46 | AGATGCCAGATAAACGAACTAGG | − | |
| PR162 | 1-32 | SacI | CGCGCGAGCTCTAATACGACTCACTATAGGAAATTCTGCAGGATTTCTCGACCTAGTTCG | + |
| PR163 | 47-78 | AGAAATTCTGTACGCAAAGCAACGGTAGCGC | − | |
| PR164 | 1-25 | SacI | CGCGCGAGCTCTAATACGACTCACTATAGGTTATTCTCCAGGATTTCTCGACC | + |
| PR165 | 47-71 | AGTTATTCTCTACGCAAAGCAACGG | − | |
| PR166 | 1-25 | SacI | CGCGCGAGCTCTAATACGACTCACTATAGGATTTTCTCCAGGATTTCTCGACCAGATTTTCTCTACGCAAAGCAACGG | + |
| PR167 | 47-71 | AGATTTTCTCTACGCAAAGCAACGG | − | |
| PR170 | 47-66 | TGCGCTACCGTTGCTTTGCG | + | |
| PR171 | 79-107 | CTCCATAATTATTATCTTTAGTTGTGGGG | + | |
| PHN54 | 31-78 | AGAAATTCTCGCCGCAAAGCAACGGTAGCGGCAGTCACCAGATAAACG | − | |
| PHN55 | 31-78 | AGAAATTCTCGACGCAAAGCAACGGTAGCGTCAGTCACCAGATAAACG | − | |
| PHN56 | 31-78 | AGAAATTCTCAACGCAAAGCAACGGTAGCGTCAGTCACCAGATAAACG | − | |
| PHN57 | 31-78 | AGAAATTCTCATCGCAAAGCAACGGTAGCGATAGTCACCAGATAAACG |
Coordinates correspond to those of the TBSV genome (10).
Restriction enzyme site.
Viral sequences corresponding to the coordinates shown are underlined. Restriction enzyme sites are italicized.
Refers to the sense of the oligonucleotide in reference to the positive-sense viral RNA.
Computer-aided analysis of viral RNA.
The nucleotide sequences for TBSV (NC_001554), TBSV statice isolate (AJ249740), TBSV pepper isolate (U80935), Artichoke mottled crinkle virus (X62493), Cucumber Bulgarian latent virus (AY163842), Cymbidium ringspot virus (X15511), Cucumber necrosis virus (M25270), Carnation Italian ringspot virus (X85215), Pear latent virus (AY100482), and Pothos latent virus (X87115) were obtained from the National Center for Biotechnology Information's GenBank genetic sequence database. The nucleotide sequence for Cucumber leaf spot virus has been published previously (15). RNA secondary structures were predicted at 37°C using mfold version 3.1 (13, 33).
In vitro transcription.
Viral transcripts were synthesized in vitro by transcription of SmaI-linearized DNAs with an Ampliscribe T7 RNA polymerase transcription kit (Epicenters Technologies) as described previously (17). Transcript concentrations were determined spectrophotometrically, and RNA integrity was verified using agarose gel electrophoresis.
Isolation and inoculation of protoplasts.
Protoplasts were prepared from 6- to 7-day-old cucumber cotyledons (var. Straight 8) as described previously (29). Isolated protoplasts were inoculated, using polyethylene glycol-CaCl2, as described previously (29) with uncapped viral RNA transcripts (1 μg for DI RNA and 3 μg for genomic T100 transcripts) and were incubated in a growth chamber under fluorescent lighting at 22°C (unless otherwise noted) for 22 to 24 h.
Analysis of viral RNA accumulation in vivo.
Total nucleic acids were harvested from protoplasts as described previously (29). One-fifth of the total nucleic acid was separated in denaturing 4.5% polyacrylamide gels containing 8 M urea. Gels were stained with ethidium bromide to ensure even loading of samples. Northern blot analysis of viral positive-strand accumulation was conducted by transferring total nucleic acid onto nylon, followed by hybridization with a 32P-end-labeled oligonucleotide probe (P9) complementary to the 3′-terminal 23 nucleotides of the TBSV genome. The intensity of the hybridization signal was quantified by radioanalytical scanning using an InstantImager (Packard Instrument Co.) and is represented graphically with standard error derived from results of three independent experiments unless otherwise noted. Analysis of viral negative strands was performed as described previously (22). Northern blot analysis was conducted using 32P-end-labeled oligonucleotide probes P50, PR21, PR37, PR38, PF4, PF6, PF7, P51, PR35, and PR63.
DI RNA stability assay.
Analysis of DI RNA in vivo stability was performed as described previously (21). Briefly, protoplasts were inoculated with 5 μg of DI RNA transcripts only (i.e., without T100) and subsequently treated with RNase A (final concentration, 10 μg/ml) to remove transcripts outside of the protoplasts. Total nucleic acids were extracted from protoplasts at 1, 4, 12, and 20 h postinoculation, and DI RNA levels were analyzed by Northern blotting using a 32P-end-labeled P9 probe.
RESULTS
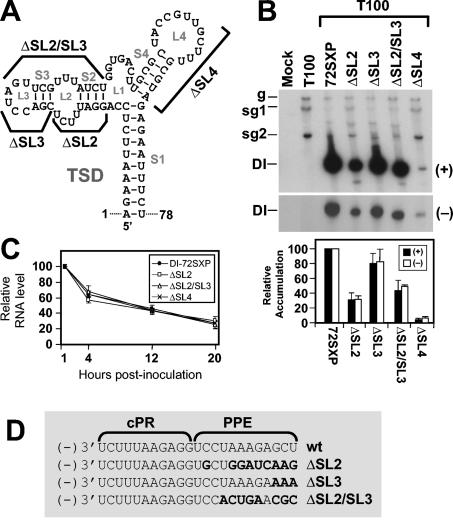
Deletion of SL2, SL3, or SL4 affects DI RNA replication.
As an initial step to investigate the roles of previously uncharacterized subelements of the TSD, SL2, SL3, SL2 and SL3, or SL4 was deleted from the prototypical DI RNA, DI-72SXP (Fig. 2A). These deletions also modified the PPE sequence in the negative strand, as indicated in Fig. 2D. In vitro-generated transcripts of the mutant DI RNAs, ΔSL2, ΔSL3, ΔSL2/SL3, and ΔSL4, were coinoculated individually with wt TBSV genome (T100) into plant protoplasts, and DI RNA accumulation levels were monitored 24 h postinoculation via Northern blot analysis. Deletion of SL4 had the most dramatic effect and virtually eliminated DI RNA accumulation (Fig. 2B). In contrast, removal of SL3 was least detrimental, allowing for accumulation of up to ∼80% that of wt DI RNA (Fig. 2B). Mutants lacking SL2 or both SL2 and SL3 showed intermediate levels of accumulation (∼30 to 40%) (Fig. 2B).
FIG. 2.
Analysis of DI RNA mutants containing deletions of various TSD subelements. (A) Depiction of TSD sequences deleted in the indicated DI RNA mutants. (B) Northern blot analysis of positive- and negative-strand DI RNA accumulation in protoplasts. Cucumber protoplasts were inoculated with transcripts of TBSV helper genome (T100) only or T100 and wt (72SXP) or mutant DI RNA transcripts as indicated above the lanes. Total nucleic acids were isolated 24 h postinoculation, separated in 8 M urea-4.5% polyacrylamide gels, transferred onto nylon membranes, and probed with 32P-end-labeled probes to detect either positive- or negative-sense DI RNA strands. Bands corresponding to TBSV genomic RNA (g), subgenomic mRNAs 1 and 2 (sg1 and sg2), and DI RNAs (DI) are indicated. For quantification of DI RNA accumulation, the level of wt DI-72SXP was set at 100% and mutant levels were normalized to this value. Relative DI RNA accumulation levels are presented as histograms. The values are means (with standard errors) of results from three separate experiments. (C) In vivo stability analysis of wt and mutant DI RNAs depicted in panel A. Protoplasts were inoculated with DI RNAs only, and total nucleic acids were extracted at 1, 4, 12, and 20 h postinoculation. DI RNA levels were determined by Northern blot analysis and quantified by radioanalytical scanning of membranes. The graph represents relative DI RNA levels as a function of time. The values are means of results from two separate experiments. (D) Effects of deletions on the sequences of the cPR and PPE. Sequence changes relative to the wt sequence are shown in bold.
To determine whether the modifications introduced were causing significant changes to the physical stabilities of the DI RNAs, in vivo RNA decay assays were performed (Fig. 2C). The profiles observed for the various mutant DI RNA transcripts showed very similar patterns, indicating that differences in physical stabilities were not contributing markedly to the varied levels of accumulation seen in replication assays. In addition, analysis of corresponding DI RNA negative-strand levels in replication assays revealed that their decreases were proportional to those observed for positive strands (Fig. 2B) and, thus, that accumulation levels of both strands were affected to similar extents.
Collectively, these data most strongly indicate increasing levels of importance for SL3, SL2, and SL4 in defects related to RNA replication. Although these results are consistent with TSD defects, the corresponding modifications in the PPE may also contribute to the defects observed. However, our results from additional analyses of the TSD and cPR-PPE elements provide persuasive evidence that the defects observed are indeed TSD related (see below and Discussion).
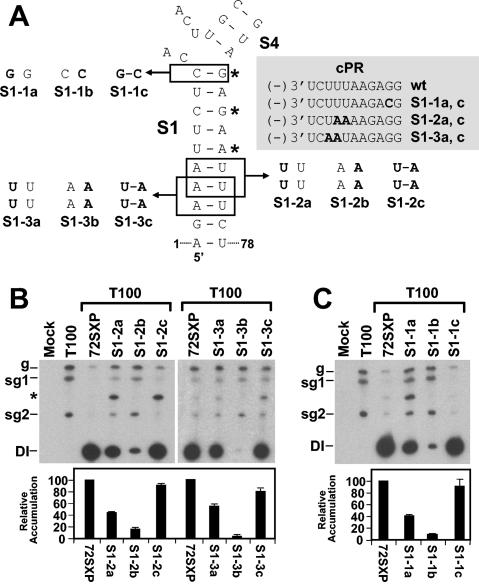
Base pairing of terminal regions of S1 is important for TSD activity.
Previous analysis of S1 established that its formation is essential for efficient DI RNA replication (32). In the previous study, three noncontiguous base pairs were targeted for compensatory mutational analysis and the mismatches introduced were predicted to effectively disrupt the entire S1 (Fig. 3A). In order to obtain more detailed information on the role of different regions of S1, we assessed the importance of base pairing at either end. The rational for this analysis was twofold. First, we proposed previously that the lower end of the S1 helix may coaxially stack on that of the adjacent helix of S5 and that this interaction may be important for TSD activity (Fig. 1C). If this proposal is correct, local disruption of the lower end of S1 would have a negative effect on the ability of S1 to coaxially stack on S5 and would lead to decreased DI RNA accumulation. Second, the upper portion of S1 forms part of the predicted 3-helix junction in the TSD. Therefore, the extent to which S1 pairs at its upper end may affect the physical and functional properties of this junction (Fig. 1C). For example, the formation of the upper C10G69 base pair may influence the possibility of S1 coaxially stacking with the predicted lower end of S4 (Fig. 1C).
FIG. 3.
Analysis of DI RNA mutants containing compensatory mutations in S1. (A) Depiction of S1 mutants where boxes show the residues modified and substitutions are shown in bold. The asterisks show the base pairs targeted for compensatory mutational analysis in a previous study (32). The grey box shows the effects of the substitutions on the sequences of the cPR and PPE. Sequence changes relative to the wt sequence are shown in bold. (B and C) Northern blot analysis of DI RNAs and quantification of their accumulation levels. An asterisk indicates the position of DI RNA head-to-tail dimers. sg1 and sg2, subgenomic mRNAs 1 and 2; g, genomic mRNA.
Two different sets of tandem substitutions (S1-2 and S1-3) that were designed to disrupt (mutants S1-2a, S1-3a, S1-2b, and S1-3b) and then restore (mutants S1-2c and S1-3c) base pairing were introduced into the lower region of S1 (Fig. 3A). The two 5′-terminal residues were not targeted, as they are known to be important for efficient positive-strand synthesis (16, 19). The a and c mutants of each series also contained modifications in their cPRs (Fig. 3A). For both sets of mutants, disruptive substitutions reduced DI RNA accumulation noticeably, whereas compensatory substitutions led to near-wt levels of recovery (Fig. 3B). Interestingly, the tandem UU mismatches were less debilitating than the tandem AA mismatches. This result is consistent with (i) reports that tandem UU mismatches can be quite stable (12) and (ii) the observation that a naturally occurring tandem UU mismatch exists in S1 of the TSD in cymbidium ringspot virus satellite RNA (32; O. A. Cheryesheva and K. A.White, unpublished data). In addition, the more biologically active tandem UU mismatch-containing mutants also had modifications in their cPRs. The higher level of biological activity observed for these mutants than for the tandem AA mismatch-containing mutants with no modification to their cPRs suggests that the identities of two substitutions in the cPR are less important than the base pairing of S1 in this region (Fig. 3B). The near-wt levels of recovery observed for the compensatory mutants S1-1c and S1-2c—also containing the cPR modifications—further indicate the greater importance of base pairing in the lower portion of S1.
Similar compensatory mutational analysis was carried out with the upper terminal base pair of S1 (Fig. 3A and C). A strong correlation between decreased base pair stability and reduced DI RNA levels was observed, with the CC mismatch in S1-1b being more detrimental than the GG mismatch in S1-1a (Fig. 3C). The compensatory mutant S1-1c showed near-wt recovery, confirming a central role for the uppermost base pair in S1. In addition, the high level of recovery observed for S1-1c, which also contained a G-to-C substitution in the cPR, indicates no major effect on the activity of this promoter (Fig. 3A and C).
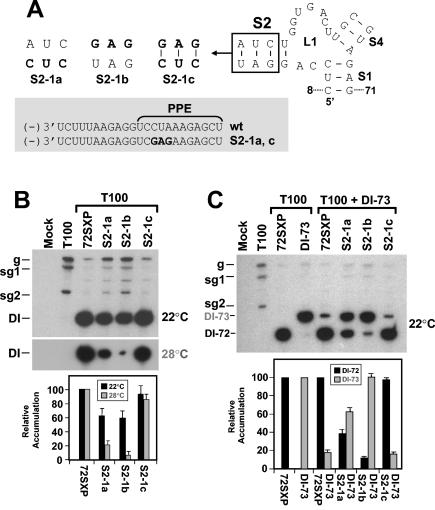
S2 is important for DI RNA fitness.
The deletion analyses described earlier in this work implicated SL2 as a determinant of DI RNA replication efficiency (Fig. 2). To determine whether formation of SL2 was biologically relevant, disruptive and compensatory mutations were introduced into this helix. The disruptive mutations in S2-1a and S2-1b reduced DI RNA accumulation to ∼60% of wt levels, and the restorative mutations in S2-1c yielded near-wt levels (Fig. 4A and B). These results at 22°C support our ideas about the formation and importance of S2; however, the decreases observed were much less pronounced than those seen for S1 disruption (compare Fig. 3B and C with Fig. 4B). In an attempt to verify that these more modest reductions were indeed meaningful, the incubation temperature for the S2 mutant infections was raised from 22 to 28°C. This increase was predicted to provide more thermodynamically stringent conditions for testing the functional relevance of RNA secondary structures. At the higher temperature, the importance of S2 formation was unmistakable (Fig. 4B).
FIG. 4.
Analysis of DI RNA mutants containing compensatory mutations in S2. (A) Depiction of S2 mutants where the box shows the residues modified and substitutions are shown in bold. The grey box shows the effects of the substitutions on the sequences of the cPR and PPE. Sequence changes relative to the wt sequence are shown in bold. (B and C) Northern blot analysis of DI RNAs and quantification of their accumulation in protoplasts at either 22 or 28°C (B) or under competitive conditions at 22°C (C). DI-73 was used as the competitor DI RNA in coinoculations. The positions of DI-72SXP, labeled as DI-72, and DI-73 are indicated. sg1 and sg2, subgenomic mRNAs 1 and 2; g, genomic mRNA.
In addition, clear differences in accumulation levels were also observed when the same mutants were tested under competitive conditions at 22°C (Fig. 4C). When both wt DI-72SXP and the larger DI-73 (see Fig. 1B) were coinoculated in equimolar amounts, the more competitive DI-72SXP dominated and suppressed accumulation of DI-73 (29) (Fig. 4C). In contrast, disruptions in S2 in DI-72SXP resulted in decreased competitiveness and restoration of S2 led to recovery of competitiveness (Fig. 4C). These results clearly illustrate that the maintenance of S2 contributes significantly to the fitness of DI RNAs under competitive conditions. These effects can be attributed to S2 activity, as corresponding modifications of the PPE in S2-1a and S2-1c did not appear to affect function markedly (Fig. 4).
S4 formation markedly stimulates TSD activity.
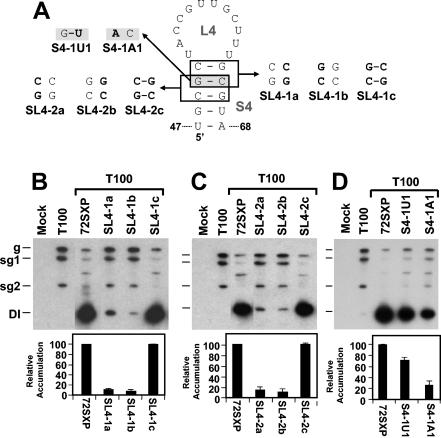
Previous studies of the 5′ UTR have shown that sequences in the TSD that map to the loop of the predicted SL4 interact with 3′-proximal residues in the DSD to form a functionally relevant pseudoknot, PK-TD1 (23) (Fig. 1C). If S4 does indeed form, it would facilitate presentation of the loop 4 (L4) residues that form PK-TD1. SL4s with stems ranging from 4 to 6 bp in length have been predicted previously for different tombusviruses by the program mfold (13, 32, 33). This comparative sequence analysis also revealed that the upper regions of the different S4s were variable (i.e., the base pairs covaried) but that in the lower regions the two terminal base pairs G48U67 and U47A68 were absolutely conserved (Fig. 1C). This analysis suggested that maintenance of S4 secondary structure is important but that the identities of the two base pairs at the bottom of this stem may also be of functional significance.
The importance of base pairing in the upper portion of S4 in TBSV was well supported by the analysis of two sets of compensatory mutations (i.e., the SL4-1 and SL4-2 series) (Fig. 5A, B, and C). These results indicate that efficient DI RNA accumulation requires S4 formation but does not require specific nucleotides within the upper portion of S4. The presence of the conserved G48U67 base pair suggests that S4 formation is relevant in the positive strand, since this base pair would correspond to a less stable AC mismatch in the negative strand. This concept is also consistent with the model that PK-TD1 forms in the positive strand (23) (Fig. 1C). To distinguish more definitively between possible positive- and negative-strand functions of S4, substitutions were introduced into the variable upper portion of S4 that would preferentially destabilize this helix in either the negative strand (mutant S4-1U1) or the positive strand (mutant SL4-1A1) (Fig. 5A). Accumulation levels for S4-1U1 were ∼70% of those for the wt, whereas a more dramatic decrease to ∼30% was observed for S4-1A1 (Fig. 5D). The greater sensitivity of S4 to reduced positive-strand stability is consistent with its function in the positive strand and its modeling as an important subelement of the TSD (Fig. 1C).
FIG. 5.
Analysis of DI RNA mutants containing compensatory mutations and strand-specific disruptions in the upper portion of S4. (A) Depiction of S4 mutants where boxes (or shading) show the residues modified and substitutions are shown in bold. (B, C, and D) Northern blot analysis of DI RNAs and quantification of their accumulation levels. sg1 and sg2, subgenomic mRNAs 1 and 2; g, genomic mRNA.
The conserved residues in S4 facilitate TSD activity.
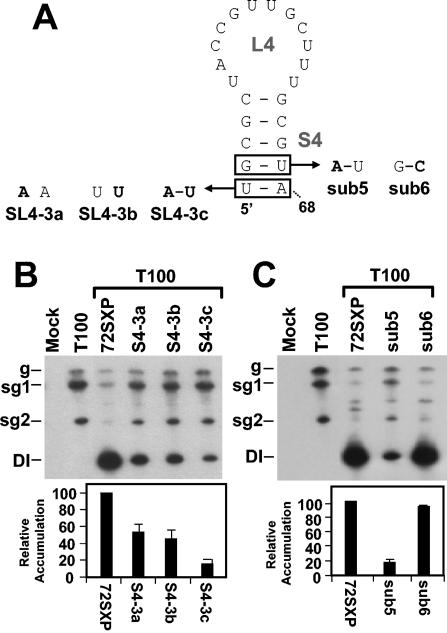
While the results described above and comparative sequence analysis support a primarily structural role for the upper portion of S4, the absolute conservation of the lower two base pairs in tombusviruses suggests that their identities may be important. The latter concept was supported by compensatory mutational analysis of the terminal U47A68 base pair (Fig. 6A and B). The results showed an approximately twofold decrease in accumulation for both mutants containing disruptions, SL4-3a and SL4-3b, and an even greater 10-fold reduction for the mutant SL4-3C, in which the base pair was restored (Fig. 6A and B). Thus, the identity of this terminal base pair is relevant to function.
FIG. 6.
Analysis of DI RNA mutants containing compensatory mutations in the lower portion of S4. (A) Depiction of S4 mutants where boxes show the residues modified and substitutions are shown in bold. (B and C) Northern blot analysis of DI RNAs and quantification of their accumulation levels. sg1 and sg2, subgenomic mRNAs 1 and 2; g, genomic mRNA.
The significance of identity in the adjacent base pair G48U67 was also investigated (Fig. 6A and C). The replacement of this base pair with a G48C67 base pair had no major effect, whereas conversion of this base pair into an A48U67 base pair markedly reduced DI RNA accumulation—implicating the guanylate as important. Considered along with the results for the adjacent A47U68 base pair, these findings suggest that S4 function relies to some degree on the identities of the lower two conserved base pairs. Thus, both maintenance of base pairing and nucleotide identity seem to be important for S4 activity.
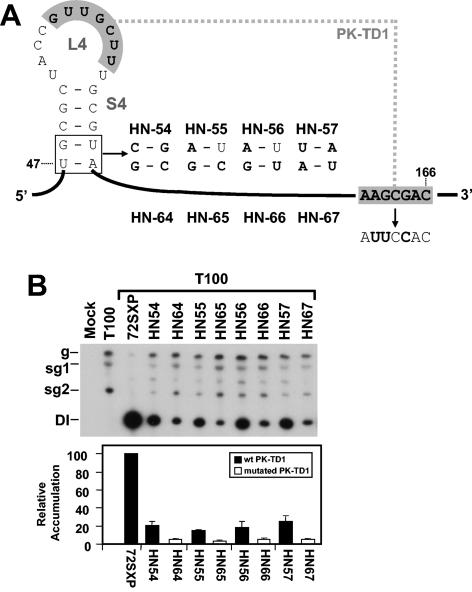
An alternative explanation for these results may be that the substitutions introduced into lower S4 led to misfolding of the SL4 region. To address this possibility, four additional mutants with substitutions in both conserved base pairs were tested (Fig. 7A). These modifications were predicted by the mfold program to maintain SL4 and TSD structure (data not shown). In the four mutants tested, HN54, HN55, HN56, and HN57, significant reductions in DI RNA levels were observed (Fig. 7B). These lower levels support the idea that nucleotide identity in lower S4 is important.
FIG. 7.
Analysis of DI RNA mutants containing substitutions in the lower portion of S4 with or without disruption of PK-TD1. (A) Depiction of S4 mutants where the box shows the residues modified in S4 and substitutions are shown in bold. Mutants HN54 through HN57 contain only the S4 mutations, whereas mutants HN64 through HN67 contain the S4 mutations and a PK-TD1-disrupting mutation (shown to the right), respectively. (B) Northern blot analysis of DI RNAs and quantification of their accumulation levels. sg1 and sg2, subgenomic mRNAs 1 and 2; g, genomic mRNA.
Although misfolding of these mutants is not predicted by mfold, we sought additional evidence to preclude it as the cause of the defects observed. We reasoned that, if the substitutions introduced did indeed cause misfolding of SL4, such structural changes would likely interfere with the presentation of L4 residues and the formation of PK-TD1. If this were the case, disrupting PK-TD1 in these mutants would not further reduce their accumulation. Conversely, if the SL4s were folding properly and promoting PK-TD1 formation, then disruption of PK-TD1 in these mutants should cause further reductions in their accumulation. The latter result would support the identity hypothesis and indicate a function independent of PK-TD1 formation.
To test this concept, PK-TD1-disrupting modifications were introduced into the four S4 mutants, thereby generating corresponding double mutants HN64, HN65, HN66, and HN67 (Fig. 7A). The substitutions added in the s8 region of the DSD (Fig. 1C and 7A) were shown previously to effectively disrupt PK-TD1 and cause a fivefold decrease in DI RNA accumulation levels (23). Thus, if PK-TD1 was still forming efficiently in HN54, HN55, HN56, and HN57, its disruption in HN64, HN65, HN66, and HN67 would be predicted to cause corresponding ∼5-fold decreases in the levels of accumulation in the latter mutants. When these double mutants were tested, they were found to have ∼5-fold lower levels of accumulation than their single mutant counterparts (Fig. 7B). These results indicate that these mutants were still PK-TD1 dependent. Accordingly, the defects caused by modifications of lower S4 were likely not due to misfolding of SL4 and instead are better explained by an identity requirement at these positions.
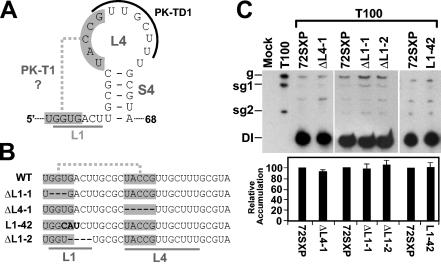
L1 sequences and a putative L1-L4 pseudoknot do not contribute notably to TSD activity.
Examination of the TSD secondary structure revealed the possibility of a tertiary base-pairing interaction between L4 and L1 resulting in a putative pseudoknot tentatively termed PK-T1 (Fig. 8A). Deletion of residues in the two segments within L1 and L4 that would form PK-T1 (mutants ΔL1-1 and ΔL4-1, respectively) did not notably alter DI RNA accumulation (Fig. 8B and C), suggesting no central role for this putative pseudoknot. Similarly, deletion of a more 3′-proximal segment of L1 did not affect DI RNA accumulation nor did replacement of a highly conserved UGA sequence in L1 that may potentially act as a UNR-type U-turn motif (mutants ΔL1-2 and L1-42, respectively) (Fig. 8B and C). Taken together, these data indicate no major role for the L1 sequences analyzed or for the 5′ portion of L4 in facilitating DI RNA accumulation.
FIG. 8.
Analysis of DI RNA mutants containing mutations in sequences corresponding to L1 and L4 of the TSD. (A) Secondary structure model for SL4 showing a potential second pseudoknot (PK-T1) involving sequences (shaded) in L1 and L4. L4 sequences that participate in the formation of PK-TD1 are also indicated (curved black line), as are L1 sequences (straight grey line). (B) Linear representation of L1 and SL4 (underlined) and the potential PK-T1 (shaded). Deletions are shown as hyphens, and substitutions are in bold. (C) Northern blot analysis of DI RNAs and quantification of their accumulation levels. sg1 and sg2, subgenomic mRNAs 1 and 2; g, genomic mRNA.
DISCUSSION
The present study has defined additional subelements within the TSD that are relevant to DI RNA replication. Of these, SL4 was identified as the most important, followed by S2 and lastly S3. Further analyses of the TSD revealed additional structural properties that facilitate its activity. Below, these findings are considered in relation to the TBSV reproductive cycle and in comparison with replication mechanisms utilized by other viral systems.
TSD versus cPR-PPE activities.
An interesting feature of the TBSV 5′ UTR is that it contains important RNA replication elements in both its positive and negative strands, namely, the TSD (32) and the cPR-PPE (18, 19), respectively. Several of the TSD deletion mutants analyzed also contained deletions in the PPE. For these mutants, the reduction in DI RNA positive and negative strands was found to be proportional. The absence of strand-preferential effects in our mutants suggests that the PPE activities were not affected markedly and that this element may be able to tolerate considerable modification without noticeable compromise of its function. This idea is supported further by in vitro analyses with partially purified tombusvirus RdRp that revealed that AU-rich sequences can functionally replace the PPE (18). All considered, the defects associated with our deletion mutants are most readily explained by modifications that affect TSD function.
The 5′ half of S1 is complementary to the cPR, and both of these elements have important activities in the positive and negative strands, respectively (19, 32). However, the structural requirements of each are quite distinct, with S1 functioning through secondary structure and the cPR operating as a linear element (19, 32). The lower six base pairs in S1 are absolutely conserved among tombusviruses, as are the corresponding six 3′-terminal nucleotides in the cPR (32). This conservation may be related to cPR function. However, our results from compensatory analysis of the lower region of S1 indicate no strict sequence requirement at positions 3 through 5 in the cPR, as changing its sequence from (−)3′UCUUUAAGAGG to (−)3′UCUAAAAGAGG (mutant S1-1c) or (−)3′UCAAUAAGAGG (mutant S1-2c; substitutions are in bold) did not significantly affect DI RNA accumulation. In contrast, this analysis showed clearly that substitutions disrupting S1 secondary structure were very detrimental. The sensitivity of S1 to helical disruption in its lower region may be related to the proposed coaxial stacking of the base of S1 with that of S5 (23). It was proposed previously that this interaction may act to further stabilize S1 and/or protect the uncapped 5′ terminus of the RNA from exonuclease attack (23). Overall, our results suggest that base pairing at these lower positions in S1 is more important than sequence identity in the cPR.
S1 functions optimally as a 10-bp-long helix.
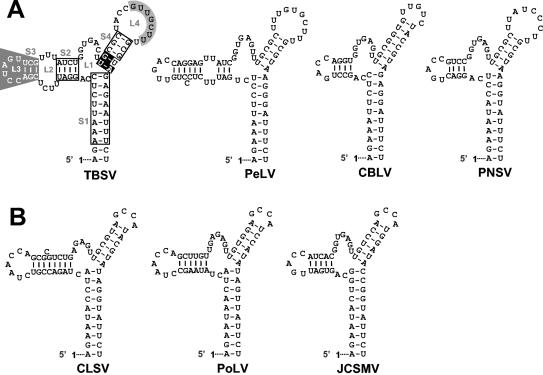
Previous studies have demonstrated the importance of S1 formation in the positive strand (32); however, the upper boundary of helical structure required for its activity was not investigated. Compensatory mutation analysis of the uppermost canonical base pair predicted in S1, C10G69, revealed a clear requirement for pairing. The necessity for this terminal base pair may be relevant for two reasons, which may not be mutually exclusive. First, as part of the 3-helix junction, it may contribute to functionally relevant organization. For example, it has been proposed previously that S1 may coaxially stack on S2 or S4 and the closing C10G69 base pair of S1 would be integral to either of these interactions (27, 32). Second, this terminal base pair defines S1 as a 10-bp-long helix. This additional property may be functionally relevant as all S1s analyzed to date have the potential to form 10-bp-long helices. This is also true for the S1s in tombusvirus satellite RNAs (32) and newly sequenced tombusviruses (Fig. 9A). In addition, members of Aureusvirus (15, 24)—the genus in the family Tombusviridae whose members are most closely related to tombusviruses—also possess potential TSDs with S1s that are 10-bp long (Fig. 9B). The relevance of conservation of an almost complete helical turn is unclear but may be related to a specific size requirement for positioning of other associated RNA elements and/or for recognition by other cis- or trans-acting factors.
FIG. 9.
RNA secondary structure comparison of the TBSV TSD with those from recently sequenced tombusviruses (A) and aureusviruses (B). In panel A, the nucleotides in open boxes indicate positions where compensatory mutational analyses from this and previous (32) studies have indicated that secondary structure is important. S4 nucleotides, in white on black, are entirely conserved within the genera Tombusvirus and Aureusvirus, and their identities are important for TSD activity. L4 nucleotides that interact with the adjacent DSD to form PK-TD1 are in black on grey, while SL3 nucleotides involved in a long-distance RNA-RNA interaction with the 3′-proximal translational enhancer are in white on grey (6, 23). For the aureusvirus TSDs shown in panel B, corresponding local pseudoknot and long-distance interactions have been predicted and presented elsewhere (6, 23). The virus abbreviations are as follows: PeLV, Pear latent virus; CBLV, Cucumber Bulgarian latent virus; PNSV, Pelargonium necrotic spot virus; CLSV, Cucumber leaf spot virus; PoLV, Pothos latent virus; and JCSMV, Johngrass chlorotic stripe mosaic virus.
Accessory role for SL2 and dispensability of SL3 and L1.
A major component of the 3-helix junction is L1. Despite its central location in the TSD and a significant degree of conservation among tombusviruses (32), the L1 sequence was dispensable for wt levels of DI RNA accumulation. This expendability rules out any major function for L1 related to the potential pseudoknot, PK-T1, or the 3-helix junction. This finding does not, however, preclude possible functions for it in other viral processes.
Our results indicate that SL3 is readily dispensable for productive viral RNA replication. However, previously, SL3 was shown to be essential for facilitating efficient cap- and poly(A) tail-independent translation of TBSV proteins (6) (Fig. 9A). In this capacity, L3 residues pair with those in the loop sequence of an RNA hairpin present in the 3′-proximal translational enhancer (6)—which is located in the 3′ UTR of the TBSV genome (31). This defined role in translation helps to explain SL3's expendability for efficient DI RNA replication. Furthermore, the dual roles of the TSD in facilitating both translation and RNA replication suggest that it may also participate in the coordinated regulation of these processes (6).
A central role for SL4 in TSD activity.
Our evidence for SL4 formation and activity in the positive strand validates the proper assignment of SL4 in the TSD secondary structure model and is consistent with the participation of SL4 in the formation of PK-TD1 (23, 32). The upper portion of S4 is sequence independent and likely facilitates presentation of L4 for PK-TD1 formation. In contrast, the lower portion of S4 shows a clear preference for particular residues. These conserved residues adjoin the 3-helix junction and may represent specificity determinants within this context. Alternatively, or additionally, the formation of these lower base pairs may also allow for coaxial stacking of S4 with S1—a concept supported indirectly by the importance of the uppermost base pair in S1. It is possible that such coaxial stacking is favored by the presences of certain base pairs at these positions in the S4 helix. In addition to these putative functions, the G48U67 and U47A68 base pairs would contribute to the overall stability of S4 and thus aid in the presentation of L4 for PK-TD1 formation. Taken together, these data establish S4 as a complex subelement of the TSD that contributes to both intra- and interdomain functions.
Role of TSD in viral RNA replication.
We have further defined important structural features of the TSD that contribute to its activity (Fig. 9A). This and other studies point to a function for the TSD in viral RNA replication (23, 32). Its contribution may be at any of several steps in the replication process, including template targeting to replication complexes, facilitating of negative-strand synthesis, and/or enhancing of positive-strand synthesis. Examples of 5′ UTR RNA elements mediating each of these types of activities have been reported for different positive-strand RNA viruses. For instance, targeting of Brome mosaic virus RNA2 to replication complexes requires a hairpin in its 5′ UTR (4). In Poliovirus (1, 11), Sindbis virus (7), and Alfalfa mosaic virus (26), 5′ UTR elements promote negative-strand synthesis, and for Potato virus X (14), an RNA structure in its 5′ UTR facilitates positive-strand synthesis. For TBSV, analyses of positive- and negative-strand DI RNA accumulation levels in this and other studies have not revealed any strand-specific functions for the TSD, SL5, or the DSD (23). Additionally, previous studies have shown that TBSV DI RNAs lacking the entire 5′ UTR (i.e., RI) can still replicate, albeit at 10% of wt levels (30). The latter finding indicates that TBSV RNAs can still be targeted to replication centers and copied into negative and then positive strands without the presence of the TSD, SL5, and the DSD. Although none of the RNA elements in the 5′ UTR are essential in cis for RNA replication, this region clearly facilitates one or more steps in the RNA replication process. Further studies will be required to elucidate the precise mechanism(s) by which these RNA elements confer their activities. It is anticipated that the mode(s) by which the TBSV TSD functions will also be applicable to all tombusviruses and other viral replicons that possess TSDs—namely tombusvirus satellite RNAs (32) and aureusvirus genomes.
Acknowledgments
We thank members of our laboratory for reviewing the manuscript. We also thank O. Nadel and M. Pritchert for constructing mutants sub5 and sub6 and O. Chernysheva for technical assistance.
This work was supported by NSERC and PREA.
REFERENCES
- 1.Barton, D. J., B. J. O'Donnell, and J. B. Flanegan. 2001. 5′ cloverleaf in poliovirus RNA is a cis-acting replication element required for negative-strand synthesis. EMBO J. 20:1439-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buck, K. W. 1996. Comparison of the replication of positive-stranded RNA viruses of plants and animals. Adv. Virus Res. 47:159-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang, Y. C., M. Borja, H. B. Scholthof, A. O. Jackson, and T. J. Morris. 1995. Host effects and sequences essential for accumulation of defective interfering RNAs of cucumber necrosis and tomato bushy stunt tombusviruses. Virology 210:41-53. [DOI] [PubMed] [Google Scholar]
- 4.Chen, J., A. Noueiry, and P. Ahlquist. 2001. Brome mosaic virus protein 1a recruits viral RNA2 to RNA replication through a 5′ proximal RNA2 signal. J. Virol. 75:3207-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fabian, M. R., H. Na, D. Ray, and K. A. White. 2003. 3′-terminal RNA secondary structures are important for accumulation of tomato bushy stunt virus DI RNAs. Virology 313:567-580. [DOI] [PubMed] [Google Scholar]
- 6.Fabian, M. R., and K. A. White. 5′-3′ RNA-RNA interaction facilitates cap- and poly(A) tail-independent translation of tomato bushy stunt virus mRNA: a potential common mechanism for Tombusviridae. J. Biol. Chem. 279:288862-288872. [DOI] [PubMed]
- 7.Frolov, I., R. Hardy, and C. M. Rice. 2001. Cis-acting RNA elements at the 5′ end of Sindbis virus genome RNA regulate minus- and plus-strand RNA synthesis. RNA 7:1638-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Havelda, Z., and J. Burgyan. 1995. 3′ terminal putative stem-loop structure required for the accumulation of cymbidium ringspot viral RNA. Virology 214:269-272. [DOI] [PubMed] [Google Scholar]
- 9.Havelda, Z., T. Dalmay, and J. Burgyan. 1995. Localization of cis-acting sequences essential for cymbidium ringspot tombusvirus defective interfering RNA replication. J. Gen. Virol. 76:2311-2316. [DOI] [PubMed] [Google Scholar]
- 10.Hearne, P. Q., D. A. Knorr, B. I. Hillman, and T. J. Morris. 1990. The complete genome structure and synthesis of infectious RNA from clones of tomato bushy stunt virus. Virology 177:141-151. [DOI] [PubMed] [Google Scholar]
- 11.Herold, J., and R. Andino. 2001. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol. Cell 7:581-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lietzke, S. E., C. L. Barnes, J. A. Berglund, and C. E. Kundrot. 1996. The structure of an RNA dodecamer shows how tandem U-U base pairs increase the range of stable RNA structures and the diversity of recognition sites. Structure 4:917-930. [DOI] [PubMed] [Google Scholar]
- 13.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters provides robust prediction of RNA secondary structure. J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 14.Miller, E. D., C. A. Plante, K. H. Kim, J. W. Brown, and C. Hemenway. 1998. Stem-loop structure in the 5′ region of potato virus X genome required for plus-strand RNA accumulation. J. Mol. Biol. 284:591-608. [DOI] [PubMed] [Google Scholar]
- 15.Miller, J. S., H. Damude, M. A. Robbins, R. D. Reade, and D. M. Rochon. 1997. Genome structure of cucumber leaf spot virus: sequence analysis suggests it belongs to a distinct species within the Tombusviridae. Virus Res. 52:51-60. [DOI] [PubMed] [Google Scholar]
- 16.Nagy, P. D., and J. Pogany. 2000. Partial purification and characterization of Cucumber necrosis virus and Tomato bushy stunt virus RNA-dependent RNA polymerases: similarities and differences in template usage between Tombusvirus and Carmovirus RNA-dependent RNA polymerases. Virology 276:279-288. [DOI] [PubMed] [Google Scholar]
- 17.Oster, S. K., B. Wu, and K. A. White. 1998. Uncoupled expression of p33 and p92 permits amplification of tomato bushy stunt virus RNAs. J. Virol. 72:5845-5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Panavas, T., Z. Panaviene, J. Pogany, and P. D. Nagy. 2003. Enhancement of RNA synthesis by promoter duplication in tombusviruses. Virology 310:118-129. [DOI] [PubMed] [Google Scholar]
- 19.Panavas, T., J. Pogany, and P. D. Nagy. 2002. Analysis of minimal promoter sequences for plus-strand synthesis by the Cucumber necrosis virus RNA-dependent RNA polymerase. Virology 296:263-274. [DOI] [PubMed] [Google Scholar]
- 20.Pogany, J., M. R. Fabian, K. A. White, and P. D. Nagy. 2003. A replication silencer element in a plus-strand RNA virus. EMBO J. 22:5602-5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ray, D., and K. A. White. 1999. Enhancer-like properties of an RNA element that modulates Tombusvirus RNA accumulation. Virology 256:162-171. [DOI] [PubMed] [Google Scholar]
- 22.Ray, D., and K. A. White. 2003. An internally located RNA hairpin enhances replication of Tomato bushy stunt virus RNAs. J. Virol. 77:245-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ray, D., B. Wu, and K. A. White. 2003. A second functional RNA domain in the 5′ UTR of the Tomato bushy stunt virus genome: intra- and interdomain interactions mediate viral RNA replication. RNA 9:1232-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rubino, L., and M. Russo. 1997. Molecular analysis of the pothos latent virus genome. J. Gen. Virol. 78:1219-1226. [DOI] [PubMed] [Google Scholar]
- 25.Russo, M., J. Burgyan, and G. P. Martelli. 1994. Molecular biology of tombusviridae. Adv. Virus Res. 44:381-428. [DOI] [PubMed] [Google Scholar]
- 26.Vlot, A. C., and J. F. Bol. 2003. The 5′ untranslated region of alfalfa mosaic virus RNA 1 is involved in negative-strand RNA synthesis. J. Virol. 77:11284-11289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walter, A. E., and D. H. Turner. 1994. Sequence dependence of stability for coaxial stacking of RNA helixes with Watson-Crick base paired interfaces. Biochemistry 33:12715-12719. [DOI] [PubMed] [Google Scholar]
- 28.White, K. A. 1996. Formation and evolution of tombusvirus defective interfering RNAs. Semin. Virol. 7:409-416. [Google Scholar]
- 29.White, K. A., and T. J. Morris. 1994. Nonhomologous RNA recombination in tombusviruses: generation and evolution of defective interfering RNAs by stepwise deletions. J. Virol. 68:14-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu, B., and K. A. White. 1998. Formation and amplification of a novel tombusvirus defective RNA which lacks the 5′ nontranslated region of the viral genome. J. Virol. 72:9897-9905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu, B., and White, K. A. 1999. A primary determinant of cap-independent translation is located in the 3′-proximal region of the tomato bushy stunt virus genome. J. Virol. 73:8982-8988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu, B., W. B. Vanti, and K. A. White. 2001. An RNA domain within the 5′ untranslated region of the tomato bushy stunt virus genome modulates viral RNA replication. J. Mol. Biol. 305:741-756. [DOI] [PubMed] [Google Scholar]
- 33.Zuker, M., D. H. Mathews, and D. H. Turner. 1999. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide, p. 11-43. In J. Barciszewski and B. F. C. Clark (ed.), RNA biochemistry and bio/technology. Kluwer Academic Publishers, Dordrecht, The Netherlands.