Abstract
Peripheral blood samples obtained from patients during an outbreak of Ebola virus (Sudan species) disease in Uganda in 2000 were used to phenotype peripheral blood mononuclear cells (PBMC), quantitate gene expression, measure antigenemia, and determine nitric oxide levels. It was determined that as the severity of disease increased in infected patients, there was a corresponding increase in antigenemia and leukopenia. Blood smears revealed thrombocytopenia, a left shift in neutrophils (in some cases degenerating), and atypical lymphocytes. Infected patients who died had reduced numbers of T cells, CD8+ T cells, and activated (HLA-DR+) CD8+ T cells, while the opposite was noted for patients who survived the disease. Expression levels of cytokines, Fas antigen, and Fas ligand (TaqMan quantitation) in PBMC from infected patients were not significantly different from those in uninfected patients (treated in the same isolation wards), nor was there a significant increase in expression compared to healthy volunteers (United States). This unresponsive state of PBMC from infected patients despite high levels of circulating antigen and virus replication suggests that some form of immunosuppression had developed. Ebola virus RNA levels (virus load) in PBMC specimens were found to be much higher in infected patients who died than patients who survived the disease. Similarly, blood levels of nitric oxide were much higher in fatal cases (increasing with disease severity), and extremely elevated levels (≥150 μM) would have negatively affected vascular tone and contributed to virus-induced shock.
Ebola viruses are enveloped, single-stranded RNA viruses that cause severe infections in human and nonhuman primates (28). This group of viruses contains some of the most pathogenic hemorrhagic fever viruses, and all are categorized as biosafety level 4 agents. These viruses are taxonomically classified in the family Filoviridae, and four species are currently recognized: Zaire (Ebola-Z), Sudan (Ebola-S), Reston (Ebola-R), and Ivory Coast (Ebola-IC) (International Committee on Taxonomy of Viruses, http://www.ncbi.nlm.nih.gov/ICTVdb/Ictv/ICTVindex.htm). Strains of the Ebola-Z and Ebola-S viruses have caused most (>99%) of the documented human cases and cause 60 to 90% and 50% mortality, respectively. These species of Ebola virus were first identified following concurrent outbreaks in 1976 (38, 39) in the northern region of the Democratic Republic of the Congo (formerly Zaire) (318 cases) and the extreme southern region of Sudan (284 cases). In 1979, 34 cases of Ebola-S were identified in the same location as the first outbreak, and in the autumn of 2000, Ebola-S virus caused the largest known outbreak of Ebola virus disease (425 presumptive cases) (9). This episode occurred in northern Uganda, primarily in and around the city of Gulu, which is near the area of Sudan where the two prior outbreaks took place.
The pathogenesis of Ebola virus infections in humans is largely uncharacterized because opportunities to study such cases are rare and it is usually quite difficult to safely collect and process specimens in the remote African areas where outbreaks have occurred. However, some human specimens have been obtained, and they, along with specimens that were derived from experimentally infected nonhuman primates, have revealed some aspects of the disease (1, 2, 3, 5, 13, 18, 19, 37). Ebola virus replication is particularly prominent in the liver, spleen, lungs, and vasculature system; hepatocytes, mononuclear phagocytic cells, and endothelial cells support virus replication (15, 16, 30, 31, 40). Monocytes and macrophages are thought to be infected early in the disease and to convey the virus to other areas of the body (30). Infections are also characterized by an immunosuppressed state, which is evidenced by a lack of inflammation in affected tissues and a delayed humoral immune response (3, 28).
The development of a strong cell-mediated immune response is key to surviving and clearing Ebola infections, while the humoral response appears to play a lesser role during the acute phase of the infection (3, 28, 34, 35). Immunologic studies of blood samples obtained during recent human outbreaks of Ebola-Z in the Democratic Republic of the Congo and Gabon (1, 37) determined that infection leads to an increase in serum cytokine levels including interleukin-2 (IL-2), IL-10, tumor necrosis factor alpha (TNF-α), alpha interferon (IFN-α), and IFN-γ, but mRNA levels indicated a decrease in T-cell functions (CD3, CD8, perforin, and IFN-γ), possibly due to apoptosis (3, 2). Infection of nonhuman primates with Ebola-Z or Ebola-R virus induces the production of proinflammatory cytokines and the accumulation of nitric oxide (NO) in the blood (18).
To further characterize the effects of Ebola virus infections in humans, we analyzed blood specimens taken during an outbreak in northern Uganda in the autumn of 2000. Samples were collected and processed by a team from the Centers for Disease Control and Prevention (CDC) participating in an international response to the outbreak. A testing laboratory was set up at facilities in the Saint Mary's Lacor Hospital, where the first cases were seen and many of the patients were hospitalized. Blood samples were obtained from patients with fatal and nonfatal disease at this site and also from a government hospital in the city of Gulu. Peripheral blood mononuclear cell (PBMC) specimens were isolated from patients with fatal and nonfatal Ebola-S hemorrhagic fever as well as hospitalized patients who were later determined to be uninfected. These specimens were used to determine the status of immunocompetent cells and levels of virus replication over the course of the disease. In addition, plasma samples from the same patients were tested for levels of virus antigen and NO, and a limited set of blood smears were also examined. The results of these studies are the subject of this paper.
MATERIALS AND METHODS
Cells and viruses.
Peripheral blood specimens were collected from Ebola-S virus-infected and uninfected patients (ill and treated in the same isolation wards but tested negative for virus antigen and antibody). Blood was collected primarily in heparinized Vacutainer tubes, but a small number of EDTA tubes were also used. Blood samples from healthy volunteers (members of the Special Pathogens Branch, CDC) were obtained after the outbreak and used as controls in the studies. Blood smears from a handful of specimens from infected and uninfected patients were prepared in the field and processed for leukocyte differential staining (Wright's stain) at the CDC (Atlanta, Ga.). PBMC were isolated by Ficoll-Hypaque gradient separation, with cells pelleted from whole blood and resuspended in phosphate-buffered saline. Cells were maintained and shipped to the CDC in a frozen state (−80°C freezer and dry ice) and stored in a nitrogen vapor freezer. Virus stock cultures of the Gulu strain of Ebola-S virus, isolated from a Ugandan specimen sent to the CDC early in the outbreak, was prepared from cultures of infected Vero E6 cells (ATCC CRL-1586) or human embryonic kidney 293 cells (ATCC CRL-1573). All work with infectious materials was performed under biosafety level 4 containment.
PBMC phenotyping.
PBMC specimens were retrieved from LN2 storage and thawed at room temperature, and then 250 μl was stained separately by directly pipetting the cells into 1.5-ml polypropylene Eppendorf tubes containing either 10 μl of MultiTEST CD3-FITC/CD16+CD56-PE/CD45-PerCP/CD19-APC (Becton Dickinson, catalog no. 340500) or 10 μl of MultiTEST CD8-fluoresceinisothiocyanate/CD38-phycoerythrin/CD3-peridinin chlorophyll protein/anti-HLA-DR-allophycocyanin (Becton Dickinson, catalog no. 340572) reagents. Cells were incubated for 30 min at room temperature and then 1 ml of 1× fluorescence-activated cell sorting lysing solution (Becton Dickinson, catalog no. 349202) was added, and the mixture was incubated for 10 min. The cell suspensions were then transferred to polystyrene snap-cap tubes (12 mm by 100 mm) containing 100 μl of 10% formalin (Ted Pella, Inc., catalog no. 18505). After an overnight incubation, the cells were assayed for specific staining with a FACSCalibur four-color flow cytometer (BD Biosciences). Counting was based on side scatter/CD45+ or side scatter/CD3+ gating depending on the MultiTEST kit used; for each assay, a minimum of 15,000 total cells were counted.
Antigen detection.
Patients in the acute phase of disease were identified by use of an antigen capture enzyme-linked immunosorbent assay to detect Ebola-S virus antigen in blood samples as previously described (22). Testing of 1:4, 1:16, 1:64, and 1:256 dilutions of patient plasma was performed in 96-well plates, with antigen levels calculated by adding the optical density readings for all four wells (sum optical density). All assays were performed at St. Mary's Lacor Hospital on fresh specimens.
Quantitative PCR.
PBMC specimens were thawed and centrifuged in a microcentrifuge for 2 min at 6,000 rpm at room temperature. Total RNA was extracted from pelleted cells by use of a QIAGEN RNeasy Mini kit and then frozen in an LN2 freezer. Conversion of RNA to cDNA was performed with a commercial kit, the SuperScript First-Strand Synthesis System for reverse transcription-PCR (Invitrogen, Life Technologies). All first-strand cDNA reactions were performed in 0.2-ml thin-walled PCR tubes with a Perkin-Elmer 9600 thermocycler.
A quantitative PCR assay system (TaqMan) was used to determine relative levels of gene expression (transcription). The following sequences were targeted for amplification and detection with the primers shown in Table 1: glyceraldehyde-3-phosphate dehydrogenase (GAPDH), TNF-α, IFN-γ, IL-6, IL-8, IL-10, MCP-1, Fas, Fas ligand (FasL), and the glycoprotein gene (GP) of the Gulu strain of Ebola-S virus. Primer sequences for detecting human mRNAs were derived from GenBank submissions (see Table 1). Primers for detection of the Gulu strain of Ebola-S virus were derived from the GP gene sequence, which was determined by direct sequencing of reverse transcription-PCR amplicons generated from virion RNA preparations. Selection of primer sequences suitable for quantitative PCR assays was aided by the use of the Primer Express software package (Applied Biosystems, version 1.5).
TABLE 1.
Synthetic deoxyoligonucleotide primers used in quantitative PCR assaysa
| Target sequence (GenBank no.) | Primers (forward, reverse, and probe) |
|---|---|
| Ebola-S virus GPb (AY316199) | 5′ GGCACTAACGGCAACCATATG |
| 5′ ACTCGGGATTTGGCTGGAG | |
| 5′ (FAM)-AGATCTCCACCATCGGGATAAGACCGA-(QSY7) | |
| TNF-α (Z15026) | 5′ GCCCAGGCAGTCAGATCATCT |
| 5′ TTGAGGGTTTGCTACAACATGG | |
| 5′ (FAM)-TCG AAC CCC GAG TGA CAA GCC TGT-(QSY7) | |
| IFN-γ (XM_006883) | 5′ CAAGGAAGACATGAATGTCAAGTT |
| 5′ ATTCAAGTCAGTTACCGAATAATTAG | |
| 5′ (FAM)-ATAGCAACAAAAAGAAACGAGATGACTTCGAAAAGC-(QSY7) | |
| IL-6 (Y00081) | 5′ GCAACACCAGGAGCAGCC |
| 5′ AACTCCTTCTCCACAAGCGC | |
| 5′ (FAM)-CAGGGAGAAGGCAACTGGACCGAA-(QSY7) | |
| IL-8 (M28130) | 5′ AGCTCTGTCTGGACCCCAAG |
| 5′ GAATTCTCAGCCCTCTTCAAAAAC | |
| 5′ (FAM)-AAAACTGGGTGCAGAGGGTTGTGGAGA-(QSY7) | |
| IL-10 (U16720) | 5′ CGGCGCTGTCATCGATTT |
| 5′ GGCATTCTTCACCTGCTCCA | |
| 5′ (FAM)-TTCCCTGTGAAAACAAGAGCAAGGCCG-(QSY7) | |
| MCP-1 (Y18933) | 5′ GCAGAGGCTCGCGAGCT |
| 5′ ACAATGGTCTTGAAGATCACAGC | |
| 5′ (FAM)-TAGAAGAATCACCAGCAGCAAGTGTCCCA-(QSY7) | |
| Fas (X63717) | 5′ TCCACTAATTGTTTGGGTGAAGAG |
| 5′ GATTCATGAGAACCTTGGTTTTCC | |
| 5′ (FAM)-AAGGAAGTACAGAAAACATGCAGAAAGCACAGAA-(QSY7) | |
| FasL (Z96050) | 5′ TGGCCCATTTAACAGGCAA |
| 5′ AATTCCATAGGTGTCTTCCCATTC | |
| 5′ (FAM)-TCCAACTCAAGGTCCATGCCTCTGG-(QSY7) | |
| GAPDH (J04038) | 5′ CTCAAGATCATCAGCAATGCCT |
| 5′ AAGTTGTCATGGATGACCTTGG | |
| 5′ (FAM)-CTGCACCACCAACTGCTTAGCACCC-(QSY7) |
All probes were synthesized with a 5′ FAM (6-carboxyfluorescein) and a 3′ dark quencher (QSY7).
Gulu (2000) strain of Ebola-S virus; primer sequences are specific for this strain.
All reactions were performed in a final volume of 25 μl containing a onefold final concentration of Platinum Quantitative PCR SuperMix-UDG (Invitrogen, Life Technologies) and 5 μl of cDNA (template). Forward and reverse primers were used at a concentration of 200 nM, and the primer probe concentration was 100 nM. Reactions were prepared in 96-well optical reaction plates, and quantitation was performed with an ABI Prism 7700 sequence detector (Applied Biosystems). Wells were heated at 50°C for 2 min and then at 95°C for 2 Min, and target sequences were amplified by 40 rounds of denaturing at 95°C for 15 s and extension at 60°C for 1 min. Maximum ramping between temperatures was used in thermocycling. A comparative CT method was used to quantitate relative levels of expression; assays were validated for linearity across a range of target sequence concentrations. GAPDH mRNA served as the endogenous reference molecule in calculating relative expression, and values were derived from the averages of duplicate reactions, with the calibrator 2−ΔΔCT, where ΔΔCT is equal to the difference in threshold cycles for the target and reference molecules in assays having approximately equal efficiencies (ABI Prism 7700 User Bulletin #2).
NO assays.
NO levels in patient blood specimens were quantitated with a colorimetric assay in which NO is converted to nitrite and then reacted with Griess reagent. Assays were performed in flat-bottomed, 96-well microtiter plates (Immulon 2HB; Thermo Labsystems). To each well was added 80 μl of assay buffer (50 mM morpholinepropanesulfonic acid, 1 mM EDTA, pH 7.0), 5 μl of sample (in triplicate wells), and 10 μl of 2 mM NADPH (Sigma-Aldrich, catalog no. N1630); 5 μl of nitrate reductase (Sigma-Aldrich, catalog no. N7265) in assay buffer (2 U/ml) was added to two of the three sample wells, while the third well received 5 μl of assay buffer (background well). Reactions were incubated at room temperature for 20 min with gentle agitation, and then 100 μl of Griess reagent was added to the duplicate wells containing enzyme and 100 μl of 1.5 N HCl was added to the third well (minus enzyme). Griess reagent was prepared by mixing equal volumes of freshly prepared 0.1% 1-naphthylethylenediamine dihydrochloride (Sigma-Aldrich, catalog no. N5889) in deionized water and 1% sulfanilamide (Sigma-Aldrich, catalog no. S9251) in 3 N HCl. Optical density readings for wells were obtained with a Tecan SpectraFluor (540-nm measurement filter, 620-nm reference filter, three flashes). Optical density readings for duplicate wells were averaged, and the value of the background well was subtracted to eliminate absorbance due to variations in plasma or serum turbidity and normalized to optical density values obtained from deionized water. NO levels were calculated from a standard curve obtained from potassium nitrate solutions ranging in concentration from 10 to 1000 μM (in deionized water).
RESULTS
White blood cells.
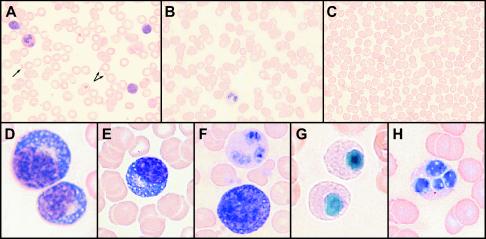
In the course of processing blood specimens for PBMC, it was noted that leukopenia developed as the severity of the disease increased, especially in fatal cases. This was largely determined from observing the intensity of PBMC bands from sequential samples, which decreased as the disease progressed (nearly disappearing during the most acute phase of disease); leukopenia was also observed in blood smears (Fig. 1). In nonfatal cases, the intensity of the PBMC band returned to normal (or increased) as patients recovered. Differential staining of blood smears from patients during the acute phase of disease showed thrombocytopenia, a left shift in neutrophils (sometimes with toxic granulation), and atypical lymphocytes (Fig. 1). These atypical lymphocytes appeared to be lymphoblasts and plasmacytoid lymphocytes, and plasma cells were observed in a few cases. Degenerating neutrophils were observed in some cases; these neutrophils resembled those found in patients exhibiting Pelger-Huët syndrome (pseudo-Pelger cells; nuclei condensed, pyknotic, and hyposegmented).
FIG. 1.
Differential staining of peripheral blood smears (Wright's stain). Shown on the top row of images are low-magnification pictures of blood cells from an uninfected patient (A), and Ebola-S virus-infected patients with nonfatal (B) and fatal (C) disease outcomes (samples taken during the acute phase). The arrows in A point to platelets (dramatically reduced or absent in B and C). The bottom row shows higher-magnification images of differentiating and abnormal cells seen in acute-phase blood smears from Ebola-S patients: (D) plasmacytoid lymphocytes, (E) plasma cell, (F) normal neutrophil (top) and lymphoblast (bottom), (G) pseudo-Pelger cells, and (H) neutrophil with fragmented nucleus.
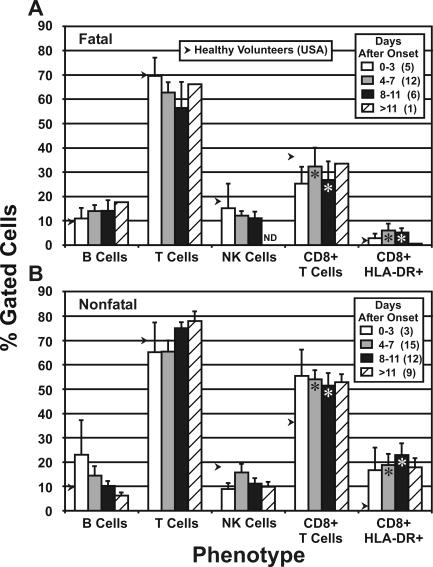
Phenotyping of PBMC.
Figure 2 shows the results of phenotyping of patient lymphocytes obtained at various times after disease onset; flow cytometry was used to enumerate surface-stained cells. A difference in profiles for fatal and nonfatal patients is evident from this analysis. In fatal cases, the percentage of T cells decreased as the disease progressed to the mean time of death (8 days) (9), with a corresponding rise in B cells. In contrast, nonfatal cases showed an increase in T cells as the disease progressed to the mean time of death, while B cells appeared to decrease during this time. The only fatal case-patient from whom a sample was obtained >11 days after onset (actually 28 days) was likely a recovering patient who died from complications or some underlying condition. No difference in NK cell numbers was evident between fatal and nonfatal cases. The number of CD8+ T cells in fatal case-patients was greatly reduced compared to the number in case-patients who survived, especially at 8 to 11 days after onset. Further examination of the CD8+ T-cell populations showed that the level of activation (HLA-DR+ staining) in patients who survived was much greater than in those who did not.
FIG. 2.
Flow cytometry (lymphocyte phenotyping) of PBMC samples obtained from fatal (A) and nonfatal (B) cases of Ebola-S disease. Average values were plotted as a percentage of gated cells (side scatter × CD24+ for B, T, and CD8+ T cells; side scatter × CD3+ for CD8+ HLA-DR+ and CD8+ CD38+). The numbers of specimens for each group are shown in parentheses in the legend. Arrows indicate the average values derived from four samples taken from healthy volunteers (Special Pathogens Branch workers). ND, not done. Asterisks indicate statistically significant differences between fatal and nonfatal cases (t < 0.05). The bars on the graph indicate standard errors.
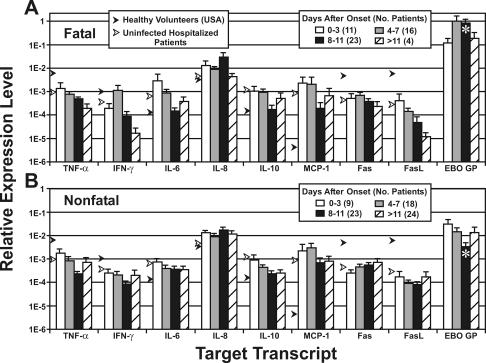
Quantitation of gene expression in PBMC specimens.
Quantitative PCR results for relative levels of expression of TNF-α, IFN-γ, IL-6, IL-8, IL-10, MCP-1, Fas, FasL, and Ebola-S virus GP genes in PBMC specimens are shown in Fig. 3. Differences in cellular gene expression between surviving patients and those who died were not significant, nor was there a substantial difference when the values were compared to those of hospitalized patients determined not to be infected with Ebola-S virus. A slight decrease in TNF-α expression was seen in Ebola-S-infected patients, and reduced IFN-γ and FasL expression may have occurred in patients who died. IL-6 was slightly elevated in patients who died 0 to 3 days after onset, but levels decreased to normal at the mean time of death. The level of MCP-1 in all patients (Ebola virus infected or uninfected) was elevated compared with levels in samples from healthy volunteers. The only statistically significant finding was seen in differences in virus load (measured by Ebola-S virus GP gene expression) between patients with fatal and nonfatal infections. The mean difference in GP expression increased from 0.5 log 0 to 3 days postonset to over 2 logs at 8 to 11 days after onset. This indicates that the virus load at or near the mean time of death can be 100-fold greater in fatal cases compared to nonfatal. Also, survivor patients appeared to clear the virus more efficiently, as levels of GP expression began declining 3 days after onset of symptoms.
FIG. 3.
Relative expression of cytokines, Fas antigen, FasL, and Ebola-S virus GP. Average values were determined from quantitative PCR (TaqMan) assays performed on samples from 48 patients with fatal disease (A) and 70 patients with nonfatal disease (B). Asterisks indicate statistically significant differences between fatal and nonfatal cases (t < 0.05). Black arrows indicate levels for 25 uninfected patients, and white arrow indicate levels for five healthy volunteers (Special Pathogens Branch workers). The bars on the graph indicate standard errors.
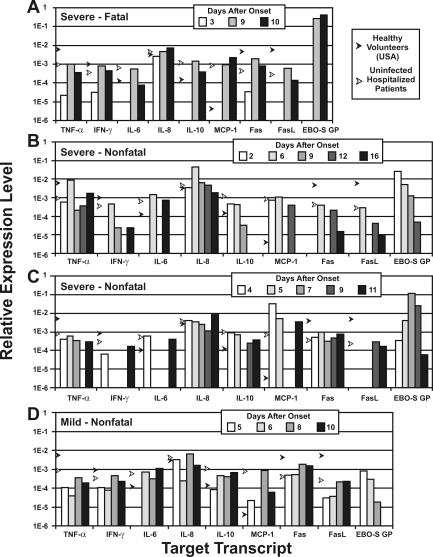
Expression data for serial PBMC specimens from patients exhibiting severe to mild disease are shown in Fig. 4. In a severe, fatal case (Fig. 4A), 3 days after onset of symptoms there was a diminished expression of inflammatory cytokines (TNF-α and IL-6), IFN-γ, IL-10, MCP-1, Fas, and FasL before virus was detected in the blood. A return to nearly normal levels was seen with the appearance of high levels of virus RNA. Patients with severe, nonfatal disease (Fig. 4B and C) also showed decreased expression, but expression of certain genes (IFN-γ, IL-6, MCP-1, Fas, and FasL) showed what might have been a cycling of decreased and increased transcription. The patient with a mild infection (Fig. 4D) showed less fluctuation in cellular gene expression levels but also showed decreases at 5 to 6 days after onset of symptoms. In these patients, there was a correlation between an increase in virus load (GP expression) and an increase in the severity of disease.
FIG. 4.
Relative expression of cytokines, Fas, FasL, and Ebola-S virus GP determined for three patients with severe Ebola-S disease (fatal and nonfatal) (A, B, and C) and one with mild disease (D). Black arrows indicate levels for 25 uninfected patients, and white arrows indicate levels for five healthy volunteers (Special Pathogens Branch workers).
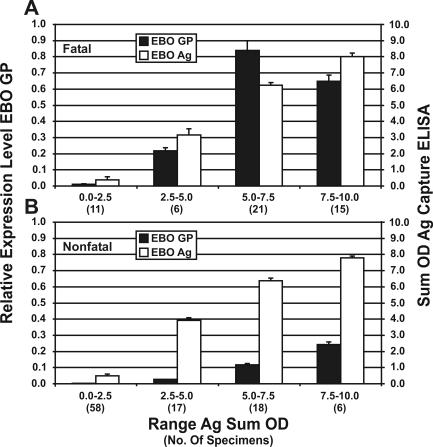
Ebola-S virus antigen level data (capture enzyme-linked immunosorbent assay sum optical density) were compared to TaqMan GP expression results (Fig. 5) that were obtained for the same blood samples and showed a correlation between rising antigen and Ebola-S virus GP expression (virus replication). However, the levels of GP expression seen in fatal and nonfatal infections differed. Fatal cases had much higher levels than nonfatal cases, which may reflect the relative numbers of infected monocytes in the blood (virus load). In fatal cases with the highest antigen levels (range of sum optical densities = 7.5 to 10.0), there appeared to be a decrease in GP expression, possibly due to depletion of infected cells and/or reduced transcription within these cells.
FIG. 5.
Ebola-S virus antigen levels in blood compared with levels of relative Ebola-S virus GP gene expression in PBMC samples. Comparisons were made only on the basis of antigen levels (antigen sum optical density) regardless of time after disease onset. The bars on the graph indicate standard errors. Differences between fatal and nonfatal cases were found not to be statistically significant (t > 0.05).
NO levels in Ebola-S virus-infected patients.
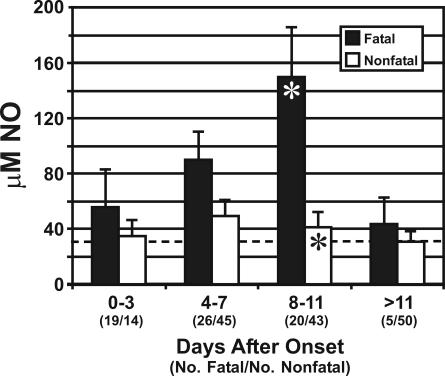
Figure 6 shows the results of NO quantitation assays performed on plasma from patients who died and those who survived. In patients who died, the level of NO in the peripheral blood was elevated, most prominently at or near the mean time of death (150 μM at 8 to 11 days after onset), while patients who survived showed only a slight increase, which was highest 4 to 7 days after the onset of symptoms (≈50 μM).
FIG. 6.
NO levels in peripheral blood of Ebola-S virus-infected patients. A comparison of average NO concentrations determined for fatal and nonfatal cases is shown. The dashed line indicates a “normal” level, determined from the serum of five healthy volunteers (Special Pathogens Branch workers). Asterisks indicate statistically significant difference between fatal and nonfatal cases (t < 0.05). The bars on the graph indicate standard errors.
DISCUSSION
Effects of Ebola-S virus infection on blood cells.
The development of leukopenia in Ebola-S virus-infected patients is characteristic of Ebola virus infections (28), but it is unclear if it is a natural response to infection or a sign of a compromised immune system. Depletion of lymphocytes may be due to apoptosis, which has been described for Ebola-Z virus infections in humans and experimentally infected nonhuman primates (3, 2, 13). The appearance of atypical lymphocytes in patient blood smears indicates a developing immune response to virus antigen. However, the appearance of pseudo-Pelger cells (neutrophils with abnormal function) in smears (Fig. 1), which were initially described following the first Ebola-S outbreak in 1976 (11), may stem from an aberrant inflammatory response involving cytokines or a cytotoxicity associated with the infection. Cytotoxicity could develop along with liver damage, because blood samples with high antigen titers also contained elevated liver enzymes and usually showed signs of hemolysis.
Phenotyping of lymphocytes in PBMC samples.
Flow cytometry of PBMC specimens showed that as the severity of Ebola-S virus disease increased in fatal cases, the percentage of T cells and CD8+ T cells decreased below normal levels, while in nonfatal cases the numbers of these cells increased (Fig. 2). Similarly, activation of CD8+ T cells (HLA-DR+ staining) is lacking in fatal cases and very prominent in nonfatal infections. These findings indicate that a vigorous cell-mediated, cytotoxic T-cell (Th1) response is associated with survival in Ebola-S virus infections. The presence of activated CD8+ cells early in the course of the disease (0 to 3 days after onset) suggests that surviving patients are more efficient at generating cytotoxic T cells and thus more capable of clearing virus-infected cells. The proportion of NK cells did not increase appreciably in either fatal or nonfatal cases, so their role in virus clearance may not be critical in the acute phase of infection, although their cytotoxic activity against Ebola-S virus-infected cells was not determined in this study. Similarly, B cells do not appear to have a significant role in surviving Ebola-S virus disease.
While plasma cells were observed in blood smears, patients who died of acute infection were serologically negative for antibody, and surviving patients usually developed detectable immunoglobulin M well after antigen levels started to decline and immunoglobulin G after antigen had been cleared from the blood (unpublished data). Also, the large amount of virus antigen circulating in the blood and expressed in various organs during the acute phase would likely tie up virus-specific antibodies in immune complexes. Specific antibody may actually augment virus infectivity; a recent study determined that convalescent-phase sera from humans infected with Ebola-Z virus enhances virus infectivity and that the responses were mediated by antibodies to the virion glycoprotein and complement (35).
Quantitation of PBMC and virus gene expression.
Quantitative PCR quantitation of cytokine transcripts in PBMC from Ebola-S virus-infected patients showed levels of expression similar to those seen in uninfected hospitalized patients (Fig. 3). Aside from MCP-1 expression, these cells appeared to be in an unresponsive state despite ongoing virus replication. The observed normal to reduced levels of Fas and FasL expression in PBMC might have been due to removal of apoptotic lymphocytes (Fas high-density cells) or to some form of virus-induced inhibition. Examination of serial specimens from individual patients revealed that the relative expression of IFN-γ, IL-6, IL-10, MCP-1, Fas, and FasL fluctuated between normal and reduced or undetectable levels (Fig. 4) in what appears to be a cycling pattern. These changes in expression levels may be due to regulation of gene expression and the appearance of naïve lymphocytes (replacing those removed through apoptosis).
In an earlier study of Ebola-Z cases (1995 Democratic Republic of the Congo outbreak), it was determined that patients had increased TNF-α and IFN-α mRNAs in blood clot specimens as well as high serum levels of cytokines. Analysis of serum cytokine levels for Ebola-S virus-infected patients (2000 Uganda outbreak) showed no elevation in TNF-α (or IFN-γ) but marked increases in MCP-1, IL-10, IL-8, IL-6, IL-1b, IFN-α, and RANTES were found, particularly in fatal cases (K. Hutchinson, unpublished data). Differences in TNF-α levels in Ebola-Z and Ebola-S virus-infected patients may be due to differences in disease progression, since the disease in Ebola-Z cases usually develop much more rapidly.
The unresponsiveness of PBMC from Ebola-S virus-infected patients may stem from immune inactivation following prolonged exposure to virus antigen (prior to and during the acute phase, when antigen levels are extremely high) that results in overstimulation of immunocompetent cells (12, 20). Alternatively, this unresponsive state could be the direct or indirect result of virus replication in host cells, such as those of the mononuclear phagocytic system, which could lead to impaired T-cell functions (such as T-cell receptor-CD3 signaling, IL-2 receptor signaling, and regulation of immune responses by suppressor T cells). In vitro studies of Ebola-Z virus revealed that suppression of immune responses can take place within infected cells (4, 5, 15, 16, 17) and that direct contact of virion particles with T cells can render them unresponsive (10). In addition, dendritic cells infected with Ebola-Z virus have been shown to be functionally impaired and are poor stimulators of T cells (24), as has also been described for measles virus (29, 32) and lymphocytic choriomeningitis virus (33). Since dendritic cells are key components in adaptive immune responses, it is reasonable to assume that the degree to which these cells are infected or impaired has a direct impact on antigen processing and the recruitment and activation of effector cells.
Levels of virus RNA in PBMC were found to be higher in patients who died, as determined by quantitative PCR analysis (Fig. 3 to 5) of PBMC specimens; similar results have been obtained from quantitative PCR assays targeting virus RNA in plasma and serum specimens (36). The higher virus load is likely due to an inability to control the replication of virus in target organs and cells of the mononuclear phagocytic system. We have determined from a genetic study of a subset of Ebola-S patients that certain major histocompatibility complex class I alleles are associated with either fatal or nonfatal outcomes (unpublished data). This suggests that the efficiency with which cell-mediated (cytotoxic T lymphocyte) responses are generated can influence the virus burden and severity of disease in Ebola-S virus-infected patients. Suppression of major histocompatibility complex class I expression in endothelial cells infected with Ebola-Z virus has been demonstrated in vitro (17), and reduced expression in dendritic cells would also contribute to impaired function.
Role of NO in Ebola virus pathogenesis.
NO is important in biological signaling and is an extremely potent effector molecule in the homeostasis of the cardiovascular system and a mediator of the immune system. Constitutive expression of NO synthase by endothelial cells functions to maintain vascular tone and arterial pressure through NO vasodilation (countering catecholamine constriction). Expression of inducible NO synthase occurs in a variety of other cells (e.g., leukocytes, Küpffer cells, and hepatocytes), and elevated blood NO can result from antimicrobial and antitumor responses of immunocompetent cells (8). The excessively high levels of NO seen in fatal Ebola-S hemorrhagic fever cases (Fig. 6) may result from host responses to higher virus loads, as has been shown in human immunodeficiency virus-infected patients (14), and could be produced following cytokine-induced overexpression of inducible NO synthase in mononuclear phagocytic cells in the liver, lung, and spleen. These high NO levels are similar to those produced as a result of a systemic inflammatory response syndrome that develops when endotoxin is administered (sepsis) (8). However, the lack of increased levels of TNF-α in Ebola-S virus-infected patients suggests that the primary source of NO production is another cell type, possibly hepatocytes (15, 21, 23), and NO levels could reflect the extent of infection and pathology in the liver.
High levels of NO in the blood (comparable to those seen in Ebola-S virus-infected patients who died) are associated with cardiac distress and heart failure and may have an important role in the shock syndrome seen in many fatal cases. Hypotension is a characteristic of Ebola virus infections (especially during the most acute phase of disease), and elevated NO levels would contribute to this condition. Extremely high NO levels may also lead to the formation of toxic molecules, such as peroxynitrite (potent oxidant associated with many pathologies), that further stress the patient (6, 7). Aside from intravenous fluid administration and blood transfusions, intervention therapies have not been attempted in severe cases of Ebola virus infections. If reduction of blood NO or its effects can lessen patient stress, then the administration of scavenging compounds (25, 26, 27) and/or vasoconstrictors (catecholamines or epinephrine/norepinephrine) could be a useful treatment for sustaining patients through critical periods of Ebola virus infection.
In conclusion, we have shown that during the acute phase of Ebola-S hemorrhagic fever, patients have an impaired immune system, seen as development of leukopenia and unresponsive PBMC. We have also associated fatal outcomes with a high virus load (infected monocytes) and elevated NO levels, while survival is linked to the maintenance and activation of CD8+ lymphocytes. Further studies of host and virus factors that influence the severity of Ebola virus infections will help not only in the characterization of disease processes but also in the development of vaccines and therapeutic treatments.
Acknowledgments
We acknowledge the Ugandan Ministry of Health and the World Health Organization Response Team for their assistance and counsel. We especially thank Piero Corti, Maria Di Santo, Franca Cian, Elio Croce, Yoti Zebulon, and the entire staff of St. Mary's Lacor Hospital for their support and many efforts, without which this study would not have been possible. We are also very grateful to John O'Connor for his editorial contributions.
REFERENCES
- 1.Baize, S., E. M. Leroy, A. J. Georges, M. C. Georges-Courbot, M. Capron, I. Bedjabaga, J. Lansoud-Soukate, and E. Mavoungou. 2002. Inflammatory responses in Ebola virus-infected patients. Clin. Exp. Immunol. 128:163-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baize, S., E. M. Leroy, E. Mavoungou, and S. P. Fisher-Hoch. 2000. Apoptosis in fatal Ebola infection. Does the virus toll the bell for immune system? Apoptosis 5:5-7. [DOI] [PubMed] [Google Scholar]
- 3.Baize, S., E. M. Leroy, M.-C. Georges-Courbot, M. Capron, J. Lansoud-Soukate, P. Debré, S. P. Fisher-Hoch, J. B. McCormick, and A. J. Georges. 1999. Defective humoral responses and extensive intravascular apoptosis are associated with fatal outcome in Ebola virus-infected patients. Nat. Med. 5:423-426. [DOI] [PubMed] [Google Scholar]
- 4.Basler, C. F., X. Wang, E. Mühlberger, V. Volchkov, J. Paragas, H.-D. Klenk, A. Garcia-Sastre, and P. Palese. 2000. The Ebola virus VP35 protein functions as a type I IFN antagonist. Proc. Natl. Acad. Sci. USA 97:12289-12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Basler, C. F., A. Mikulasova, L. Martinez-Sobrido, J. Paragas, E. Mühlberger, M. Bray, H.-D. Klenk, P. Palese, and A. Garcia-Sastre. 2003. The Ebola virus VP35 protein inhibits activation of interferon regulatory factor 3. J. Virol. 77:7945-7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beckman, J. S., and W. H. Koppenol. 1996. Nitric oxide, superoxide, and peroxynitrite: the good, the bad, and the ugly. Am. J. Physiol. 271:C1424-1437. [DOI] [PubMed] [Google Scholar]
- 7.Bian, K., and F. Murad. 2001. Diversity of endotoxin-induced nitrotyrosine formation in macrophage-endothelium-rich organs. Free Radic. Biol. Med. 31:421-429. [DOI] [PubMed] [Google Scholar]
- 8.Burgner, D., K. Rockett, and D. Kwiatkowski. 1999. Nitric oxide and infectious diseases. Arch. Dis. Child. 81:185-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Centers for Disease Control and Prevention. 2001. Outbreak of Ebola hemorrhagic fever—Uganda, August 2000-January 2001. Morb. Mortal. Wkly. Rep. 50:73-77. [PubMed] [Google Scholar]
- 10.Chepurnov, A. A., M. N. Tuzova, V. A. Ternovoy, and I. V. Chernukhin. 1999. Suppressive effect of Ebola virus on T-cell proliferation in vitro is provided by a 125-kDa GP viral protein. Immunol. Lett. 68:257-261. [DOI] [PubMed] [Google Scholar]
- 11.Deitrick, M., H. H. Schumacher, D. Peters, and J. Knobloch. 1978. Human pathology of Ebola (Maridi) virus infection in the Sudan, p. 37-41. In S. R. Pattyn (ed.), Ebola virus haemorrhagic fever. Elsevier/North-Holland Biomedical Press, New York, N.Y.
- 12.Fuller, M. J., and A. J. Zajac. 2003. Ablation of CD8 and CD4 T-cell responses by high viral loads. J. Immunol. 170:477-486. [DOI] [PubMed] [Google Scholar]
- 13.Geisbert, T. W., L. E. Hensley, T. R. Gibb, K. E. Steele, N. K. Jaax, and P. B. Jahrling. 2000. Apoptosis induced in vitro and in vivo during infection by Ebola and Marburg viruses. Lab. Investig. 80:171-186. [DOI] [PubMed] [Google Scholar]
- 14.Groeneveld, P. H., F. P. Kroon, P. H. Nibbering, S. M. Bruisten, P. van Swieten, and R. van Furth. 1996. Increased production of nitric oxide correlates with viral load and activation of mononuclear phagocytes in HIV-infected patients. Scand. J. Infect. Dis. 28:341-345. [DOI] [PubMed] [Google Scholar]
- 15.Gupta M., S. Mahanty, and R. Ahmed, and P. E. Rollin. 2001. Monocyte-derived human macrophages and peripheral blood mononuclear cells infected with Ebola virus secrete MIP-1alpha and TNF-alpha and inhibit poly-IC-induced IFN-alpha in vitro. Virology 284:20-25. [DOI] [PubMed] [Google Scholar]
- 16.Harcourt, B. H., A. Sanchez, and M. K. Offermann. 1998. Ebola virus inhibits induction of genes by double-stranded RNA in endothelial cells. Virology 252:179-188. [DOI] [PubMed] [Google Scholar]
- 17.Harcourt, B. H., A. Sanchez, and M. K. Offermann. 1999. Ebola virus selectively inhibits responses to interferons, but not to interleukin-1beta, in endothelial cells. J. Virol. 73:3491-3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hensley, L. E., H. A. Young, P. B. Jahrling, and T. W. Geisbert. 2002. Proinflammatory response during Ebola virus infection of primate models: possible involvement of the tumor necrosis factor receptor superfamily. Immunol. Lett. 80:169-179. [DOI] [PubMed] [Google Scholar]
- 19.Hutchinson, K. L., F. Villinger, M. E. Miranda, T. G. Ksiazek, C. J. Peters, and P. E. Rollin. 2001. Multiplex analysis of cytokines in the blood of cynomolgus macaques naturally infected with Ebola virus (Reston serotype). J. Med. Virol. 65:561-566. [PubMed] [Google Scholar]
- 20.Jooss, K., B. Gjata, O. Danos, H. von Boehmer, and A. Sarukhan. 2001. Regulatory function of in vivo anergized CD4(+) T cells. Proc. Natl. Acad. Sci. USA 98:8738-8743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kandemir, O., A. Polat, and A. Kaya. 2002. Inducible nitric oxide synthase expression in chronic viral hepatitis and its relation with histological severity of disease. J. Viral Hepatol. 9:419-423. [DOI] [PubMed] [Google Scholar]
- 22.Ksiazek, T. G., P. E. Rollin, P. B. Jahrling, E. Johnson, D. W. Dalgard, and C. J. Peters. 1992. Enzyme immunosorbent assay for Ebola virus antigens in tissues of infected primates. J. Clin. Microbiol. 30:947-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lake-Bakaar, G., D Sorbi., and V. Mazzoccoli. 2001. Nitric oxide and chronic HCV and HIV infections. Dig. Dis. Sci. 46:1072-1076. [DOI] [PubMed] [Google Scholar]
- 24.Mahanty, S., K. Hutchinson, S. Agarwal, M. McRae, P. E. Rollin, and B. Pulendran. 2003. Cutting edge: impairment of dendritic cells and adaptive immunity by Ebola and Lassa viruses. J. Immunol. 170:2797-2801. [DOI] [PubMed] [Google Scholar]
- 25.Millar, C. G., and C. Thiemermann. 2002. Carboxy-PTIO, a scavenger of nitric oxide, selectively inhibits the increase in medullary perfusion and improves renal function in endotoxemia. Shock 18:64-68. [DOI] [PubMed] [Google Scholar]
- 26.Reiter, C. D., X. Wang, J. E. Tanus-Santos, N. Hogg, R. O. Cannon, A. N. Schechter, and M. T. Gladwin. 2002. Cell-free hemoglobin limits nitric oxide bioavailability in sickle-cell disease. Nat. Med. 8:1383-1389. [DOI] [PubMed] [Google Scholar]
- 27.Roza, A. M., M. Cooper, G. Pieper, G. Hilton, K. Dembny, C. S. Lai, P. Lindholm, R. Komorowski, C. Felix, C. Johnson, and M. Adams. 2000. NOX 100, a nitric oxide scavenger, enhances cardiac allograft survival and promotes long-term graft acceptance. Transplantation 69:227-231. [DOI] [PubMed] [Google Scholar]
- 28.Sanchez, A., A. S. Khan, S. R. Zaki, G. J. Nabel, T. G. Ksiazek, and C. J. Peters. 2001. Filoviridae: Marburg and Ebola viruses, p. 1279-1304. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 29.Schneider-Schaulies, S., I. M. Klagge, and V. ter Meulen. 2003. Dendritic cells and measles virus infection. Curr. Top. Microbiol. Immunol. 276:77-101. [DOI] [PubMed] [Google Scholar]
- 30.Schnittler, H. J., and H. Feldmann. 1998. Marburg and Ebola hemorrhagic fevers: does the primary course of infection depend on the accessibility of organ-specific macrophages? Infect. Dis. 27:404-406. [DOI] [PubMed] [Google Scholar]
- 31.Schnittler, H. J., and H. Feldmann. 2003. Viral hemorrhagic fever-a vascular disease? Thromb. Haemost. 89:967-972. [PubMed] [Google Scholar]
- 32.Server-Delprat, C., P.-O. Vidalain, H. Valentin, and C. Rabourdin-Combe. 2003. Measles virus and dendritic cell functions: how specific response cohabits with immunosuppression. Curr. Top. Microbiol. Immunol. 276:103-123. [DOI] [PubMed] [Google Scholar]
- 33.Sevilla, N., S. Kunz, and M. B. A. Oldstone. 2003. Infection of dendritic cells by lymphocytic choriomeningitis virus. Curr. Top. Microbiol. Immunol. 276:125-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sullivan, N., Z.-Y. Yang, and G. J. Nabel. 2003. Ebola virus pathogenesis: implications for vaccines and therapies. J. Virol. 77:9733-9737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takada, A., H. Feldmann, T. G. Ksiazek, and Y. Kawaoka. 2003. Antibody-dependent enhancement of Ebola virus infection. J. Virol. 77:7539-7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Towner, J. S., P. E. Rollin, D. G. Bausch, A. Sanchez, S. M. Crary, M. Vincent, W. F. Lee, C. F. Spiropoulou, T. G. Ksiazek, M. Lukwiya, F. Kaducu, R. Downing, and S. T. Nichol. 2004. Rapid diagnosis of Ebola hemorrhagic fever by reverse transcription-PCR in an outbreak setting and assessment of patient viral load as a predictor of outcome. J. Virol. 78:4330-4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Villinger, F., P. E. Rollin, S. S. Brar, N. F. Chikkala, J. Winter, J. B. Sundstrom, S. R. Zaki, R. Swanepoel, A. A. Ansari, and C. J. Peters. 1999. Markedly elevated levels of interferon (IFN)-gamma, IFN-alpha, interleukin (IL)-2, IL-10, and tumor necrosis factor-alpha associated with fatal Ebola virus infection. J. Infect. Dis. 179:S188-191. [DOI] [PubMed] [Google Scholar]
- 38.World Health Organization. 1978. Ebola haemorrhagic fever in Sudan, 1976. Report of a W.H.O./International Study Team. Bull. W.H.O. 56:247-270. [PMC free article] [PubMed] [Google Scholar]
- 39.World Health Organization. 1978. Ebola haemorrhagic fever in Zaire, 1976. Bull. W.H.O. 56:271-293. [PMC free article] [PubMed] [Google Scholar]
- 40.Zaki, S. R., W. J. Shieh, P. W. Greer, C. S. Goldsmith, T. Ferebee, J. Katshitshi, F. K. Tshioko, M. A. Bwaka, R. Swanepoel, P. Calain, A. S. Khan, E. Lloyd, P. E. Rollin, T. G. Ksiazek, and C. J. Peters. 1999. A novel immunohistochemical assay for the detection of Ebola virus in skin: implications for diagnosis, spread, and surveillance of Ebola hemorrhagic fever. J. Infect. Dis. 179(Suppl. 1):S36-S47. [DOI] [PubMed] [Google Scholar]