Abstract
Virion infectivity factor (Vif) protein of human immunodeficiency virus type 1 (HIV-1) is essential for the productive infection of primary human CD4 T lymphocytes and macrophages. Vif overcomes the HIV-inhibitory effects of cellular factor APOBEC3G, which has cytidine deaminase activity. We previously reported the isolation of a Vif-interacting ring finger protein, Triad 3, from a human leukocyte cDNA library, using the yeast two-hybrid system. The full-length cellular protein homologue of Triad 3 has been recently identified as the zinc finger protein inhibiting NF-κB (ZIN). Sequence analysis indicates that Triad 3 protein contains all four major ring-like motifs of ZIN. We report here that ZIN binds to purified Vif in vitro and that Triad 3/ZIN interacts with HIV-1 Vif in transfected human 293T cells, as demonstrated by coimmunoprecipitation. To test the biological relevance of this interaction, we produced infectious HIV-1 NL4.3 in the presence or absence of cotransfected ZIN. HIV-1 NL4.3 virus stocks produced in the presence of exogenously expressed ZIN were twofold less infectious in a single-cycle infectivity assay than virus produced in the absence of exogenous ZIN. It was further shown that cells infected with HIV NL4.3 virus stocks produced in the presence of exogenously expressed ZIN were impaired in viral DNA synthesis by twofold. The impairment in viral reverse transcription and the reduction in single-cycle viral infectivity were both shown to be dependent on the presence of Vif in the virus producer cells. The possible mechanisms by which ZIN interferes with the early events of HIV-1 replication are discussed.
Human immunodeficiency virus type 1 (HIV-1) Vif (virion infectivity factor) is a phosphorylated 23-kDa protein conserved among all primate and most other lentiviruses. Vif is expressed during the late stage of virus replication, is present in the cell nucleus and cytoplasm, and colocalizes with the major HIV structural protein Gag (2, 9, 10, 29, 32, 35). In cell culture, Vif-defective HIV-1 is able to replicate in some T-lymphoblastoid cell lines (termed permissive cells), such as CEM-ss and SupT1, but not in other cell types (termed nonpermissive), which include H9 and HUT-78 cells (8, 24, 33, 41). Human primary CD4 T cells and macrophages, the target cell types for HIV infection in vivo, are all nonpermissive cells and require Vif function for productive viral infection. The existence of permissive and nonpermissive cell types for Vif-defective HIV strongly suggests the involvement of cellular factor(s) in Vif related functions. Early experiments had suggested the presence of an inhibitor(s) of HIV replication in nonpermissive cells that could be overcome or neutralized by Vif during viral replication (24, 33). More recently a cellular factor, CEM15, was identified. CEM15 is present only in nonpermissive cell types and when transfected and expressed in permissive cells rendered them nonpermissive, consistent with the action of an inhibitor of HIV replication (30). CEM15 is identical to apolipoprotein B mRNA editing enzyme catalytic polypeptide-like 3G (APOBEC3G), a cytidine deaminase that introduces C-to-U changes into the minus strand of newly synthesised HIV-1 viral DNA in infected target cells in the absence of functional Vif (13, 22). Mariani et al. further demonstrated that Vif binds to APOBEC3G in producer cells and prevents APOBEC3G incorporation into HIV-1 virions, thereby preventing G-to-A mutations in newly synthesized viral DNA in infected cells (27). Additionally Vif also induces degradation of APOBEC3G through interaction with the host Skp1-Cullin-F-box (SCF)-like ubiquitin ligase complex (31, 42). Thus Vif interacts with an important cell factor, APOBEC3G, thereby suppressing the host inhibitory effects on HIV replication. Vif, however, has other properties, such as binding viral RNA (19, 31) and NC (16), and has been shown to affect the stability of the viral nucleoprotein core (15, 28, 36). Despite earlier controversial reports, recent evidence suggests that Vif is incorporated into virions (17, 18, 23). These properties of Vif implicate other potential functions independent of APOBEC3G. Indeed a number of cellular factors have been shown to interact with Vif. These include tyrosine kinase Hck (14), an ATP-binding protein, HP68 (45), and lymphocyte-specific nuclear body protein Sp140 (25). However, the biological significance of these cellular factors in relation to their association with Vif is not clear.
By using a high-stringency yeast two-hybrid screening, we previously identified a Vif-interacting protein termed Triad 3 that was derived from a human leukocyte cDNA library (21). Triad 3 is member of a family of proteins possessing a novel ring finger structure that is thought to mediate protein-protein interactions (40). Triad 3 has also been identified as the partial protein sequence of a recently reported cellular protein named zinc finger protein inhibiting NF-κB (ZIN). ZIN is a cytoplasmic protein in mammalian cells that colocalizes with a serine/threonine protein kinase receptor-interacting protein (RIP) and inhibits RIP-mediated NF-κB activation and tumor necrosis factor alpha (TNF-α) signal transduction (5). In the present study, we report that ZIN directly interacts with purified HIV-1 Vif in vitro and that ZIN interacts with Vif in human 293T cells expressing both proteins. We further show that HIV NL4.3 virus stocks produced in the presence of exogenously expressed ZIN are less infectious in a single-cycle viral infectivity assay and are impaired in the early stages of viral replication before and/or at reverse transcription in newly infected cells. The impairment in viral DNA synthesis and reduction in single-cycle viral infectivity were shown to be dependent on the presence of Vif in the virus producer cells.
MATERIALS AND METHODS
Cells.
293T, HeLa, and COS-7 cells were grown in Dulbecco's modified Eagle's medium (DMEM); and H9, CEM-ss, A3.01 and HUT-78 cells in RPMI 1640 medium supplemented with 10% fetal calf serum, 2 mM glutamine, and antibiotics. HeLa-CD4-long terminal repeat-β-galactosidase indicator cells (hereafter, HeLa-CD4-LTR-β-gal cells) were obtained from the National Institutes of Health AIDS Research and Reference Reagents Program and were maintained in DMEM with 10% fetal calf serum (FCS) and antibiotics supplemented with 200 μg of G418 and 100 μg of hygromycin per ml. Human peripheral blood lymphocytes (PBLs) were isolated from fresh blood of healthy donors with Lymphoprep (Vital Diagnostics), and monocytes were separated from peripheral blood mononuclear cells by their adherence to plastic, cultured in vitro, and matured to monocyte-derived macrophages as previously described (3). All cells were cultured at 37°C in an atmosphere containing 5% CO2.
DNA constructs.
The mammalian expression plasmid expressing Triad 3 (pHA-Triad 3) was produced by PCR amplification of Triad 3 cDNA isolated from a human leukocyte cDNA library (Clontech), using the flanking sequence-specific primer pair TrE (5′-CACACACGAATTCGAGGGGAATTTCCATTCGAG 3′) and TrX (5′-CACACACCTCGAGAGTGGGATCGGTCCAGAGAG 3′) and cloning it downstream of the hemagglutinin (HA) epitope tag sequence and cytomegalovirus (CMV) immediate-early promoter of pCMV-HA (Clontech). Likewise NVBP-1 cDNA (21) was PCR amplified from a human leukocyte cDNA library (Clontech), cloned into pCMV-HA, and named pHA-NVBP-1. Full-length ZIN cDNA was amplified by PCR with primer pair ZnE-F (5′-CACACACGAATTCTATGGCCGACTTCAAAGTGC-3′) and ZnX-R (5′-CACACACGGATCCTCAGAAGCGATGCCGCGGCT-3′) from first-strand cDNA synthesized from total RNA extracted from A3.01 cells by using a poly(A)-specific primer (Smart RACE cDNA amplification kit; Clontech). This was cloned downstream of the Flag epitope sequence and CMV promoter into p3XFlag-CMV-10 (Sigma) to generate the expression plasmid encoding ZIN (pFlag-ZIN). The expression plasmid expressing HIV Vif (pMyc-Vif) was generated by cloning the full-length vif gene, amplified by PCR from pNL4.3 HIV-1 plasmid DNA, into the protein expression plasmid pCMV-Myc (Clontech), downstream of the c-Myc epitope tag sequence. In addition, the vif gene was cloned into the plasmid pGEX 2T (Amersham Biosciences) downstream of the glutathione S-transferase (GST) sequence, hereafter called pGST-Vif, for GST-Vif fusion protein expression in Escherichia coli under the control of the chemically inducible tac promoter. The cDNA sequences of all the above constructs were confirmed by sequencing with ABI Prism BigDye terminator sequencing reaction kits (PE Applied Biosystems) and plasmid-specific forward and reverse primers. Homology search of nucleotide and deduced protein sequences was performed with the GenBank database.
RNA extraction and Northern blot analysis.
Total RNA extracts were made from different cell types with the TriZol reagent (Invitrogen). The RNA was separated on 1% agarose-formaldehyde gels, transferred to an N+ membrane (Amersham Pharmacia), and analyzed by Northern blot hybridization with a 32P-Megaprimer (Amersham Biosciences)-labeled Triad3 cDNA probe in Prehyb/Hyb solution NorthernMax (Ambion). 32P-labeled bands were visualized and quantified with a PhosporImager (Molecular Dynamics).
Transient cell transfection.
For DNA transfection, 100-mm-diameter dishes were seeded with human 293T cells at a density of 2 × 106 cells in 12 ml of DMEM cell culture medium and maintained at 37°C and 5% CO2. The following day, separate cultures were singly transfected with 5 μg of plasmid DNA of either pMyc-Vif or pHA-Triad3 or pFlag-ZIN by using SuperFect transfection reagent (QIAGEN) according to the manufacturer's recommendations. Separate cultures were cotransfected with 5 μg each of pMyc-Vif and pHA-Triad3 plasmid DNA or 5 μg each of pMyc-Vif and pFlag-ZIN plasmid DNA. Each transfected cell culture was reseeded into two 100-mm-diameter dishes at 24 h after transfection and incubated for a further 24 h before harvesting. Production of ZIN protein for use in the GST fusion protein pull-down assay was achieved in a similar manner by transfecting 10 μg of pFlag-ZIN plasmid DNA into 293T cells contained in a 100-mm-diameter dish.
Coimmunoprecipitation and Western blot analysis.
Forty-eight hours after transfection, cells were harvested and lysed in 800 μl of lysis buffer containing 1% Nonidet P-40, 50 mM Tris-HCl (pH 8.0), 100 mM NaCl, and 1 mM EDTA. The lysates were sonicated for three separate 10-s pulses at setting no. 3 in an Ultrasonic cell disruptor (Microson), followed by centrifugation at 13,500 rpm at 4°C in a Biofuge 17 RS centrifuge (Hereaus) for 10 min (min) to remove cell debris. Clarified cell lysates were immunoprecipitated by adding 1.8 μg of HA-specific rabbit polyclonal antiserum (Clontech), 2 μg of HA-specific monoclonal antibody (no. 262K; Cell Signaling Technology), or 2 μg of Flag-specific monoclonal antibody (Sigma) and then incubating for 12 h at 4°C. Next 100 μl of 50% suspension of protein A-Sepharose CL-4B beads (Pharmacia) was added, and the reaction mixtures were incubated for a further 1 h. Sepharose CL-4B beads containing immunoprotein complexes were pelleted, washed three times in lysis buffer, and then boiled in denaturing sodium dodecyl sulfate (SDS) protein loading buffer. Proteins were resolved on an SDS-PAGE (12% polyacrylamide) gel and then electroblotted onto Hybond-C membrane (Amersham). Protein blots were blocked in TBST (50 mM Tris [pH 7.4], 135 mM NaCl, 0.1% Tween 20) containing 5% milk for 30 to 40 min before incubation with mouse anti-Myc antibody (no. 9E10; Zymed) or Vif-specific rabbit antiserum (no. 2221; NIH AIDS Research and Reagents Program). The secondary immunoreaction was performed with horseradish peroxidase-conjugated goat anti-mouse (Pierce) or goat anti-rabbit immunoglobulin G (Pierce) and detected by using SuperSignal West Dura extended duration substrate (Pierce).
GST fusion protein pull-down assay.
Plasmids pGST and pGST-Vif were transformed into E. coli BL21 cells by the CaCl2 method. At an optical density at 600 nm (OD600) of 0.8, the cells were induced with isopropyl-β-d-thiogalactopyranoside (IPTG; 0.2 mM final concentration) at 30°C for 3 h and then lysed in buffer containing 25 mM Tris/HCl (pH 7.4), 150 mM NaCl, 10% glycerol, 1 mM dithiothreitol (DTT), protease inhibitor cocktails (Complete Mini; Roche), and 0.05% Triton X-100, followed by three 10-s pulses of sonication as described above. Glutathione-Sepharose CL-4B beads (Pharmacia Biotech) were incubated with the lysates for 1 h, and the beads containing the bound GST-Vif fusion protein or the bound GST protein were pelleted and extensively washed with lysis buffer. Protein contents were quantified against known amounts of bovine serum albumin on Coomassie blue-stained SDS-PAGE gels. To express ZIN, 293T cells were transfected with the pFlag-ZIN expression plasmid and lysed in the same lysis buffer and sonicated as described above, followed by centrifugation in a Biofuge 17 RS centrifuge at 13,500 rpm at 4°C for 10 min to remove cell debris. Fresh glutathione-Sepharose CL-4B beads were added to the pFlag-ZIN-transfected cell lysate for 1 h and then removed by centrifugation in order to preclear nonspecific glutathione-protein interactions in the lysate. Equal quantities of GST-Vif fusion protein or GST protein, bound to the glutathione Sepharose CL-4B beads, were separately added to the precleared pFlag-ZIN cell lysates and incubated overnight at 4°C. The bound proteins from the ZIN-expressing cell lysates (pull-down assays), were pelleted with the beads at 2,000 rpm in a microcentrifuge, washed, and separated on SDS-PAGE gels. The separated proteins were then analyzed by Western blotting with anti-Flag antibody to detect the pulled-down ZIN, followed by anti-GST antibody to confirm the presence of GST protein or GST-Vif fusion protein in samples as appropriate, and finally by anti-Vif polyclonal antibody to confirm Vif in the GST-Vif fusion protein samples. This was performed sequentially by using the same filter with successive stripping of the previous antibody.
Single-cycle viral infectivity assay.
293T cells were cotransfected with either pNL4.3 together with pFlag-ZIN or control plasmid pFlag, or with pNL4.3(Δvif) together with pFlag-ZIN or control plasmid pFlag. The culture medium from each transfected cell culture was collected 48 h after transfection and filtered with a 0.2-μm filter. Virus p24 levels in the culture medium were then measured in triplicate from serial dilutions of culture medium. For the single-cycle viral infectivity assay, infection was carried out in HeLa-CD4-LTR-β-gal cells as described previously (20). Briefly, individual wells of a 48-well plate were preseeded with 1.5 × 104 HeLa-CD4-LTR-β-gal cells 24 h before infection with 10−1 to 10−3 dilutions of 400-ng/ml p24 virus stock and incubation in a 100-μl virus inoculum for 2 h. Following addition of fresh culture medium, cells were incubated for a further 30 h and then washed in PBS and fixed in 1% formaldehyde in PBS for 10 min. Cells were again washed three times in PBS before incubation in X-Gal (5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid) staining solution (5 mM potassium ferrocyanide, 5 mM potassium ferricyanide, 2 mM MgCl2, 1-mg/ml X-Gal) for exactly 50 min. The reaction was stopped by removing the staining solution and washing the cells twice in PBS. Finally blue cells were counted under a microscope and scored in wells corresponding to the virus dilution in which at least 80 to 100 blue colonies per well were present.
HIV DNA levels following infection.
To measure the levels of reverse transcription in newly infected HUT-78 cells, equal amounts of virus inoculum, measured as 10 ng of HIV p24 in the culture medium from 293T cells cotransfected with either pNL4.3 and pFlag-ZIN, pNL4.3 and pFlag, pNL4.3(Δvif) and pFlag-ZIN, or pNL4.3(Δvif) and pFlag, were separately added to HUT-78 cells, and the virus was adsorbed by centrifugal enhancement for 1 h at 37°C (39). Cells were then washed twice in fresh medium and divided into duplicate wells. The wells were harvested 16 and 24 h postinfection, and extrachromosomal DNA was extracted by the Hirt method as previously described (38, 39). HIV DNA levels present in the Hirt supernatant were quantified against pretitrated HIV copy number standards (39) by real-time PCR with a primer pair, SS1 (5′-CTAACTAGGGAACCCACTGC-3′) and SS2b (5′-GGGTCTGAGGGATCTCTAGT-3′), specific for strong-stop DNA (U5-U5) and a primer pair, NL4-F (5′-AGATCTCTCGACGCAGGACT-3′) and NL4-R (5′-TCTCTCTCCTTCTAGCCTCC-3′), specific for the U5-Gag region reverse transcription product.
RESULTS
Vif-interacting proteins Triad 3 and ZIN.
To identify potential Vif-interacting cellular factors, we previously screened a human leukocyte cDNA library and found eight expressed cDNA clones that interacted with HIV-1 Vif in yeast cells (21). One clone encoded a 218-amino-acid peptide with complete homology to a ring finger protein called Triad 3. More recently, a full-length version of Triad 3 has been identified and described as zinc finger protein inhibiting NF-κB protein, or ZIN (5). Sequence analysis of Triad 3 demonstrates complete homology to part of ZIN which contains all four ring-like domains (RLDs) in the central portion of the full-length protein (GenBank accession no. NP_061884) (Fig. 1). In order to further investigate the association of full-length Triad 3 (subsequently referred to as ZIN) with Vif, the complete protein coding region of ZIN was amplified by PCR, using cDNA synthesized from RNA extracted from A3.01 cells and primers complementary to the published ZIN mRNA sequence (GenBank accession no. AY062174). A single product of 1,464 bp was obtained and cloned into the pFlag protein expression vector. Analysis of the cloned cDNA sequence showed that it encoded a 488-amino-acid protein that was identical to the published ZIN protein sequence except for a leucine instead of a phenylalanine at position 30. The Triad 3 sequence we isolated has a single proline-to-alanine amino-acid change at position 219 (according to the ZIN sequence numbering), which is outside the four RLDs and the proline-rich domain. ZIN protein contains a central segment with four RLDs, a C-terminal proline-rich domain, and an amino-terminal sequence of unknown function. The Triad 3 sequence maps to the central part of ZIN, which includes the four RLDs but not the N- and C-terminal sequences (Fig. 1). We therefore expected that most structural/functional properties of Triad 3, including its interaction with Vif, would likely be shared with ZIN, but not all functions of ZIN could be performed by Triad 3.
FIG. 1.
Sequence analysis of Triad 3 and ZIN. Deduced protein sequences of the 488-amino-acid ZIN and the 218-amino-acid Triad 3 sequences are shown. These sequences were determined in the present study. The GenBank accession no. for the previously published ZIN protein sequence is NP_061884. Triad 3 amino acid residues that are identical to the ZIN sequence are marked with asterisks. The two amino acid residues different from the published ZIN sequence are circled (see text). RLDs are enclosed by open boxes. A proline-rich domain (PRD) is underlined.
Triad 3/ZIN mRNA is expressed in both permissive and nonpermissive cells.
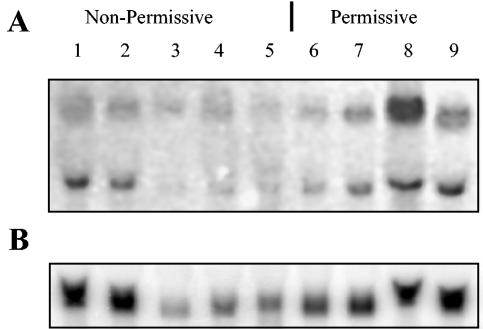
To investigate whether Triad 3/ZIN is expressed in nonpermissive and permissive cells, total cellular RNA was extracted from a variety of different cell types and the levels of Triad 3 mRNA determined by Northern blot analysis. RNA was extracted in triplicate experiments from freshly isolated primary human PBLs, macrophages, and other nonpermissive cell lines, including H9, A3.01, and HUT-78 cells, as well as from permissive cell lines 293T, CEM-ss, COS-7, and HeLa cells. A typical Northern blot is shown (Fig. 2A). The relative mRNA levels were compared by normalization against GAPDH mRNA levels detected in the same sample (Fig. 2B). Triad 3/ZIN mRNA was detected in all cell types in various amounts, as two alternatively spliced species approximately 3.0 and 6.0 kb in size. The Triad 3 cDNA probe used spans the entire central domain of ZIN and would detect all ZIN mRNA species. Detection of two distinct ZIN mRNA species has also been reported by Chen et al. (5). Estimation of the relative mRNA levels in all cell types suggests that Triad 3/ZIN mRNA levels were similar in all cell types regardless of their permissiveness for replication of Δvif HIV.
FIG. 2.
Triad 3/ZIN mRNA is expressed in permissive and nonpermissive cells. Northern blot hybridization of total RNA extracted from nonpermissive HUT-78, H9, PBLs, macrophages, and A3.01 cells (lanes 1 to 5) and permissive 293T, CEM-ss, Cos-7, and HeLa cells (lanes 6 to 9). The filter was first probed with 32P-labeled Triad 3 cDNA (A) and then stripped and reprobed with 32P-labeled GAPDH cDNA (B).
ZIN interacts with purified Vif in vitro.
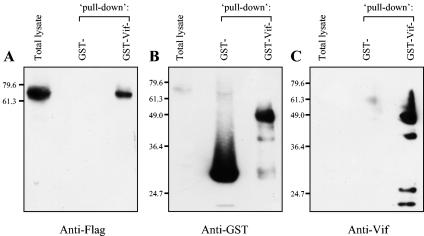
To investigate whether ZIN interacts with Vif, glutathione Sepharose CL-4B beads were complexed with equal quantities of either E. coli-expressed GST-Vif fusion protein or GST protein itself, washed to remove bacterial contaminants, and incubated with precleared 293T cell lysates expressing Flag-ZIN fusion protein. The glutathione Sepharose CL-4B beads together with associated proteins (which include GST protein or GST-Vif fusion protein and any pulled-down proteins) were then pelleted from 293T cell lysates, and proteins associated with the beads were analyzed by SDS-PAGE gel electrophoresis and Western blotting. The GST-Vif fusion protein, but not GST protein itself, was able to pull down Flag-tagged ZIN protein expressed in the 293T cells. This was shown by Western blot detection of Flag-tagged ZIN protein, approximately 61 kDa in size, from total pFlag-ZIN-transfected 293T cell lysate as well as from complexes pulled down by GST-Vif, but not by GST itself (Fig. 3A). This indicates that the pull down of Flag-ZIN by GST-Vif was specifically dependent on an interaction with the Vif part of the GST-Vif fusion protein. After stripping off the antibody and reprobing the same filter with a GST-specific antibody, a GST-Vif fusion protein of approximately 49 kDa was detected as expected in Sepharose beads complexed with GST-Vif fusion protein. Beads complexed with GST protein showed the expected 26-kDa GST protein band (Fig. 3B). This indicates there were no loading or other irregularities among the three lanes of the filter. The identity of the 49-kDa GST-Vif fusion protein responsible for the pull down of Flag-ZIN was further confirmed by stripping and reprobing the filter with Vif-specific antibody. The polyclonal anti-Vif antibody detected the 49-kDa GST-Vif fusion protein and several bands with lower molecular mass, probably representing Vif products cleaved from the fusion protein (Fig. 3C). The pulling down of ZIN by bacterially expressed and purified GST-Vif suggests a direct binding between Vif and ZIN.
FIG. 3.
Vif binds ZIN in vitro. Western blot analysis of total protein in pFlag-ZIN-transfected 293T cell lysate or protein pulled down by GST protein-bound glutathione Sepharose CL-4B beads or by GST-Vif fusion protein-bound beads. The same filter was analyzed by Western blot successively probed with monoclonal anti-Flag antibody (A), anti-GST antibody (B), and polyclonal anti-Vif antiserum (C).
Triad 3/ZIN interacts with HIV Vif in mammalian cells.
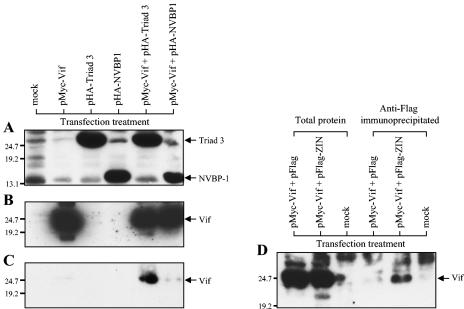
To investigate whether HIV-1 Vif interacts with Triad 3/ZIN in human cells, 293T cells were singly transfected or cotransfected with Vif- and Triad 3-expressing plasmids. After checking correct protein expression, lysates from the transfected cells were immunoprecipitated with various antibodies, and the precipitated and coprecipitated proteins were analyzed by SDS-PAGE gel followed by Western blotting. 293T cells were cotransfected with pMyc-Vif and pHA-Triad 3 or pMyc-Vif and pHA-NVBP 1. NVBP 1, like Triad 3, was selected on the basis of its interaction with Vif in yeast cells but had not been confirmed to interact with Vif in other systems (21). Single plasmids were also individually transfected into the same batch of 293T cells to control for nonspecific protein-protein interactions. Analysis by Western blot of total protein expression in transfected cells demonstrated abundant expression of HA-tagged Triad 3 and HA-tagged NVPB 1, using the rabbit anti-HA antibody probe (Fig. 4A), and abundant expression of Myc-Vif, using the mouse anti-Myc antibody probe (Fig. 4B). Figure 4C shows that after immunoprecipitation with rabbit anti-HA antibody, Vif was coprecipitated with HA-Triad 3, but not with HA-NVBP 1. The interaction between Vif and Triad 3, but not NVBP-1, was further confirmed by using an alternative mouse anti-HA antibody to immunoprecipitate HA-Triad 3 and HA-NVBP 1. Similarly, Vif protein was detected in only the Triad 3 immunoprecipitates, but not the NVBP 1 immunoprecipitates, by Western blotting using rabbit anti-Vif antibody (data not shown). This suggests that the interaction between Vif and Triad 3 is genuine in both yeast and human cells, while the interaction between Vif and NVBP 1 is considerably weaker or nonexistent in human cells.
FIG. 4.
Coimmunoprecipitation of Vif and Triad 3/ZIN. (A, B, and C) Western blot analysis of total protein (A and B) and protein immunoprecipitated by HA-specific rabbit polyclonal antiserum (C) from transfected 293T cell lysate probed with HA-specific rabbit polyclonal antiserum (A) or monoclonal mouse anti-Myc antibody (B and C). (D) Total protein and protein immunoprecipitated with mouse anti-Flag antibody from transfected 293T cells analyzed by Western blot probed with polyclonal rabbit anti-Vif antiserum. Plasmid DNAs used in different transfection treatments are labeled on top of the lanes.
To examine whether ZIN also interacts with Vif in human cells, 293T cells were cotransfected with pMyc-Vif and pFlag-ZIN or pMyc-Vif and control pFlag. Cell lysates were immunoprecipitated with Flag-specific antibody, and the presence of coprecipitated Vif was determined by Western blot with rabbit anti-Vif antiserum. Despite the abundant expression of Vif in both types of cotransfected cells, precipitation with anti-Flag antibody led to coprecipitation of Vif only from cell lysates containing Flag-ZIN protein, and not from lysates containing Flag protein tag alone (Fig. 4D). These results clearly demonstrate that the specific interaction between Triad 3/ZIN and Vif we have characterized in vitro also occurs in human cells.
HIV-1 virus produced in the presence of exogenous ZIN is less infectious.
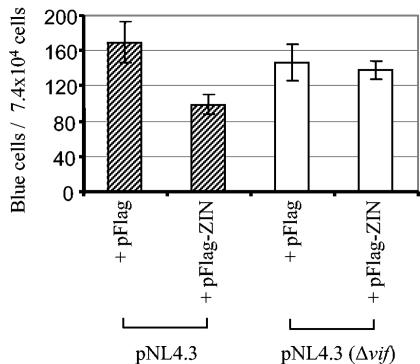
Having shown that Triad 3/ZIN interacts with HIV-1 Vif in transfected cells, we next examined the biological significance of this Vif-ZIN interaction for HIV-1 replication in virus infection experiments. These experiments were performed by using virus produced in cells in the presence or absence of cotransfected ZIN, as Vif was known to function in the late stage of viral assembly in the viral producer cells. Human 293T cells were cotransfected with HIV-1 pNL4.3 and pFlag-ZIN (or control plasmid pFlag), or pNL4.3(Δvif) and pFlag-ZIN (or control plasmid pFlag). The four types of transfection cell culture media were harvested 48 h postinfection, and their p24 levels were quantified and subsequently used as virus inocula in infection experiments. For the single-cycle viral infectivity assay, 10-fold dilutions of 400-ng/ml p24 of each virus stock were used to infect HeLa-CD4-β-gal indicator cells as described in Materials and Methods. To minimize secondary infection, cells were fixed and stained 30 h postinfection and blue cells were scored. The results (Fig. 5) showed that the wild-type HIV NL4.3 produced in the presence of cotransfected ZIN was twofold less infectious than NL4.3 produced in the absence of ZIN. The twofold difference in infectivity is significant as it is calculated based on counting nine replica wells for infected (blue) cells 30 h postinfection, without resorting to multiround viral replication. In contrast, the infectivity of Vif-deletion NL4.3 was not affected by the coexpression of ZIN (Fig. 5). This suggests that the suppressing effect of cotransfected ZIN on virus infectivity was dependent on the presence of Vif in virus producer cells.
FIG. 5.
Virus produced in cells overexpressing ZIN has reduced single-cycle infectivity. HIV virus stocks were produced from 293T cells cotransfected with pNL4.3 (or pNL4.3(Δvif)) and either pFlag or pFlag-ZIN. HeLa-CD4-LTR-β-gal cells were infected with a 100-μl inoculum made of 10-fold dilutions of 40 ng of p24 of each of the four virus stocks in 48-well plates. At 30 h postinfection, cells were fixed and stained as described previously. Blue cells were counted in each well infected at a virus dilution of 10−2 (0.4 ng of p24/well) in which there are minimum of 80 blue cells. Error bars were obtained from infections performed in nine replica wells.
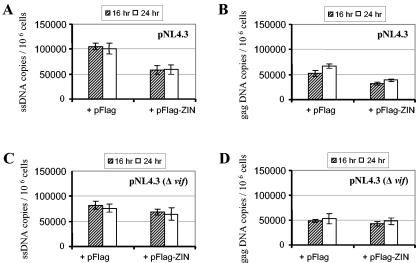
Vif has been suggested to specifically function in the early stage of HIV reverse transcription in newly infected target cells (6, 11, 37, 41). To investigate whether the above-observed decrease in virus infectivity was due to interference with early steps of the viral replication cycle (up to and including reverse transcription), HUT-78 cells were infected with virus stocks produced in the presence or absence of exogenously expressed ZIN. The newly reverse-transcribed DNA levels were measured 16 and 24 h postinfection. Equivalent amounts (10 ng of HIV p24) of each of four virus stocks produced from 293T cells cotransfected with pNL4.3 and pFlag-ZIN, pNL4.3 and pFlag, pNL4.3(Δvif) and pFlag-ZIN, or pNL4.3(Δvif) and pFlag were used in the infection experiments to investigate (i) whether the negative effects ZIN had on virus infectivity were acting before or at the stage of viral reverse transcription and (ii) whether this was dependent on the interaction between ZIN and Vif. Viral reverse transcription was monitored by HIV DNA synthesis using primers detecting early strong-stop HIV DNA or detecting later viral DNA spanning the U5/Gag region. Vif-deleted virus produced in permissive 293T cells is known to undergo a single round of replication in nonpermissive cells (2). To avoid secondary infection by progeny virus, cells were harvested for viral DNA analysis within the first 24 h of infection. Figure 6A shows that at 16 and 24 h postinfection, there was approximately a 50% decrease in early strong-stop HIV DNA synthesis in cells infected with HIV virus produced in the presence of cotransfected pFlag-ZIN compared with control. The same difference in DNA levels was also evident for HIV gag region DNA (Fig. 6B). There was no difference, however, when HIV DNA synthesis was compared in cells infected with Vif-deleted HIV NL4.3(Δvif) produced in the presence or absence of coexpressed ZIN (Fig. 6C and D). These results are clearly consistent with the decrease in viral infectivity measured by using the HeLa-CD4-β-gal indicator cells (Fig. 5). Furthermore, the DNA synthesis data indicate that ZIN expressed in virus-producing cells interferes with an early step of viral replication.
FIG. 6.
Virus produced in cells overexpressing ZIN has impaired reverse transcription in newly infected cells: Levels of HIV-1 strong-stop DNA (ssDNA) (A and C) and U5/Gag DNA (B and D) in 106 HUT-78 cells infected with 10 ng of p24 of HIV virus stocks 16 and 24 h postinfection. Virus stocks were produced from 293T cells cotransfected with pNL4.3 and pFlag or pFlag-ZIN (A and B) or pNL4.3(Δvif) and pFlag or pFlag-ZIN (C and D). Viral DNA synthesis was quantified by real-time PCR. DNA copy number values represent the average of four PCR measurements from duplicate infected wells.
DISCUSSION
While the function of HIV Vif in regulating viral infectivity has long been recognized, the biochemical mechanisms by which Vif functions remain to be fully elucidated. A role for cellular factors in Vif function has been implied in numerous studies (24, 33, 34). Recently, cellular factor APOBEC3G has been shown to directly inhibit HIV replication in nonpermissive cell types by deamination of newly synthesized minus-strand DNA during reverse transcription. Vif acts to suppress this inhibitory effect of APOBEC3G in nonpermissive cells by inducing degradation of APOBEC3G and preventing it from being packaged into virions (22, 26, 27, 30, 31, 44).
We had previously identified a partial clone from a human leukocyte cDNA library which encoded a protein that interacted with HIV Vif in yeast cells (21). The deduced amino acid sequence of this cDNA clone was shown to be homologous to a then hypothetical protein containing novel ring finger motifs, called Triad 3. In the present study, we have demonstrated that Triad 3 shares exact homology with the central portion, including all four RLDs, of the recently identified protein ZIN. mRNA encoding Triad 3/ZIN was shown to be present in both permissive and nonpermissive cell types as two alternatively spliced 3- and 6-kb transcripts consistent in size with what has been described elsewhere (5). The comparable levels of mRNA shown in this study and detection of ZIN protein in both permissive and nonpermissive human cell lines as shown by Chen et al., suggest that ZIN normally functions in most cell types (5). We further demonstrated that ZIN protein expressed in transfected 293T cells interacts with purified Vif protein in vitro using a GST fusion protein pull-down assay and that ZIN can be coprecipitated from 293T cell lysate by Vif-specific antibody, indicating a direct interaction between the two proteins in human cells. Immunofluorescent staining and confocal microscopy analysis also indicated concurrent nuclear localization of Vif and ZIN in cotransfected cells, lending support to intracellular Vif-ZIN interaction (data not shown).
Using HeLa-CD4-LTR-β-gal as indicator cells, ZIN was shown to reduce HIV infectivity in a Vif-dependent manner when overexpressed in 293T virus producer cells. Since activation of HeLa-CD4-LTR-β-gal reporter cells was dependent on virus entry and completion of reverse transcription and integration by the infecting virus, this suggested that the reduction in infectivity of wild-type virus produced in ZIN-expressing cells occurred during these early steps of HIV replication. It was subsequently shown that in HUT-78 cells infected with virus produced in cells expressing cotransfected ZIN, levels of newly reverse-transcribed viral DNA were reduced twofold compared with infection using virus produced in the absence of ZIN. Again, this reduction was shown to depend on Vif expression in virus producer cells. Since virus producer 293T cells used n the current study are permissive, these results suggest that the decrease in virus infectivity is probably unrelated to APOBEC3G, which is packaged into virions only from nonpermissive viral producer cells. Taken together, our results show that an interaction between ZIN and Vif in virus producer cells interferes with an early step in HIV replication in newly infected target cells and this interference cannot be interpreted by the known mechanism of APOBEC3G inhibition of viral DNA synthesis. Therefore the inhibitory effect that ZIN mediates through its interaction with Vif alludes to other roles of Vif in HIV infectivity.
In the late stages of HIV infection, Vif interacts with viral RNA and NC proteins during virus assembly and recent evidence also suggests that Vif is incorporated into virions (18, 28). Dettenhofer et al. demonstrated that Vif contributed to enhancement of tRNA3Lys annealing to RNA template for efficient initiation of reverse transcription (7). They also suggested that the initial point of contact between Vif and genomic RNA might take place in the nucleus where Vif has been shown to localize (4, 12, 35), before becoming associated with the Gag-assembling complexes in the cytoplasm. It has also been reported that deletion of the SLQYL motif from Vif, which is highly homologous to that of the double-stranded RNA binding protein XlrbpA from Xenopus laevis, resulted in abolition of Vif function (7, 19). The RNA binding function of Vif has also been shown by Zhang et al. to be important for virus replication in nonpermissive H9 cells (43). Thus the interaction of Vif with genomic RNA and its role on primer tRNA binding, the Vif interaction with viral Gag proteins during virus assembly, and the role of Vif in maintaining the stability of the virus core (28) are potentially other biologically relevant functions of Vif. Additionally, Vif has been shown to function within virions to regulate virus infectivity. In virions, Vif is proteolytically cleaved and has been shown to positively correlate with virus infectivity, but only at proper levels of Vif expression (1, 19). Overexpressed ZIN in our experimental settings may compete and interfere with any of the above interactions, and this would be consistent with the defect in reverse transcription we observed following HIV infection. We have preliminary evidence that suggests ZIN influences the packaging of Vif into virus particles (data not shown). How ZIN may affect the levels and the role of intravirion Vif are currently under investigation.
The biological significance of endogenous ZIN in HIV infection is not clear. One function of ZIN as a component of TNF receptor-mediated signal transduction pathway is to down-regulate TNF-α-induced NF-κB activation, which is critical to many aspects of cellular and viral (including HIV) transcription (5). Potential interference in this pathway by Vif, for example, may have implications for viral transcription in response to stimuli. Interestingly, ZIN shares sequence homology with E3 ubiquitin ligases, which ubiquinate target proteins regulating their activity or induce their degradation by the proteosome. It is therefore interesting to postulate that ZIN may influence Vif function and HIV replication by virtue of a similar activity. In any case, greater understanding of normal ZIN function and its interaction with Vif will be valuable in defining further the mechanisms of Vif function and the role of cellular factors in HIV infection.
Acknowledgments
We are grateful to Chris Bagley for assistance in analysis of protein sequence homology and protein function databases. We thank Jennifer Clarke and Ghafer Sarvestani and Jane Arthur for advice and discussion of confocal microscopy work, Kelly Cheney for p24 analysis, and all members in the HIV Research laboratory for their support.
This work was supported by the Australian Commonwealth AIDS Research Grant Programme and the Australian National Health & Medical Research Council.
REFERENCES
- 1.Akari, H., M. Fujita, S. Kao, M. A. Khan, M. Shehu-Xhilaga, A. Adachi, and K. Strebel. 2004. High level expression of human immunodeficiency virus type-1 Vif inhibits viral infectivity by modulating proteolytic processing of the Gag precursor at the p2/nucleocapsid processing site. J. Biol. Chem. 279:12355-12362. [DOI] [PubMed] [Google Scholar]
- 2.Blanc, D., C. Patience, T. F. Schulz, R. Weiss, and B. Spire. 1993. Transcomplementation of VIF- HIV-1 mutants in CEM cells suggests that VIF affects late steps of the viral life cycle. Virology 193:186-192. [DOI] [PubMed] [Google Scholar]
- 3.Carr, J. M., H. Hocking, P. Li, and C. J. Burrell. 1999. Rapid and efficient cell-to-cell transmission of human immunodeficiency virus infection from monocyte-derived macrophages to peripheral blood lymphocytes. Virology 265:319-329. [DOI] [PubMed] [Google Scholar]
- 4.Chatterji, U., C. K. Grant, and J. H. Elder. 2000. Feline immunodeficiency virus Vif localizes to the nucleus. J. Virol. 74:2533-2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, D., X. Li, Z. Zhai, and H. B. Shu. 2002. A novel zinc finger protein interacts with receptor-interacting protein (RIP) and inhibits tumor necrosis factor (TNF)- and IL1-induced NF-kappa B activation. J. Biol. Chem. 277:15985-15991. [DOI] [PubMed] [Google Scholar]
- 6.Courcoul, M., C. Patience, F. Rey, D. Blanc, A. Harmache, J. Sire, R. Vigne, and B. Spire. 1995. Peripheral blood mononuclear cells produce normal amounts of defective Vif− human immunodeficiency virus type 1 particles which are restricted for the preretrotranscription steps. J. Virol. 69:2068-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dettenhofer, M., S. Cen, B. A. Carlson, L. Kleiman, and X. F. Yu. 2000. Association of human immunodeficiency virus type 1 Vif with RNA and its role in reverse transcription. J. Virol. 74:8938-8945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gabuzda, D. H., K. Lawrence, E. Langhoff, E. Terwilliger, T. Dorfman, W. A. Haseltine, and J. Sodroski. 1992. Role of vif in replication of human immunodeficiency virus type 1 in CD4+ T lymphocytes. J. Virol. 66:6489-6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garrett, E. D., L. S. Tiley, and B. R. Cullen. 1991. Rev activates expression of the human immunodeficiency virus type 1 vif and vpr gene products. J. Virol. 65:1653-1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goncalves, J., P. Jallepalli, and D. H. Gabuzda. 1994. Subcellular localization of the Vif protein of human immunodeficiency virus type 1. J. Virol. 68:704-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goncalves, J., Y. Korin, J. Zack, and D. Gabuzda. 1996. Role of Vif in human immunodeficiency virus type 1 reverse transcription. J. Virol. 70:8701-8709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goncalves, J., B. Shi, X. Yang, and D. Gabuzda. 1995. Biological activity of human immunodeficiency virus type 1 Vif requires membrane targeting by C-terminal basic domains. J. Virol. 69:7196-7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harris, R. S., K. N. Bishop, A. M. Sheehy, H. M. Craig, S. K. Petersen-Mahrt, I. N. Watt, M. S. Neuberger, and M. H. Malim. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803-809. [DOI] [PubMed] [Google Scholar]
- 14.Hassaine, G., M. Courcoul, G. Bessou, Y. Barthalay, C. Picard, D. Olive, Y. Collette, R. Vigne, and E. Decroly. 2001. The tyrosine kinase Hck is an inhibitor of HIV-1 replication counteracted by the viral vif protein. J. Biol. Chem. 276:16885-16893. [DOI] [PubMed] [Google Scholar]
- 15.Hoglund, S., A. Ohagen, K. Lawrence, and D. Gabuzda. 1994. Role of vif during packing of the core of HIV-1. Virology 201:349-355. [DOI] [PubMed] [Google Scholar]
- 16.Huvent, I., S. S. Hong, C. Fournier, B. Gay, J. Tournier, C. Carriere, M. Courcoul, R. Vigne, B. Spire, and P. Boulanger. 1998. Interaction and co-encapsidation of human immunodeficiency virus type 1 Gag and Vif recombinant proteins. J. Gen. Virol. 79:1069-1081. [DOI] [PubMed] [Google Scholar]
- 17.Kao, S., H. Akari, M. A. Khan, M. Dettenhofer, X.-F. Yu, and K. Strebel. 2003. Human immunodeficiency virus type 1 Vif is efficiently packaged into virions during productive but not chronic infection. J. Virol. 77:1131-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khan, M. A., C. Aberham, S. Kao, H. Akari, R. Gorelick, S. Bour, and K. Strebel. 2001. Human immunodeficiency virus type 1 Vif protein is packaged into the nucleoprotein complex through an interaction with viral genomic RNA. J. Virol. 75:7252-7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khan, M. A., H. Akari, S. Kao, C. Aberham, D. Davis, A. Buckler-White, and K. Strebel. 2002. Intravirion processing of the human immunodeficiency virus type 1 Vif protein by the viral protease may be correlated with Vif function. J. Virol. 76:9112-9123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kimpton, J., and M. Emerman. 1992. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated β-galactosidase gene. J. Virol. 66:2232-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lake, J. A., J. Carr, F. Feng, L. Mundy, C. Burrell, and P. Li. 2003. The role of Vif during HIV-1 infection: interaction with novel host cellular factors. J. Clin. Virol. 26:143-152. [DOI] [PubMed] [Google Scholar]
- 22.Lecossier, D., F. Bouchonnet, F. Clavel, and A. J. Hance. 2003. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science 300:1112. [DOI] [PubMed] [Google Scholar]
- 23.Liu, H., X. Wu, M. Newman, G. M. Shaw, B. H. Hahn, and J. C. Kappes. 1995. The Vif protein of human and simian immunodeficiency viruses is packaged into virions and associates with viral core structures. J. Virol. 69:7630-7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Madani, N., and D. Kabat. 1998. An endogenous inhibitor of human immunodeficiency virus in human lymphocytes is overcome by the viral Vif protein. J. Virol. 72:10251-10255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Madani, N., R. Millette, E. J. Platt, M. Marin, S. L. Kozak, D. B. Bloch, and D. Kabat. 2002. Implication of the lymphocyte-specific nuclear body protein Sp140 in an innate response to human immunodeficiency virus type 1. J. Virol. 76:11133-11138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mangeat, B., P. Turelli, G. Caron, M. Friedli, L. Perrin, and D. Trono. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99-103. [DOI] [PubMed] [Google Scholar]
- 27.Mariani, R., D. Chen, B. Schrofelbauer, F. Navarro, R. Konig, B. Bollman, C. Munk, H. Nymark-McMahon, and N. R. Landau. 2003. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114:21-31. [DOI] [PubMed] [Google Scholar]
- 28.Öhagen, Å., and D. Gabuzda. 2000. Role of Vif in stability of the human immunodeficiency virus type 1 core. J. Virol. 74:11055-11066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwartz, S., B. K. Felber, and G. N. Pavlakis. 1991. Expression of human immunodeficiency virus type 1 vif and vpr mRNAs is Rev-dependent and regulated by splicing. Virology 183:677-686. [DOI] [PubMed] [Google Scholar]
- 30.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646-650. [DOI] [PubMed] [Google Scholar]
- 31.Sheehy, A. M., N. C. Gaddis, and M. H. Malim. 2003. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 9:1404-1407. [DOI] [PubMed] [Google Scholar]
- 32.Simon, J. H., E. A. Carpenter, R. A. M. Fouchier, and M. H. Malim. 1999. Vif and the p55Gag polyprotein of human immunodeficiency virus type 1 are present in colocalizing membrane-free cytoplasmic complexes. J. Virol. 73:2667-2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simon, J. H., N. C. Gaddis, R. A. Fouchier, and M. H. Malim. 1998. Evidence for a newly discovered cellular anti-HIV-1 phenotype. Nat. Med. 4:1397-1400. [DOI] [PubMed] [Google Scholar]
- 34.Simon, J. H., D. L. Miller, R. A. Fouchier, M. A. Soares, K. W. Peden, and M. H. Malim. 1998. The regulation of primate immunodeficiency virus infectivity by Vif is cell species restricted: a role for Vif in determining virus host range and cross-species transmission. EMBO J. 17:1259-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simon, J. H. M., R. A. M. Fouchier, T. E. Southerling, C. B. Guerra, C. K. Grant, and M. H. Malim. 1997. The Vif and Gag proteins of human immunodeficiency virus type 1 colocalize in infected human T cells. J. Virol. 71:5259-5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simon, J. H. M., and M. H. Malim. 1996. The human immunodeficiency virus type 1 Vif protein modulates the postpenetration stability of viral nucleoprotein complexes. J. Virol. 70:5297-5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sova, P., and D. J. Volsky. 1993. Efficiency of viral DNA synthesis during infection of permissive and nonpermissive cells with vif-negative human immunodeficiency virus type 1. J. Virol. 67:6322-6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vandegraaff, N., R. Kumar, C. J. Burrell, and P. Li. 2001. Kinetics of human immunodeficiency virus type 1 (HIV) DNA integration in acutely infected cells as determined using a novel assay for detection of integrated HIV DNA. J. Virol. 75:11253-11260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vandegraaff, N., R. Kumar, H. Hocking, T. R. Burke, Jr., J. Mills, D. Rhodes, C. J. Burrell, and P. Li. 2001. Specific inhibition of human immunodeficiency virus type 1 (HIV-1) integration in cell culture: putative inhibitors of HIV-1 integrase. Antimicrob. Agents Chemother. 45:2510-2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van der Reijden, B. A., C. A. Erpelinck-Verschueren, B. Lowenberg, and J. H. Jansen. 1999. TRIADs: a new class of proteins with a novel cysteine-rich signature. Protein Sci. 8:1557-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.von Schwedler, U., J. Song, C. Aiken, and D. Trono. 1993. Vif is crucial for human immunodeficiency virus type 1 proviral DNA synthesis in infected cells. J. Virol. 67:4945-4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu, X., Y. Yu, B. Liu, K. Luo, W. Kong, P. Mao, and X. F. Yu. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302:1056-1060. [DOI] [PubMed] [Google Scholar]
- 43.Zhang, H., R. J. Pomerantz, G. Dornadula, and Y. Sun. 2000. Human immunodeficiency virus type 1 Vif protein is an integral component of an mRNP complex of viral RNA and could be involved in the viral RNA folding and packaging process. J. Virol. 74:8252-8261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang, H., B. Yang, R. J. Pomerantz, C. Zhang, S. C. Arunachalam, and L. Gao. 2003. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424:94-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zimmerman, C., K. C. Klein, P. K. Kiser, A. R. Singh, B. L. Firestein, S. C. Riba, and J. R. Lingappa. 2002. Identification of a host protein essential for assembly of immature HIV-1 capsids. Nature 415:88-92. [DOI] [PubMed] [Google Scholar]