Abstract
The human gammaherpesviruses Epstein-Barr virus and Kaposi Sarcoma-associated herpesvirus both contain a glycoprotein (gp350/220 and K8.1, respectively) that mediates binding to target cells and has been studied in great detail in vitro. However, there is no direct information on the role that these glycoproteins play in pathogenesis in vivo. Infection of mice by murid herpesvirus 4 strain 68 (MHV-68) is an established animal model for gammaherpesvirus pathogenesis and expresses an analogous glycoprotein, gp150. To elucidate the in vivo function of gp150, a recombinant MHV-68 deficient in gp150 production was generated (vgp150Δ). The productive viral replication in vitro and in vivo was largely unaffected by mutation of gp150, aside from a partial defect in the release of extracellular virus. Likewise, B-cell latency was established. However, the transient mononucleosis and spike in latently infected cells associated with the spread of MHV-68 to the spleen was significantly reduced in vgp150Δ-infected mice. A soluble, recombinant gp150 was found to bind specifically to B cells but not to epithelial cells in culture. In addition, gp150-deficient MHV-68 derived from mouse lungs bound less well to spleen cells than wild-type virus. Thus, gp150 is highly similar in function in vitro to the Epstein-Barr virus gp350/220. These results suggest a role for these analogous proteins in mononucleosis and have implications for their use as vaccine antigens.
Murid herpesvirus 4 is an endogenous pathogen of free-living rodents of the Apodemus genus, e.g., wood mice (5). Infection of laboratory mice by murid herpesvirus 4 strain 68 (MHV-68; also called γHV-68) is an amenable model system for the study of gammaherpesvirus pathogenesis and for the development of therapeutic strategies against these viruses (4, 13, 28, 33, 34, 48). After intranasal inoculation of mice with MHV-68, a productive infection occurs in the lung (39). This is cleared around day 10 to 14 postinfection (p.i.) by CD8+ T cells (17), although the virus persists in a latent form in epithelial cells at this site (38). MHV-68 spreads to the spleen, where it also becomes latent, predominantly within B lymphocytes but also in macrophages and dendritic cells (19, 40, 45, 52).
Spread to the spleen and the establishment of latency is associated with a marked splenomegaly, an increase in cell numbers in the spleen (splenic mononucleosis or lymphocytosis) (44), and a subsequent peripheral mononucleosis that is reminiscent that caused by primary infection of humans by Epstein-Barr virus (EBV) (42). Splenomegaly and splenic mononucleosis, which peak at day 14 p.i., are driven by CD4+ T cells (17, 44) and are dependent on MHV-68-infected B cells in the spleen (45, 51). Concomitant with the splenic mononucleosis is a sharp rise in the number of latently infected B cells, the resolution of which to a relatively constant baseline level is achieved by CD8+ T cells (17, 41, 51). CD8+ T cells, along with antibody (23, 38), are important in the long-term control of persistent infection (9, 38, 51).
The infection of cells by herpesviruses is complex and involves the interaction between glycoproteins embedded in the virion membrane and cell surface ligands (for a review, see reference 31). Within the gammaherpesvirus subfamily, the process has been studied in greatest detail with EBV. Here, an interaction between the virion glycoprotein gp350/220 and complement receptor type 2 (CR2) on B cells is a critical first step in B-cell infection by EBV (18). Attachment to epithelial cells (that lack CR2) is mediated by glycoprotein H (gH) and entry requires a gH/gL complex (27). However, entry into B cells requires a complex of gH, gL, and gp42, a glycoprotein unique to EBV that uses HLA class II as a coreceptor (50). Although there is no direct sequence homologue of gp350/220, each gammaherpesvirus contains a gene encoding an analogous glycoprotein in the same relative genomic location. This positional analog is called gene 51 in herpesvirus saimiri (the prototypic gamma-2-herpesvirus) and K8.1 in Kaposi's sarcoma-associated herpesvirus (KSHV) (29). It appears that this glycoprotein may have a conserved function in binding to cellular receptors since K8.1 has been shown to bind to heparan sulfate and mediate entry into cells (3, 49).
Our previous work has shown that MHV-68 contains a glycoprotein (gp150) that is a major constituent of the virion membrane (35). Further, vaccination with a recombinant vaccinia virus expressing gp150 protected mice from mononucleosis but not lung infection after intranasal challenge with MHV-68 (37). MHV-68 gp150 is analogous to the KSHV K8.1 glycoprotein and EBV gp350/220. Both gp350/220 and K8.1 have been well characterized and are known to be virion membrane glycoproteins that are involved in the binding of virus to cells. However, other than virus attachment, little is known about alternative roles of gp350/220 and K8.1 in the virus life cycle in vivo. The aim of the present study was to study the role of MHV-68 gp150 in virus infection in the host and to use the conclusions to shed light on the role of gp350/200 and K8.1.
MATERIALS AND METHODS
Cell culture and virus stocks.
Baby hamster kidney cells (BHK-21) were maintained in Glasgow modified minimal essential medium supplemented with 10% newborn calf serum, 10% tryptose-phosphate broth, 2 mM l-glutamine, 70 μg of penicillin/ml, and 10 μg of streptomycin/ml. The mouse B-cell line, A20, was maintained in RPMI medium containing 10% fetal calf serum, 50 μM 2-mercaptoethanol, 100 μg of streptomycin/ml, 100 U of penicillin/ml, and 25 mM HEPES. MHV-68 was originally isolated during field studies from the bank vole Clethrionomys glareolus (6) and was subsequently plaque purified on BHK-21 cells to obtain clone g2.4 as described previously (16). Viruses were propagated and titrated by using BHK-21 cells as described previously (39).
Generation of a gp150 mutant.
A recombinant virus with a mutation in the open reading frame (ORF) encoding gp150 was constructed by using MHV-68 cloned as bacterial artificial chromosome (BAC) as recently described (1). To delete a part of the gp150 ORF, a linear PCR fragment containing an FLP recombinase target-flanked tetracycline resistance cassette was generated from vector pCP16 with a primer pair that contained 24 nucleotides for amplification of the tetracycline resistance gene and an additional 50 nucleotides homologous to the sequences flanking the region to be deleted, corresponding to nucleotide positions 69472 to 69521 and 70855 to 70904 (47), respectively. This PCR product was transferred into the MHV-68 BAC by homologous recombination with Escherichia coli strain JC8679 (ET-cloning) as described previously (1), leading to the deletion of nucleotides 69522 to 70854 of MHV-68. Recombinant BAC plasmids were transfected by electroporation into E. coli strain DH10B and characterized by restriction enzyme analysis and sequencing.
A revertant BAC plasmid was generated by a two-step replacement procedure as described previously (1, 26). For that purpose, a 5.3-kb BglII fragment of MHV-68 (nucleotide positions 67744 to 73044) was cloned into the shuttle plasmid pST76K-SR (1) and electroporated into E. coli strain DH10B that already contained the gp150 deletion BAC. Recombinant BAC plasmids were characterized by restriction enzyme analysis.
Transfection by electroporation of 2 to 3 μg of the recombinant BAC plasmids into BHK-21 cells resulted in the reconstitution of recombinant viruses expressing green fluorescent protein. To remove the BAC vector sequences, rat embryonic fibroblasts expressing recombinase Cre (1) were infected, and viral clones with the BAC vector sequences deleted were purified by limiting dilution with loss of green fluorescent protein (GFP) expression as a screening marker. DNA of all reconstituted, recombinant viruses was isolated from infected BHK-21 cells and analyzed by restriction enzyme digestion. Successful mutagenesis and the absence of the BAC vector sequences were confirmed by Southern blot analysis.
In vitro infections. Single- and multistep virus growth curves were obtained by infecting subconfluent BHK-21 cells at a multiplicity of infection (MOI) of 5 or 0.01, respectively. After the virus was allowed to adsorb for 1 h at 37°C, the cells were washed with medium to remove unbound virus, and fresh growth medium was then added. At various times p.i., the wells were harvested and infectious virus was quantified by plaque assay. All infections (per time point) and analysis of virus production by plaque assay were performed in duplicate.
Infection of mice and analysis of tissues.
All animal work was performed under UK Home Office project license numbers 60/2429 and 40/2483 and personal license number 60/6501. BALB/c mice were purchased from Bantin and Kingman (Hull, United Kingdom). Mice were infected intranasally at 4 to 6 weeks of age with 4 × 105 PFU of virus in 40 μl of sterile phosphate-buffered saline (PBS) under light anesthesia (halothane or isoflurane). In some experiments, as an alternative, mice were infected intravenously with 4 × 105 PFU MHV-68 in 0.1 ml of PBS in the tail vein. At various times p.i., mice were euthanized and tissues were harvested for analysis. Plaque assays were performed with BHK-21 cells to detect infectious virus as described previously (39). The limit of detection of the plaque assay was 10 PFU per organ. An infective center (IC) assay was used to detect latent virus as described previously (39). The data were analyzed statistically using two-way analysis of variance with Bonferroni post-tests; P values were set at a 95% confidence interval.
Microarray analysis.
RNA isolation, labeling, microarray hybridization, and data analysis were performed as described previously (15). Briefly, total RNA (25 μg) was isolated and reverse transcribed with 400 U of Superscript II (Life Technologies) in the presence of Cy3-dCTP (Amersham Pharmacia), followed by hybridization on MHV-68 75-mer oligonucleotide microarrays which included all known MHV-68 ORFs. Normalization between arrays was based on spike RNA corresponding to Bacillus subtilis genes transcribed in vitro.
Quantitative real-time PCR analysis.
DNA was extracted from spleen or lung tissue by using a Qiagen DNeasy kit and quantified by UV spectrophotometry. Quantification of viral DNA copy number was made by using an Opticon Monitor-2 real-time PCR machine (MJ Research). Amplification was performed in 20-μl reaction volumes with a DyNamo SYBR Green kit (Finnzymes). The cycling parameters consisted of a hot start at 95°C 10 min, denaturation at 94°C 10 s, annealing at 60°C 20 s, and extension at 72°C 15 s. Melting-curve analysis was carried out to confirm the specificity of the products between 65 and 95°C with 0.2°C increments. A portion (205 bases) of MHV-68 M3 gene was amplified with forward primer CCCCATCATGACTTGTCATC and reverse primer AAAACTTGCCCATGCTACT. The MHV-68 genome copy number was estimated against a standard curve constructed by serial dilution of MHV-68 BAC DNA (1). The murine ribosomal protein L8 (rpl8) gene (GenBank accession number AF091511) was used to normalize for input DNA between samples. The standard curve for rpl8 was constructed by serial dilution of a plasmid containing a 163-bp fragment of rpl8 (pCR2.1/rpl8). Amplifications of pCR2.1/rpl8 were carried out by using forward primer CAGTGAATATCGGCAATGTTTTG and reverse primer TTCACTCGAGTCTTCTTGGTCTC. Each sample was tested in triplicate. Mean viral genome copy numbers were determined from four individual mice and were expressed relative to the copy number of L8.
Protein expression and purification.
A fragment of the gp150 gene encoding the ectodomain of the mature protein (i.e., minus N-terminal signal sequence, transmembrane anchor, and endodomain) was amplified from viral DNA by PCR by using the primers 5′-GGACCATGGGATTTCTGGGGAATCACAACTTAG (sense, genome coordinate 69520) and 5′-GCTCTCGAGGGTAGAAATGTCTGGAACGG (antisense, genome coordinate 70826). This fragment was cut with NcoI and XhoI and inserted between the corresponding sites of the His6 fusion expression vector pET22b (Novagen) in frame with a sequence encoding the pel B leader sequence. The pel B periplasmic leader sequence directs the fusion protein into the periplasm of the host bacteria and is cleaved on entry. gp150-His fusion protein was expressed in E. coli BL21(DE3) bacteria containing this plasmid; protein was extracted from the bacterial periplasm and purified by using Ni2+ charged resin beads (Novagen) according to the manufacturer's instructions. To generate a GST-His fusion protein, two complementary oligonucleotides were synthesized that encoded the His6 motif, and this was inserted in frame with the sequence encoding glutathione S-transferase (GST) in the vector pGEX-2T (Pharmacia). Protein was purified directly from the bacterial cytoplasm by using Ni2+-charged resin beads (Novagen) according to the manufacturer's instructions. Protein concentrations were determined by spectrophotometry at 280 nm. For GST, the molar extinction coefficient of 5.4 × 104 M−1 cm−1 had been previously determined (30). For gp150, amino acid analysis was performed on purified gp150, and from this the concentration of a sample was determined. From this the molar extinction coefficient was determined to be 7 × 104 M−1 cm−1.
FACS analysis and binding assay.
Cells (106) were incubated in 200 μl of fluorescence-activated cell sorting (FACS) buffer (PBS, 1% bovine serum albumin, 0.2% sodium azide) for 45 min, washed, and then incubated at 37°C for 1.5 h with 50 μl of either 20 μM fusion protein preparation, 50 μl of MHV-68 virus preparation containing 107 PFU or 50 μl of BHK cell lysate, each diluted in FACS buffer. Cells were then washed three times in FACS buffer before being resuspended in 50 μl of rabbit anti-gp150/GST antibody (35), diluted 1:250 in FACS buffer, and incubated at 4°C for 60 min. Cells were washed as described above and then incubated with the swine anti-rabbit fluorescein isothiocyanate antibody (Dakopatts; 30 μl/well, diluted 1:30) for 30 min at 4°C. After this, cells were washed five times in FACS buffer and then resuspended in 200 μl of 1% Formol saline. Cells were sorted by using a FACStar (Becton Dickinson) FACS machine. Events from 10,000 cells were used for analysis. The gate for the definition of positive cells was positioned such that 1% of the cells with primary and secondary antibody alone were positive. The amounts of virus and recombinant protein used were determined by using preliminary titration experiments to give optimal signal to noise ratios. The results are presented as the mean ± the standard deviation of three separate determinations.
Virus binding and infection assay.
Virus used in these experiments was either prepared by infection of BHK-21 cells in vitro as described above or derived from homogenates of lungs harvested from mice infected with recombinant virus (vgp150Δ) or revertant virus (vgp150R) at 7 days p.i. The number of virus copies was determined by real-time PCR by the method described above. Lymphocytes were then extracted from uninfected mice and diluted to 2 × 106 cells in fresh, precooled RPMI medium and exposed to 0.1 copy per cell of either vgp150Δ or vgp150R from lung preparations. For the binding assay, the cells and virus were incubated on a shaking platform at 4°C for 3 h. For the infection assay, the cells and virus were then incubated on a shaking platform at 37°C for 1 h. The cells were washed three times in cold PBS to remove any unbound virus particles, and DNA was extracted by using a Qiagen DNeasy kit. The viral DNA copy number was determined in relation to copies of mouse rpl8 by quantitative real-time PCR analysis by using the method stated above. All experiments were carried out in triplicate, and results are expressed as mean values.
RESULTS
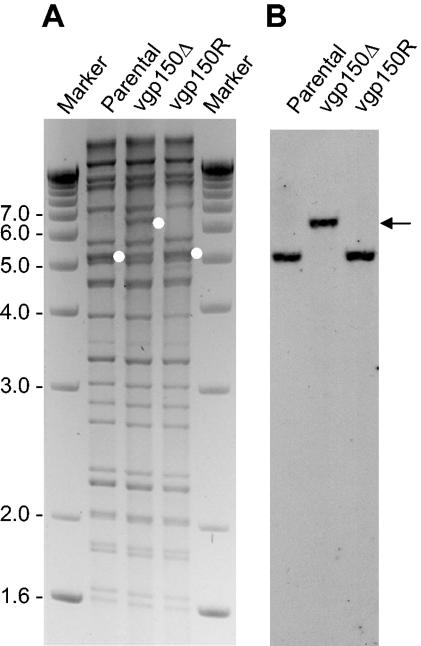
Construction of MHV-68 with a mutation in gp150. To assess directly the function of gp150 in the life cycle of murine gammaherpesvirus, we generated a recombinant virus (vgp150Δ) with a mutation in the gp150 locus by replacing part of it (coordinates 69521 to 70855) with a tetracycline resistance cassette. We performed this by using a BAC clone of the virus and the RecE/T recombination system (1). To exclude effects due to rearrangements outside gp150, a revertant virus (vgp150R) was generated with a nonmutated genomic fragment by a two-step replacement procedure. The structure of all cloned genomes was analyzed by restriction enzyme digestion with different enzymes and by sequencing the site of mutation (data not shown). Interestingly, the mutation of gp150 was found to be nonessential for virus replication in cell culture as transfection of BAC clones resulted in viral plaques. BAC sequences were removed from recombinant viruses by passage through Cre-expressing cells (1). The structure of all recombinant viruses was then analyzed by restriction enzyme digestion of viral DNA and Southern blotting. As shown in Fig. 1, a 5.3-kbp BglII fragment containing the gp150 ORF in the parental and revertant virus was replaced by a 6.3-kbp fragment in the mutant containing the tetracycline cassette.
FIG. 1.
Structural analysis of the genomes of recombinant viruses. (A) Ethidium bromide-stained agarose gel of BglII-digested DNA isolated from infected cells. Parental BAC virus (Parental), gp150 mutant (vgp150Δ), and revertant (vgp150R) are shown. (B) Southern blot analysis of the gel shown in panel A with a digoxigenin -labeled probe spanning nucleotide positions 68650 to 69407 of MHV-68, recognizing a 5.3-kbp BglII fragment in the parental BAC virus and in the revertant virus, respectively, and a 6.6-kbp BglII fragment in the gp150 mutant virus (indicated by white dots in panel A and by an arrowhead in panel B). Marker sizes (in kilobase pairs) are indicated on the left.
To confirm a lack of gp150 protein expression in vgp150Δ, we performed indirect immunofluorescence analysis on infected BHK-21 cells by using polyclonal antisera to MHV-68 gB and gp150. As seen in Fig. 2, cells infected with either vgp150Δ or the revertant vgp150R were positive for expression of the gB gene but only vgp150R expressed gp150 protein.
FIG. 2.
Immunofluorescence analysis of cells infected with vgp150Δ and vgp150R. BHK-21 cells were infected with 5 PFU of virus/cell for 18 h. Slides were then fixed with acetone before the indirect immunofluorescence staining procedure. Cells were stained with either rabbit polyclonal antibodies to gB or gp150, and fluorescence was then visualized by using a UV microscope. All panels are shown at the same magnification, and a size bar of 40 μm is indicated in the first panel.
Deletion of gp150 does not appreciably affect expression of other viral genes.
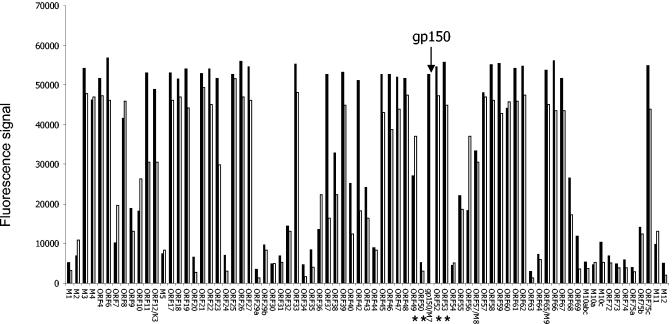
To asses whether disruption of gp150 affected expression of other MHV-68 genes, we compared global viral mRNA expression between vgp150Δ and wild-type revertant virus by microarray analysis. RNA from BHK cells 24 h p.i. was hybridized to an oligonucleotide-based DNA microarray containing all known MHV-68 ORFs and control genes as described previously (15). The results (Fig. 3) showed that, aside from gp150, there was no appreciable difference in mRNA expression between the viruses. For this, we have taken a fourfold difference as being a cutoff, based on previous comparisons between microarray and real-time analyses (10, 12). Any differences seen were far smaller than this cutoff and no greater than those observed between individual microarray analyses on wild-type virus. This was true for genes immediately surrounding gp150 (ORFs 49, 50, 52, and 53; marked by asterisks), as well as those more distant.
FIG. 3.
Transcriptome profiles of vgp150Δ and vgp150R. Cells were infected with viruses for 18 h before extraction of total RNA and analysis by DNA oligonucleotide microarray. The histogram shows median signal intensity on an arbitrary scale above background (mean plus two standard deviations of five negative control genes). The data were normalized against spiked RNA. Each datum point is the median signal intensity (n = 3). Data are shown for vgp150Δ (□) and the wild-type revertant vgp150R (▪). The positions for data pertaining to gp150 are indicated by an arrow above the profiles and those pertaining to surrounding genes by asterisks below the profiles.
gp150 is nonessential for virus replication in vitro but affects extracellular virus production.
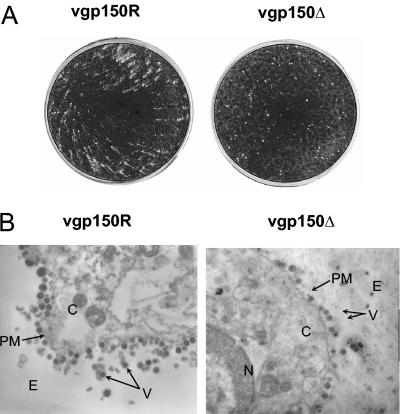
During construction of the virus recombinants we noticed that in the absence of an overlay of carboxymethyl cellulose or agarose, the plaque morphology of the gp150 mutant was restricted compared to the revertant and wild-type virus (Fig. 4A). This suggested a possible defect in release of extracellular virus. To test this hypothesis and to distinguish between virus released from the surface of infected cells and virus released by cellular lysis, infected BHK cells were examined by electron microscopic analysis. The results (Fig. 4B) showed that viral particles were released from the cell surface of infected cells by both vgp150Δ and vgp150R. Thus, the restricted plaque morphology was not due to an absolute defect in release of extracellular virus from the cell surface.
FIG. 4.
Plaque morphology and electron microscopic analysis of infected cells. (A) BHK-21 cells were infected with either vgp150R or gp150Δ, as indicated, at an MOI of 10−4. After 4 days the cells were fixed with formol saline and stained with toluidine blue. (B) BHK-21 cells were infected with either vgp150R or vgp150Δ, as indicated, at an MOI of 5 for 24 h. Cells were then fixed with glutaraldehyde, negatively stained with uranyl acetate, and examined by transmission electron microscopy. Magnification, ×10,000. Each panel shows an image of part of an individual infected cell. The positions of virus particles (V), extracellular space (E), cytoplasm (C), plasma membrane (PM), and nucleus (N) are as indicated.
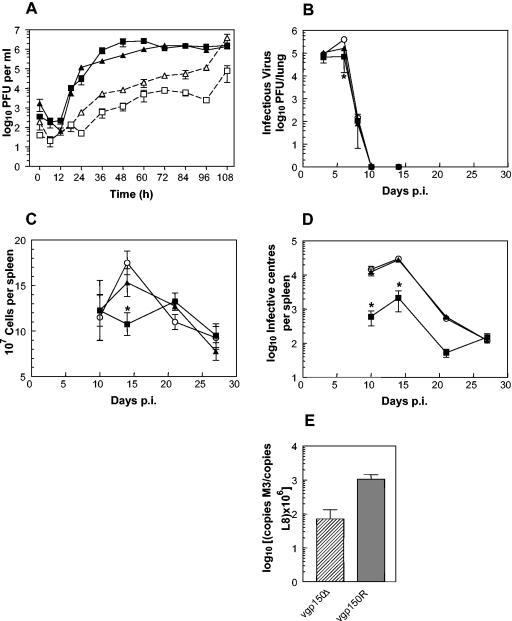
To analyze the replication of the gp150 mutant further, we performed standard in vitro growth curves. The one-step growth curves of vgp150Δ, vgp150R, and wild-type MHV-68 on BHK-21 cells (MOI of 5) were essentially identical (not shown). Likewise multistep growth curves (MOI of 0.01) between viruses were similar. We next performed multistep growth curves and analyzed cell-associated and cell-free virus separately. The results (Fig. 5A) showed that, although the curves for cell-associated virus were extremely similar, at 24 h p.i. and later the titers of extracellular vgp150Δ were markedly less (1 to 1.5 log10) than those of vgp150R.
FIG. 5.
Biological characterization of viruses. (A to D) Data were obtained from infections with vgp150Δ (squares), vgp150R (triangles), and parental wild-type virus (circles). All infections of mice (BALB/c female) were intranasal with 4 × 105 PFU of virus. In all panels, where indicated, bars represent the standard deviation from the mean, and asterisks represent a statistically significant difference from the revertant value. (A) In vitro multistep growth curve comparing replication of viruses in BHK-21 cells (MOI of 0.01). Titers were determined for both cell-associated (▪ and ▴) and cell-free (□ and ▵) virus. The data are representative of two independent experiments, each performed in duplicate. (B) Virus replication in the lung. The data shown are the mean virus titers obtained from four mice per group at each time point. (C) Total spleen cell number within infected mice. The data are the mean cell numbers obtained from four infected mice per group at each time point. (D) Latently infected cells in the spleens of infected mice as determined by IC assay. The data from four mice per group are shown at each time point. No preformed infectious virus was detected in this assay (data not shown). (E) Data obtained from real-time quantitative PCR analysis of the amount of viral DNA present relative to the amount of cellular L8 gene DNA present in spleen samples at 14 days p.i. The data were compiled from three separate analyses of four individual mice infected with either vgp150Δ or vgp150R as indicated.
These results indicated that gp150 is not essential for the production of infectious MHV-68 in vitro but that it has a role to play in the efficient release of infectious extracellular virus.
gp150 does not affect productive replication in the lung.
To assess the role of gp150 in vivo, cohorts of BALB/c mice were infected intranasally with vgp150Δ, vgp150R, and wild-type MHV-68. The degree of productive replication in the lungs was then assessed at selected time points by plaque assay. The titers of all three viruses were extremely similar and not significantly different at all time points, peaking at 6 days p.i. and being resolved below the level of detection by day 10 p.i. (Fig. 5B). Thus, gp150 does not play a role in the acute infection by MHV-68 in the lung.
gp150 is important for acute splenic mononucleosis.
To assess the role of gp150 in infection of spleen cells and establishment of latency, mice were infected intranasally as described above and analyzed at selected times p.i. First, the total spleen mononuclear cell numbers were assessed (Fig. 5C). Spleen mononuclear cell numbers in mice infected with wild-type MHV-68 and vgp150R increased to peak at day 14 p.i. and decreased to a baseline by day 28 p.i. In contrast, there was no overall significant increase in the spleen cell numbers of vgp150Δ-infected mice, and at day 14 p.i. the numbers were significantly lower than either wild-type or vgp150R-infected animals (P = <0.01). Thus, vgp150Δ was defective in its ability to cause production of the typical transient peak in splenic mononuclear cell numbers (splenic mononucleosis) seen after MHV-68 infection.
gp150 is important for the early expansion of latently infected cells in the spleen.
Next, the numbers of latently infected cells in the spleen were assessed by a virus reactivation or IC assay (Fig. 5D). The number of IC in mice infected with both vgp150R and wild-type MHV-68 were similar, peaking at day 14 p.i. and resolving to a baseline at 28 days p.i. Although IC were observed in vgp150Δ-infected mice and peaked at 14 days p.i., the number was significantly lower than with the other two viruses at days 10 (P = 0.01) and 14 (P = 0.001) p.i. In spite of the lower numbers observed at day 10 and 14 p.i., the long-term (baseline) level of IC established in vgp150Δ-infected mice was similar to that seen in mice infected with wild-type and revertant viruses. No preformed infectious virus was seen in these assays; thus, the number of IC was representative of the number of cells that can be reactivated from latency. Thus, although vgp150Δ was able to establish latency in the spleen, the initial typical spike in reactivatable latency was much reduced.
To check that there was no defect in the ability of the gp150 mutant virus to reactivate from latency, resulting in lower estimates of latently infected cells, we determined the viral genome copy number in the spleens of infected mice by quantitative real-time PCR. We have previously shown that genome copy number correlates directly with the number of latently infected cells in MHV-68-infected mice as determined by a limiting dilution reactivation assay (46). The results are shown in Fig. 5E. At day 14 p.i., it was found that viral DNA copy number in spleens obtained from mice infected with wild-type MHV-68 and vgp150R were ∼10-fold higher (1 copy in 103 copies of L8) than in vgp150Δ-infected animals (1 copy in 104 copies of L8). This finding indicated that, with all three viruses, viral DNA copy number was in good agreement with the IC assay. Thus, the reduced peak of IC at days 14 p.i. caused by vgp150Δ was due to a reduced number of latently infected cells and not a defect in reactivation.
gp150 is not involved in the spread from lungs to spleen.
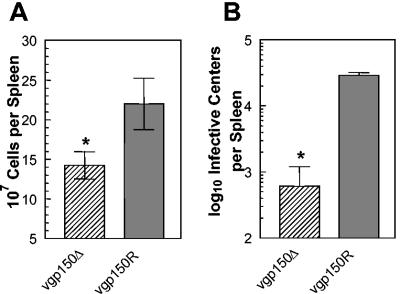
We surmised that one reason for the decreased mononucleosis and spike of latency in the spleens of mice infected with vgp150Δ was that there was a defect in its ability to traffic from the lung to the spleen. To address this, we infected mice with vgp150Δ and vgp150R via the intravenous route. This technique delivers the virus more directly to the spleen without the need to replicate in the lungs and traffic through secondary lymphoid tissue. Spleens from infected mice were analyzed for mononuclear cell number and IC at day 7 p.i., since this is the time at which the peak of splenic latency occurs in mice infected via the intravenous route (39). The results (Fig. 6) showed that the numbers of mononuclear cells and IC were extremely similar to those seen at 14 days p.i. after intranasal infection with both vgp150Δ and vgp150R. Thus, there were significantly lower numbers of spleen mononuclear cells (P = 0.005) and IC (P = 0.004) in vgp150Δ-infected mice. This indicated that gp150 did not play a role in the spread of virus from the lung to the spleen.
FIG. 6.
Characterization of latent infection in the spleen after intravenous infection. Mice (BALB/c female) were infected with 4 × 105 PFU of either vgp150Δ of vgp150R via the intravenous route for 7 days. (A) Total spleen cell number within infected mice. The mean cell numbers obtained from four infected mice per virus are shown. (B) Latently infected cells in the spleens of infected mice as determined by IC assay. Data from four mice per group are shown at each time point. No preformed infectious virus was detected in this assay; thus, the number of IC is representative of the number of latently infected cells. In both panels, bars represent the standard deviation from the mean, and asterisks represent a statistically significant difference from the revertant value.
gp150-deficient virus can still infect B cells.
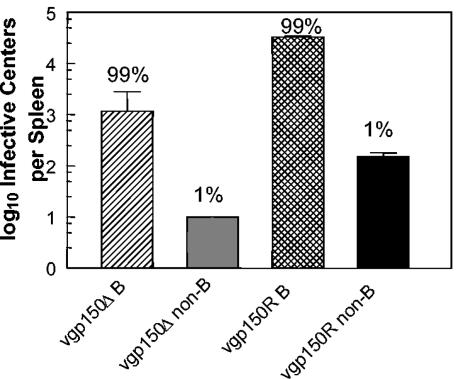
To determine whether vgp150Δ had an altered cellular tropism in the spleen, mice were infected intranasally with vgp150Δ and vgp150R, and spleens were harvested at 14 days p.i. Spleen cells were then fractionated into CD19+ (B lymphocytes) and CD19− cells (non-B cells) by using magnetic cell sorting (MACS) as described previously (14). The purity of the separated fractions was tested by FACS analysis and found to be >95%. The two cell fractions were analyzed for the presence of latent virus by using the IC assay. As shown in Fig. 7, as before, the overall numbers of IC were more than 10-fold greater in vgp150R-infected than in vgp150Δ-infected mice (3.3 × 104 versus 1.1 × 103, respectively). However, in both cases, the same proportion (99%) of IC was present within the CD19+-B-cell subpopulation. The distribution of vgp150Δ within different cell types in the spleen during mononucleosis therefore appeared to be similar to that of wild-type MHV-68 (40) in that the majority of IC were associated with B lymphocytes. This indicated that gp150 is not essential for infection of B cells and hence latency in B cells, but that its presence increases the frequency of latently infected cells during splenic mononucleosis.
FIG. 7.
Spleen type containing latently infected cells. Mice (BALB/c female) were infected intranasally with 4 × 105 PFU of either vgp150Δ or vgp150R for 14 days. Splenic B lymphocytes were then separated from other spleen cells by positive selection by MACS with CD19 microbeads. The numbers of latently infected cells were then determined by IC assay. Data from four mice per virus are shown. No preformed infectious virus was detected in this assay; thus, the number of IC is representative of the number of latently infected cells. Bars represent the standard deviation from the mean. Numbers above the bars represent the percentage of the observed mean total IC for each virus.
gp150 binds to B cells but not epithelial cells in vitro. Due to the deficit in B-cell infection, we wanted to determine whether gp150 was involved in binding selectively to B cells. To make a preliminary investigation of the role of gp150 in binding of virus to either B cells or epithelial cells, we made a soluble recombinant form of gp150 in E. coli. This consisted of the intact ectodomain of gp150 (i.e., without the N-terminal signal sequence, transmembrane anchor and carboxy-terminal domain) with the addition of a His6 fusion tag at the carboxy terminus to enable purification on Ni2+ charged resin. A bacterial system was chosen because of the high levels of protein that can be produced. Although a glycoprotein, gp150 is not highly glycosylated, containing only two potential N-linked glycosylation sites (35). Thus, we did not believe that the lack of glycosylation in E. coli would be a major problem. The purified recombinant protein (gp150-His) had an apparent molecular mass of 120 kDa on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (results not shown), which was in good agreement with that predicted for the gp150 ectodomain (35). Using this system we were consistently able to purify soluble recombinant gp150 at a level of ca. 1.3 mg/liter. As a control we produced and purified a GST-His6 (GST-His) fusion protein in a similar fashion.
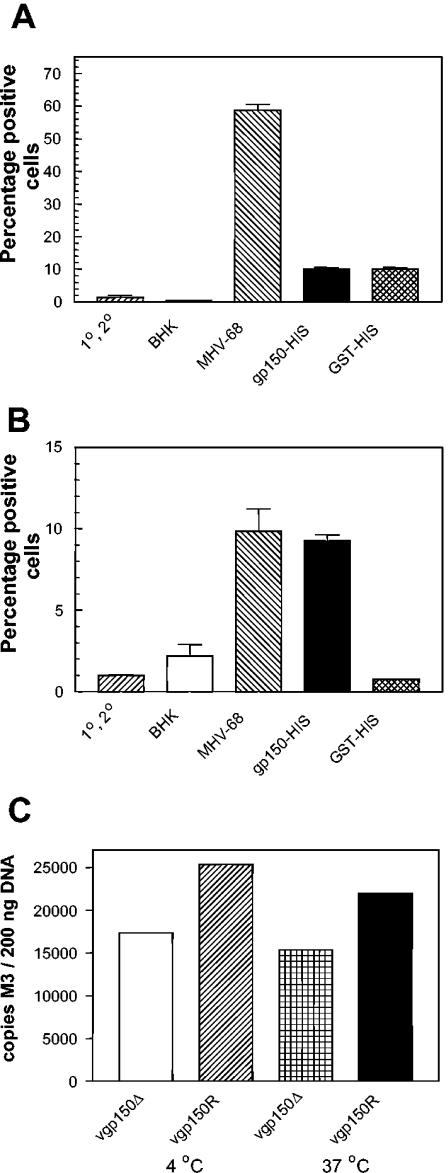
To assess binding of gp150 to cells, we used either a murine epithelial cell line, C127, that was known to support MHV-68 infection (35) or primary CD19+ murine B cells purified by MACS as described above. Recombinant gp150-His, GST-His, MHV-68 virus preparation, or control lysate from BHK-21 cells was incubated with the cells. Binding was then detected by FACS analysis with an antiserum generated in rabbits to a gp150-GST fusion protein. This reagent was known to contain high levels of antibodies to both gp150 and GST (35) and was therefore capable of detecting virus, recombinant gp150, and GST. The results of this analysis are shown in Fig. 8. As expected, MHV-68 bound in a specific fashion to epithelial cells (Fig. 8A) with ca. 60% of the cells being positive. In contrast, a much smaller percentage (ca. 10%) of cells were positive for gp150 binding, and this binding was not specific since the GST fusion protein bound to an equivalent percentage of cells. When splenic B cells were analyzed (Fig. 8B), it was found that MHV-68 again bound in a specific fashion, although the proportion of cells positive was much lower (ca. 10%). A similar proportion of cells bound recombinant gp150 protein as virus, this time in a specific fashion since only ∼1% of B cells were positive for GST-His binding. These results demonstrated that MHV-68 was able to bind to both epithelial cells and B cells, but to a higher proportion of the former. In contrast, gp150 only bound in a specific fashion to B cells.
FIG. 8.
Binding of MHV-68 and gp150 to cells. (A and B) Cells (106/well) were incubated with equimolar concentrations (20 μM) of fusion protein (gp150-His or GST-His), 107 PFU MHV-68, or BHK cell lysate (BHK) as a virus-negative control for 1.5 h at 37°C in the presence of sodium azide. Binding was detected by using a combination of rabbit anti-gp150-GST antibody, followed by swine anti-rabbit fluorescein isothiocyanate-conjugated antibody. Cells were formaldehyde fixed and analyzed by FACS. The marker for positive staining was positioned at 1% of staining with primary and secondary antibody alone. The bars denote the standard deviation of triplicate samples. (A) C127 murine epithelial cells; (B) primary B cells. (C) A total of 2 × 106 fresh mouse spleen cells were incubated with the indicated viruses at either 4°C or 37°C. After a washing step, the DNA copy number of bound virus was determined by quantitative PCR analysis. Numbers represent the mean of three determinations.
gp150 influences the binding of MHV-68 to lymphocytes in vitro.
Although the above in vivo and protein binding results strongly suggested a role for gp150 in B-cell infection, we had not formally proved this. To determine whether gp150 had a direct role in binding of virus to B cells, we compared the ability of either vgp150Δ or vgp150R to bind to either A20, a murine B-cell line, or freshly isolated mouse spleen cells. The level of binding was determined by incubating equal amounts of virus, as defined by DNA copy number, at 4°C with cells and then assessing the relative copy numbers bound by quantitative PCR.
In the first set of experiments we used virus prepared in vitro in BHK-21 cells. Surprisingly, the results showed that vgp150Δ bound substantially better than vgp150R to both A20 B cells and splenocytes (data not shown). Similar results were obtained when the relative levels of vgp150Δ and vgp150R bound and internalized after 4 h of infection at 37°C were compared (data not shown).
We surmised, based on evidence with EBV, that there may be differences in the capacity of virus to bind to lymphocytes depending on the cell type from which it was derived (8). Thus, in a subsequent set of experiments we tested infectious virus derived from the lungs of infected mice at 7 days p.i. The majority of productively infected cells at this time p.i. in the lung are alveolar epithelial cells (39). Thus, most of the virus should be derived from this cell type. The results (Fig. 8C) showed that vgp150Δ bound less well to splenocytes than did the wild-type vgp150R. Similar results were obtained when the levels of virus bound and internalized after 4 h of infection at 37°C were compared.
These results demonstrate that, although not essential, gp150 derived from infected lungs enhances the binding of MHV-68 to splenocytes. However, the cellular source of the virus is critical in determining the influence of gp150 on binding.
DISCUSSION
The human gammaherpesviruses EBV and KSHV both contain a glycoprotein (gp350/220 and K8.1, respectively) that mediates binding to target cells. However, there is no direct information on the role that these glycoproteins play in the pathogenesis of viral infection in vivo. MHV-68 particles contain an analogous glycoprotein, termed gp150 (35). The aim of the present study was to mutate the gene encoding gp150 so as to generate a virus deficient in gp150 and test the effect of this mutation on viral pathogenesis in a mouse model system. To do this, a recombinant MHV-68 was made wherein the majority of the gp150 gene (vgp150Δ) was replaced by a tetracycline resistance cassette. This virus did not express gp150, and the expression of other virus genes was not adversely affected as shown by global microarray analysis. The gp150 mutant virus replicated normally in vitro but displayed a partial defect in the extracellular release of virus. Likewise, the productive replication of vgp150Δ in the lungs of infected mice was similar to wild-type virus. The transient splenic mononucleosis and spike in latently infected cells associated with spread of MHV-68 to the spleen was significantly reduced in the absence of gp150, but the mutant virus was able to infect and become latent in B cells. We determined that a soluble, recombinant gp150 bound specifically to B cells but not to epithelial cells in culture. In line with this, virus defective in gp150 bound less well to splenocytes in vitro.
The nonessential nature of gp150 for virus infection in vitro in BHK-21 cells was not surprising since it had been shown that the analogous protein from EBV, gp350/220, was also nonessential in culture (22). The fact that the absence of gp150 did not influence the total growth kinetics in either one or multistep growth curves suggests that it does not play a fundamental role in virus binding to cells or cell-to-cell spread in BHK cells. Likewise, since intracellular infectious virus levels are similar in its absence, it does not seem likely that gp150 plays a significant role in virion maturation. However, a partial defect in cell-free virus production by vgp150Δ suggests that gp150 plays a role in release from the infected cell surface. This is a novel observation, and it is possible that the analogous EBV gp350/220 and KSHV K8.1 may fulfill similar functions. Further studies are needed to dissect the exact nature of the role of gp150 and analogous glycoproteins in extracellular virion release.
As for in vitro productive replication, gp150 was redundant for the acute replication of MHV-68 in the lungs of infected mice (Fig. 5B). Thus, gp150 is not important for the entry of virus into pulmonary epithelial cells, the primary site of acute MHV-68 productive replication (39), as well as spread within this location. Given that there was a partial defect in cell-free virus production with vgp150Δ, these results also strongly suggest that cell free virus does not play a major role in the acute pulmonary infection.
In contrast to the acute infection in the lung, the transient splenic mononucleosis and spike in latently infected cells that follows acute lung infection was significantly influenced by the lack of gp150 (Fig. 5C and D). This indicated either a defect in spread from the initial pulmonary focus to the spleen or a deficiency in the ability to infect relevant target cells in the spleen. Bypassing the lung infection by using intravenous inoculation excluded the former (Fig. 6). Although a number of cell types within the spleen are infected by MHV-68 (20, 52), infection of B cells is critical for the development mononucleosis and the spike in latency (38, 44, 45, 51). The proportions of B and non-B cells infected in the absence or presence of gp150 were similar, showing that an alternative spleen cell population was not targeted in its absence. Thus, our results strongly suggest that the observed reductions in the peak of mononucleosis and latency were most likely due to a defect in the ability to infect target B cells. A reduced mononucleosis and latency spike, followed by longer-term latency were still established by the gp150 mutant virus. Thus, although important for these functions, gp150 is not essential. This fits well with observations with the EBV analog gp350/220, which has been shown to be influential but not essential for the infection of B cells in vitro (22).
The positional analogs of MHV-68 gp150, EBV gp350/220 and KSHV K8.1, are involved in the infectious process by binding to target cells. K8.1 appears to interact with cellular heparan sulfate present on a range of target cells, but it is not yet known how essential K8.1-heparan sulfate reactions are for infection, particularly on lymphocytes (3, 49). The EBV gp350/220 binds to CD21 present on B cells (18) and is important but not essential for EBV infection of these cells (22). With both EBV and KSHV there are other glycoproteins known to be involved in infection. Specifically, the EBV gH/gL/gp42 complex binds to HLA class II (21, 24, 25, 32), the BMRF2 gene product binds integrins (43) and the KSHV gB binds to integrins (2). We show here that MHV-68 gp150 binds specifically to B cells and not epithelial cells in vitro and that gp150 enhances the binding of virus derived from lung tissue to splenocytes. Thus, gp150 would appear to be more similar in function to EBV gp350/220 than to K8.1. MHV-68 gp150, like gp350/220, enhances but is not essential for lymphocyte infection, which implicates other glycoprotein-receptor interactions. Here, complexes involving gH and gL are likely to be involved. gB is another candidate. However, the MHV-68 gB, like EBV gB, is unusual in that it does not contain the consensus [RX(R/K)RS] cleavage site present in most other herpesvirus gB molecules, and it is largely retained in the endoplasmic reticulum with only a small proportion, if any, being incorporated into virions (7, 36). Thus, MHV-68 gB is unlikely to be a major player in binding to target cells. The question of the molecules involved in MHV-68 binding to target cells is therefore complex and requires further study.
The in vitro binding data support our in vivo data showing a role for gp150 in MHV-68 pathogenesis in the spleen and not the lung. MHV-68 gp150 was shown to be important for the transient sharp increase in both spleen mononuclear cell and latently infected B-cell numbers, presumably by increasing the chances of B-cell infection in vivo. Extrapolation of these data to EBV pathogenesis suggests that in turn, gp350/220 may not be essential for infection or latency in vivo but may influence the development of clinically apparent EBV-associated mononucleosis by increasing the proportion of B cells that are infected.
Viral binding studies in vitro identified that the cellular source of MHV-68 was critical in determining the influence of gp150 on binding. Thus, whereas gp150-deficient virus derived from mouse lungs bound less well to splenocytes than did the wild type, the situation was reversed when virus derived from BHK-21 fibroblasts was used. This implies that there is a fundamental difference in the glycoprotein composition between virions derived from the two sources. Alveolar epithelial cells, not fibroblasts, are the principal natural source of infectious MHV-68, and so the binding results with this cell type are more reflective of the situation in vivo. Differences in the binding could be due to the specific type or relative proportion of glycoprotein species incorporated or the biochemical modification of glycoprotein species. There is a precedence for the former in that EBV derived from epithelial cells contains a high proportion of gH/gL complexes that contain gp42 and preferentially infect B cells, whereas virions originating from B cells contain a higher proportion of gH/gL complexes without gp42 and preferentially infect epithelial cells (8). Alternatively, we have observed differences in the glycosylation of MHV-68 gp150 depending on the context and cell type (J. P. Stewart, unpublished observations) which could affect the binding capacities of this protein. Clearly, as in EBV, the cellular source of virions can affect the binding properties of MHV-68 to target cells, and great care should taken in choosing an appropriate source for study.
While this study was under review, another study detailing a MHV-68 gp150 mutant was published (11) that confirmed many of our in vitro findings but differed fundamentally in that the gp150 mutant did not display any phenotypic difference from the wild type in vivo. The reasons for these differences are not immediately clear. However, the mutations made in gp150 by de Lima et al. would enable part of the protein to be expressed, and it is possible that this is somehow influencing the in vivo phenotype.
MHV-68 gp150 is clearly not essential in our laboratory system for all aspects of viral pathogenesis but plays a role in enhancing various aspects of the process. Since gp150 is apparently involved in the production of cell-free virus, it may be that it would naturally play a more important or essential role in spread from host to host. We have never been able to demonstrate spread from one animal to another in the laboratory, and therefore we currently do not have a system with which to test this hypothesis.
It has been suggested that EBV gp350/220 would be a good candidate antigen for a vaccine against EBV. We have previously observed that vaccination with a recombinant vaccinia virus expressing gp150 had a negative impact on acute splenic mononucleosis and latency but did not affect the initial infection in the lung or long-term latency (37). These exactly parallel the results with vgp150Δ. Taken together, this adds weight to the view that although an EBV vaccine based on gp350/220 might be expected to be effective against mononucleosis, protection against latency and subsequent tumorigenesis may require a different strategy.
Acknowledgments
We are grateful to C. Burgmeier and B. Steer for excellent technical assistance. We thank E. J. Usherwood and J. T. Sample for critical review of the manuscript.
This study was funded in part by a Royal Society of London University Research Fellowship (to J.P.S.), Biotechnology and Biological Sciences Research Council (United Kingdom) grant 15/C12782 (to J.P.S.), and Medical Research Council Studentship G78/5082 (to I.M.D.A). H.A. was supported by grants from the Deutsche Forschungsgemeinschaft, Ad 121/2-1, and from the BMBF (Nationales Genomforschungsnetz).
REFERENCES
- 1.Adler, H., M. Messerle, M. Wagner, and U. H. Koszinowski. 2000. Cloning and mutagenesis of the murine gammaherpesvirus 68 genome as an infectious bacterial artificial chromosome. J. Virol. 74:6964-6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akula, S. M., N. P. Pramod, F. Z. Wang, and B. Chandran. 2002. Integrin α3β1 (CD 49c/29) is a cellular receptor for Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) entry into the target cells. Cell 108:407-419. [DOI] [PubMed] [Google Scholar]
- 3.Birkmann, A., K. Mahr, A. Ensser, S. Yaguboglu, F. Titgemeyer, B. Fleckenstein, and F. Neipel. 2001. Cell surface heparan sulfate is a receptor for human herpesvirus 8 and interacts with envelope glycoprotein K8.1. J. Virol. 75:11583-11593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blackman, M. A., E. Flano, E. Usherwood, and D. L. Woodland. 2000. Murine gamma-herpesvirus-68: a mouse model for infectious mononucleosis? Mol. Med. Today 6:488-490. [DOI] [PubMed] [Google Scholar]
- 5.Blasdell, K., C. McCracken, A. Morris, A. A. Nash, M. Begon, M. Bennett, and J. P. Stewart. 2003. The wood mouse is a natural host for murid herpesvirus 4. J. Gen. l Virol. 84:111-113. [DOI] [PubMed] [Google Scholar]
- 6.Blaskovic, D., M. Stancekova, J. Svobodova, and J. Mistrikova. 1980. Isolation of five strains of herpesviruses from two species of free living small rodents. Acta Virol. 24:468. [PubMed] [Google Scholar]
- 7.Bortz, E., J. P. Whitelegge, Q. Jia, I. Atanasov, Z. H. Zhou, J. P. Stewart, T.-T. Wu, and R. Sun. 2004. Identification of proteins associated with murine gammaherpesvirus-68 virions. J. Virol. 78:3343-3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borza, C. M., and L. M. Hutt-Fletcher. 2002. Alternate replication in B cells and epithelial cells switches tropism of Epstein-Barr virus. Nat. Med. 8:594-599. [DOI] [PubMed] [Google Scholar]
- 9.Cardin, R. D., J. W. Brooks, S. R. Sarawar, and P. C. Doherty. 1996. Progressive loss of CD8+ T cell-mediated control of a gamma-herpesvirus in the absence of CD4+ T cells. J. Exp. Med. 184:863-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang, Y. E., and L. A. Laimins. 2000. Microarray analysis identifies interferon-inducible genes and Stat-1 as major transcriptional targets of human papillomavirus type 31. J. Virol. 74:4174-4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Lima, B. D., J. S. May, and P. G. Stevenson. 2004. Murine gammaherpesvirus 68 lacking gp150 shows defective virion release but establishes normal latency in vivo. J. Virol. 78:5103-5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Der, S. D., A. Zhou, B. R. Williams, and R. H. Silverman. 1998. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl. Acad. Sci. of the USA 95:15623-15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doherty, P. C., J. P. Christensen, G. T. Belz, P. G. Stevenson, and M. Y. Sangster. 2001. Dissecting the host response to a gamma-herpesvirus. Phil. Trans. R. Soc. London Ser. B Biol. Sci. 356:581-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dutia, B. M., J. P. Stewart, R. A. Clayton, H. Dyson, and A. A. Nash. 1999. Kinetic and phenotypic changes in murine lymphocytes infected with murine gammaherpesvirus-68 in vitro. J. Gen. Virol. 80:2729-2736. [DOI] [PubMed] [Google Scholar]
- 15.Ebrahimi, B., B. M. Dutia, K. L. Roberts, J. J. Garcia-Ramirez, P. Dickinson, J. P. Stewart, P. Ghazal, D. J. Roy, and A. A. Nash. 2003. Transcriptome profile of murine gammaherpesvirus-68 lytic infection. J. Gen. Virol. 84:99-109. [DOI] [PubMed] [Google Scholar]
- 16.Efstathiou, S., Y. M. Ho, and A. C. Minson. 1990. Cloning and molecular characterization of the murine herpesvirus 68 genome. J. Gen. Virol. 71:1355-1364. [DOI] [PubMed] [Google Scholar]
- 17.Ehtisham, S., N. P. Sunil-Chandra, and A. A. Nash. 1993. Pathogenesis of murine gammaherpesvirus infection in mice deficient in CD4 and CD8 T cells. J. Virol. 67:5247-5252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fingeroth, J. D., J. J. Weis, T. F. Tedder, J. L. Strominger, P. A. Biro, and D. T. Fearon. 1984. Epstein-Barr virus receptor of human B lymphocytes is the C3d receptor CR2. Proc. Natl. Acad. Sci. USA 81:4510-4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flano, E., S. M. Husain, J. T. Sample, D. L. Woodland, and M. A. Blackman. 2000. Latent murine gamma-herpesvirus infection is established in activated B cells, dendritic cells, and macrophages. J. Immunol. 165:1074-1081. [DOI] [PubMed] [Google Scholar]
- 20.Flano, E., I. J. Kim, J. Moore, D. L. Woodland, and M. A. Blackman. 2003. Differential gammaherpesvirus distribution in distinct anatomical locations and cell subsets during persistent infection in mice. J. Immunol. 170:3828-3834. [DOI] [PubMed] [Google Scholar]
- 21.Haan, K. M., W. W. Kwok, R. Longnecker, and P. Speck. 2000. Epstein-Barr virus entry utilizing HLA-DP or HLA-DQ as a coreceptor. J. Virol. 74:2451-2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Janz, A., M. Oezel, C. Kurzeder, J. Mautner, D. Pich, M. Kost, W. Hammerschmidt, and H. J. Delecluse. 2000. Infectious Epstein-Barr virus lacking major glycoprotein BLLF1 (gp350/220) demonstrates the existence of additional viral ligands. J. Virol. 74:10142-10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, I. J., E. Flano, D. L. Woodland, and M. A. Blackman. 2002. Antibody-mediated control of persistent gamma-herpesvirus infection. J. Immunol. 168:3958-3964. [DOI] [PubMed] [Google Scholar]
- 24.Li, Q., M. K. Spriggs, S. Kovats, S. M. Turk, M. R. Comeau, B. Nepom, and L. M. Hutt-Fletcher. 1997. Epstein-Barr virus uses HLA class II as a cofactor for infection of B lymphocytes. J. Virol. 71:4657-4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, Q., S. M. Turk, and L. M. Hutt-Fletcher. 1995. The Epstein-Barr virus (EBV) BZLF2 gene product associates with the gH and gL homologs of EBV and carries an epitope critical to infection of B cells but not of epithelial cells. J. Virol. 69:3987-3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Messerle, M., I. Crnkovic, W. Hammerschmidt, H. Ziegler, and U. H. Koszinowski. 1997. Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA 94:14759-14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Molesworth, S. J., C. M. Lake, C. M. Borza, S. M. Turk, and L. M. Hutt-Fletcher. 2000. Epstein-Barr virus gH is essential for penetration of B cells but also plays a role in attachment of virus to epithelial cells. J. Virol. 74:6324-6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nash, A. A., B. M. Dutia, J. P. Stewart, and A. J. Davison. 2001. Natural history of murine gamma-herpesvirus infection. Phil. Trans. R. Soc. London Ser. B Biol. Sci. 356:569-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raab, M. S., J. C. Albrecht, A. Birkmann, S. Yaguboglu, D. Lang, B. Fleckenstein, and F. Neipel. 1998. The immunogenic glycoprotein gp35-37 of human herpesvirus 8 is encoded by open reading frame K8.1. J. Virol. 72:6725-6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith, D. B., and K. S. Johnson. 1988. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene 67:31-40. [DOI] [PubMed] [Google Scholar]
- 31.Spear, P. G., and R. Longnecker. 2003. Herpesvirus entry: an update. J. Virol. 77:10179-10185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spriggs, M. K., R. J. Armitage, M. R. Comeau, L. Strockbine, T. Farrah, B. Macduff, D. Ulrich, M. R. Alderson, J. Mullberg, and J. I. Cohen. 1996. The extracellular domain of the Epstein-Barr virus BZLF2 protein binds the HLA-DR beta chain and inhibits antigen presentation. J. Virol. 70:5557-5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stevenson, P. G., J. M. Boname, B. de Lima, and S. Efstathiou. 2002. A battle for survival: immune control and immune evasion in murine gamma-herpesvirus-68 infection. Microbes Infect. 4:1177-1182. [DOI] [PubMed] [Google Scholar]
- 34.Stewart, J. P. 1999. Of mice and men: murine gammaherpesvirus 68 as a model. Epstein-Barr Virus Rep. 6:31-35. [Google Scholar]
- 35.Stewart, J. P., N. J. Janjua, S. D. Pepper, G. Bennion, M. Mackett, T. Allen, A. A. Nash, and J. R. Arrand. 1996. Identification and characterization of murine gammaherpesvirus 68 gp150: a virion membrane glycoprotein. J. Virol. 70:3528-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stewart, J. P., N. J. Janjua, N. P. Sunil-Chandra, A. A. Nash, and J. R. Arrand. 1994. Characterization of murine gammaherpesvirus 68 glycoprotein B (gB) homolog: similarity to Epstein-Barr virus gB (gp110). J. Virol. 68:6496-6504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stewart, J. P., N. Micali, E. J. Usherwood, L. Bonina, and A. A. Nash. 1999. Murine gamma-herpesvirus 68 glycoprotein 150 protects against virus-induced mononucleosis: a model system for gamma-herpesvirus vaccination. Vaccine 17:152-157. [DOI] [PubMed] [Google Scholar]
- 38.Stewart, J. P., E. J. Usherwood, A. Ross, H. Dyson, and T. Nash. 1998. Lung epithelial cells are a major site of murine gammaherpesvirus persistence. J. Exp. Med. 187:1941-1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sunil-Chandra, N. P., S. Efstathiou, J. Arno, and A. A. Nash. 1992. Virological and pathological features of mice infected with murine gamma-herpesvirus 68. J. Gen. Virol. 73:2347-2356. [DOI] [PubMed] [Google Scholar]
- 40.Sunil-Chandra, N. P., S. Efstathiou, and A. A. Nash. 1992. Murine gammaherpesvirus 68 establishes a latent infection in mouse B lymphocytes in vivo. J. Gen. Virol. 73:3275-3279. [DOI] [PubMed] [Google Scholar]
- 41.Tibbetts, S. A., L. F. van Dyk, S. H. Speck, and H. W. t. Virgin. 2002. Immune control of the number and reactivation phenotype of cells latently infected with a gammaherpesvirus. J. Virol. 76:7125-7132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tripp, R. A., A. M. Hamilton-Easton, R. D. Cardin, P. Nguyen, F. G. Behm, D. L. Woodland, P. C. Doherty, and M. A. Blackman. 1997. Pathogenesis of an infectious mononucleosis-like disease induced by a murine gamma-herpesvirus: role for a viral superantigen? J. Exp. Med. 185:1641-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tugizov, S. M., J. W. Berline, and J. M. Palefsky. 2003. Epstein-Barr virus infection of polarized tongue and nasopharyngeal epithelial cells. Nat. Med. 9:307-314. [DOI] [PubMed] [Google Scholar]
- 44.Usherwood, E. J., A. J. Ross, D. J. Allen, and A. A. Nash. 1996. Murine gammaherpesvirus-induced splenomegaly: a critical role for CD4 T cells. J. Gen. Virol. 77:627-630. [DOI] [PubMed] [Google Scholar]
- 45.Usherwood, E. J., J. P. Stewart, K. Robertson, D. J. Allen, and A. A. Nash. 1996. Absence of splenic latency in murine gammaherpesvirus 68-infected B cell-deficient mice. J. Gen. Virol. 77:2819-2825. [DOI] [PubMed] [Google Scholar]
- 46.Usherwood, E. J., K. A. Ward, M. A. Blackman, J. P. Stewart, and D. L. Woodland. 2001. Latent antigen vaccination in a model gammaherpesvirus infection. J. Virol. 75:8283-8288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Virgin, H. W., P. Latreille, P. Wamsley, K. Hallsworth, K. E. Weck, A. J. Dal Canto, and S. H. Speck. 1997. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J. Virol. 71:5894-5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Virgin, H. W., and S. H. Speck. 1999. Unraveling immunity to gamma-herpesviruses: a new model for understanding the role of immunity in chronic virus infection. Curr. Opin. Immunol. 11:371-379. [DOI] [PubMed] [Google Scholar]
- 49.Wang, F. Z., S. M. Akula, N. P. Pramod, L. Zeng, and B. Chandran. 2001. Human herpesvirus 8 envelope glycoprotein K8.1A interaction with the target cells involves heparan sulfate. J. Virol. 75:7517-7527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang, X., W. J. Kenyon, Q. Li, J. Mullberg, and L. M. Hutt-Fletcher. 1998. Epstein-Barr virus uses different complexes of glycoproteins gH and gL to infect B lymphocytes and epithelial cells. J. Virol. 72:5552-5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weck, K. E., M. L. Barkon, L. I. Yoo, S. H. Speck, and H. W. Virgin. 1996. Mature B cells are required for acute splenic infection, but not for establishment of latency, by murine gammaherpesvirus 68. J. Virol. 70:6775-6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weck, K. E., S. S. Kim, H. W. Virgin, and S. H. Speck. 1999. Macrophages are the major reservoir of latent murine gammaherpesvirus 68 in peritoneal cells. J. Virol. 73:3273-3283. [DOI] [PMC free article] [PubMed] [Google Scholar]