Abstract
Jaagsiekte sheep retrovirus (JSRV) is an exogenous retrovirus of sheep that induces a contagious lung cancer, ovine pulmonary adenocarcinoma. We previously showed that the gene encoding JSRV envelope protein (Env) appears to function as an oncogene, since it can transform mouse NIH 3T3 cells. The cytoplasmic tail of the Env transmembrane protein (TM) is necessary for the transformation. However, previous experiments did not exclude the involvement of the Env surface protein (SU) in transformation. In this study, we created a series of nested deletion mutants through the SU domain and assessed their ability to transform rodent fibroblasts. All SU deletion mutants downstream of the predicted signal peptide were unable to transform murine NIH 3T3 or rat 208F cells. Transport to the plasma membrane of selected deleted Env proteins was confirmed by confocal immunofluorescence microscopy of hemagglutinin-tagged versions. Additional sequential SU deletion mutants lacking 50-amino-acid (aa) blocks throughout SU also were unable to transform. Furthermore, minimal insertion mutants of two amino acids (Leu/Gln) at various positions in SU also abolished transformation. These data indicate that domains in SU facilitate efficient JSRV transformation. This could reflect a necessity of SU for appropriate configuration of the Env protein or independent activation by SU of a signaling pathway necessary for transformation. Complementation between SU and TM mutants for transformation supported the latter hypothesis. Cotransfection with ΔGP Y590F (mutant in the TM cytoplasmic tail) with ΔGP SUΔ103-352 (lacking most of SU) resulted in efficient transformation. The resulting transformants showed evidence for the presence and expression of both mutant plasmids.
Jaagsiekte sheep retrovirus (JSRV) is the etiological agent of ovine pulmonary adenomatosis (OPA), a contagious lung cancer of sheep (18, 19, 24). Secretory epithelial cells of the distal airways, most notably type II pneumocytes and Clara cells, are the targets of transformation. OPA shares similarities with the human bronchioloaveolar carcinoma, a lung cancer not strongly associated with cigarette smoke. Thus, understanding the mechanisms of oncogenesis by JSRV may provide insight into development of human lung cancer as well.
Complete and infectious molecular clones of JSRV have been isolated (6, 24). These clones allowed production of JSRV virions and the demonstration that JSRV is necessary and sufficient to cause OPA in sheep. JSRV is a betaretrovirus, most closely related to murine mammary tumor virus and Mason-Pfizer monkey virus. The JSRV genome contains open reading frames typical of other retroviruses (gag, pro, pol, and env), with an alternate reading frame (Orf-X) overlying pol.
The mechanism of oncogenic transformation by JSRV has been of great interest. JSRV is a potent oncogenic agent when inoculated intratracheally into newborn lambs. The mean time to development of end-stage OPA is 3 weeks under these conditions, with disease evident in as little as 10 days (10, 29, 30). The rapid oncogenesis would be consistent with JSRV carrying an oncogene, although inspection of the genomic sequence did not reveal any obvious oncogenes (i.e., open reading frames with homology to known cellular genes). Nevertheless, we found that transfection of JSRV DNA into NIH 3T3 fibroblasts led to the formation of foci of transformation, indicating that JSRV does indeed carry transforming potential (14, 15). Interestingly, the JSRV env gene was found to be necessary and sufficient for inducing transformation. The finding of the env gene of a replication-competent retrovirus having transforming potential is quite novel.
Investigations into the mechanism of JSRV env-induced transformation are being carried out. The env gene is expressed as a polyprotein that is glycosylated and transported through the endoplasmic reticulum to the cell surface. At the cell surface, it is cleaved into the surface (SU) protein and transmembrane (TM) protein, which are held together by disulfide linkages. After virus budding, the SU protein is on the surface of the virion, and it is anchored to the virus particle by its interaction with TM, which spans the lipid bilayer of the viral envelope. It is generally assumed that the SU and TM proteins have the same relationship to the plasma membrane of JSRV-transformed cells as for virions—i.e., SU is extracellular, and TM spans the plasma membrane. We previously showed that the short cytoplasmic tail of TM is necessary for transformation of NIH 3T3 cells (22). For instance, mutation of a YXXM motif (a putative binding site for phosphatidylinositol 3-kinase [PI3-kinase]) in the cytoplasmic tail abolished transformation. Other investigators have also found that the cytoplasmic tail of TM is necessary for transformation, although the exact requirements in different cell transformation systems may differ (2, 13, 22).
While the requirement of the TM cytoplasmic tail for JSRV transformation is well established, so far experiments have not excluded the requirement for other domains of Env as well. Indeed, it is noteworthy that the cellular receptor for JSRV has been identified as hyaluronidase-2 (Hyal-2) (16, 26). Human Hyal-2 maps to chromosome 3p21.3, a locus that is frequently deleted in human lung cancer (25, 26). Such deletions are often indicative of tumor suppressor genes, which led to the hypothesis that the JSRV Env protein might cause transformation by binding and inactivating Hyal-2 via interactions with the SU receptor binding domain. Evidence supporting this has been recently published (5, 12).
In the experiments reported here, we directly tested whether JSRV SU might play a role in transformation. A series of deletions and insertions in SU suggested that domains of SU may be important for transformation of rodent fibroblasts. Complementation experiments between nontransforming mutants in SU and the TM cytoplasmic tail strongly supported this concept.
MATERIALS AND METHODS
Generation of mutant Env expression plasmids.
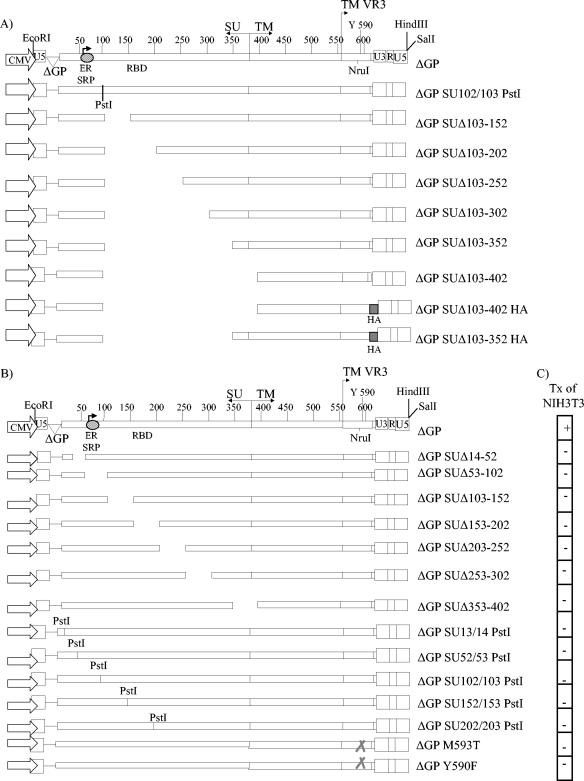
The different JSRV Env expression constructs used in this study are shown in Fig. 1 and 4. All SU deletion and insertion constructs were designed in the context of the JSRV Env expression plasmid pCMV3JS21ΔGP (ΔGP) (15). ΔGP is derived from a plasmid containing a full-length JSRV provirus, with a substitution of the upstream U3 sequences by the human cytomegalovirus immediate-early promoter, and from which the gag and pol sequences were deleted. This plasmid contains 5′ JSRV sequences corresponding to U5 through the splice donor site for env mRNA. Details for the cloning procedures will be provided on request. The nomenclature indicates the amino acid residues of Env that have been deleted or where a PstI site was inserted. As a consequence of the cloning strategy, all mutants contained a two-amino-acid (aa) insertion (Leu and Gln; CTGCAG) at the site of the deletion. Likewise, the PstI site insertions contained the same two additional amino acids.
FIG. 1.
SU deletion constructs. (A) Nested deletions through SU are shown. The nomenclature reflects amino acids deleted in the JSRV Env protein. All deletions were constructed with a common endpoint at residue 103, downstream of the predicted ER signal recognition peptide (SRP). The putative location of the receptor binding domain (RBD) is shown. The influenza HA epitope tag was fused immediately downstream of the cytoplasmic tail of the two largest SU deletion constructs. (B) Fifty-aa deletions progressing through SU are shown. The PstI insertions are a by-product of the cloning strategy to make SU deletions. (C) The transforming activities of each construct in panel B assayed in NIH 3T3 cells and scored at 4 weeks posttransfection are shown. +, >35 foci in the standard assay; −, no foci observed.
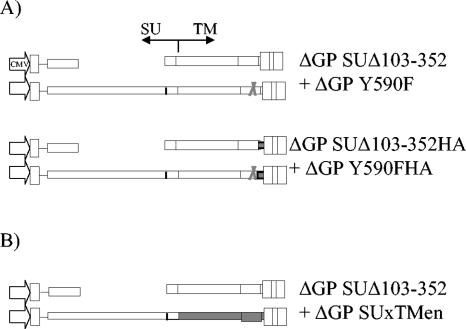
FIG. 4.
Constructs used in cotransfection experiments. (A) The two pairs of nontransforming env mutations used in experiment 1 of Table 3 are shown. (B) The pair of nontransforming env expression constructs employed in experiment 2 of Table 3 is shown.
The pCMV3JSRV21ΔGP Y590F (ΔGP Y590F) and SUxTMen constructs were described previously (22), as was ΔGP Y590FHA, which contains the influenza hemagglutinin (HA) epitope tag at the C terminus of Env (14). The ΔGP SUΔ103-352HA and ΔGP SUΔ103-402HA constructs were created by substituting the NruI-HindIII fragment containing the epitope tag from ΔGPHA (14) into the NruI and HindIII sites of the two deletion constructs. All constructs were confirmed by DNA sequencing.
Cells.
Human 293T and Rat 208F cells were both grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (208F cells were a gift from Dusty Miller). Mouse NIH 3T3 cells were grown in DMEM supplemented with 10% calf serum. All cells were cultured at 37°C, 95% humidity, 5% CO2.
Transformation assays.
Cell transformation assays with NIH 3T3 cells were performed as described previously (15), and foci of transformed cells were counted at 4 weeks. Briefly, 4 × 106 cells in a 10-cm dish were transfected with 28 μg of DNA, using the BD CalPhos Mammalian Transfection kit (BD Biosciences, Palo Alto, Calif.), and were washed with 1× phosphate-buffered saline and refed with fresh media every 3 days after DNA transfection. Transformation assays were also performed with 208F Fischer rat embryo fibroblasts, a morphologically flat cell line suitable for transformation assays (13). Transformation assays using 208F cells were carried out as described for NIH 3T3 cells, except these cultures were scored for foci beginning at 2 weeks and up to 7 weeks. Confirmation of transformation was carried out by selective trypsinization of individual transformed foci followed by reculturing. Briefly, 6- by 8-mm glass cloning rings (Corning) were placed over the focus, and the cells in the ring were trypsinized and reseeded in a single well (diameter, 35 mm) of a six-well tissue culture plate. The re-formation of transformed foci was scored 2 weeks postreseeding. For transformation assays with 208F cells that were cotransfected with two mutant Env constructs, 14 μg of each plasmid was used for a total of 28 μg of plasmid DNA. Each mutant construct was tested in at least three independent experiments, and at least two different plasmid preparations were tested for each mutant. SU mutant constructs were subjected to additional 208F transformation assays under conditions described by other investigators (3, 13). Briefly, 3 × 105 208F cells growing in 6-cm plates were transfected with 10 μg of DNA, followed by trypsinization and transfer to a 10-cm plate 24 h posttransfection; further culturing was in DMEM plus 10% fetal bovine serum medium containing 1 μM dexamethasone. The presence of dexamethasone has been reported to make the differences between transformed and nontransformed cells more evident (17).
mRNA and protein analysis.
Northern blot analysis for JSRV env RNA expression was performed on RNA from 293T cells transfected with the ΔGP mutant constructs as previously described (22, 23). Western blot analysis for JSRV Env protein expression was carried out with 293T cells transfected with the epitope-tagged constructs ΔGPHA, ΔGP SUΔ103-352HA, and ΔGP SUΔ103-352HA. Cell lysates were first immunoprecipitated with anti-HA antibody 48 h posttransfection, followed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting of the immunoprecipitates with an anti-HA antibody (14).
Immunofluorescence assays.
Confocal immunofluorescence microscopy on 293T cells transiently transfected with HA-tagged expression constructs was performed as described previously (14).
Detection of JSRV env DNA and RNA in transformed cells.
To test for the presence of JSRV DNA or RNA in JSRV-transformed cells, foci of transformed 208F cells were isolated and regrown as described above and subjected to PCR or reverse transcription (RT)-PCR analyses for JSRV env sequences. Genomic DNA from the isolated foci was obtained by using the DNeasy tissue kit (QIAGEN, Valencia, Calif.), and 500 ng of DNA was subjected to PCR amplification, using the primer set 5522SUPstI (f) and 6579TMPstI (r) (5′-GTGACACCTACTAAGCATCTTG-3′ and 5′-GCAGCTTGAATAGACTGTGCTAAAG-3′, respectively) under the following conditions: incubation at 94°C for 1 min followed by 35 cycles of 94°C for 30 s, 59°C for 1 min, and 72°C for 1 min with a final extension of 72°C for 10 min, using a Stratagene Robocycler, as previously described for the detection of JSRV DNA (21). This primer set encompasses SU sequences that have been deleted. PCR products were analyzed by electrophoresis in a 1.5% agarose gel. For RT-PCR, total RNA was extracted using the RNeasy tissue kit (QIAGEN). One microgram of total RNA was subjected to RT-PCR, using the same primer set used for the DNA PCR, with the Titan One Tube RT-PCR system (Roche, Basel, Switzerland) as recommended by the manufacturer. PCR products were analyzed on a 1.5% agarose gel.
RESULTS
Generation of nested deletions in the JSRV SU domain.
To test if domains in the JSRV Env SU protein are necessary for transformation, we designed a series of JSRV Env expression constructs that contain progressive deletions through SU (Fig. 1A). All deletion constructs were based on pCMV3JS21ΔGP (ΔGP), a cytomegalovirus promoter-driven expression plasmid for the full-length JSRV Env protein. This plasmid retains the native JSRV mRNA splice donor and splice acceptor sites. It has been suggested that the putative endoplasmic reticulum signal peptide in JSRV Env resides upstream of residue 77 (8); all deletions in SU were made downstream of this region to preserve transport of the mutant proteins to the plasma membrane (Fig. 1A). The SU deletion constructs maintained the reading frame of wild-type JSRV Env, and this was confirmed by sequence analysis. The strategy for generating the deletions involved first inserting a linker containing a PstI restriction endonuclease site at defined positions in the SU domain (see Materials and Methods). As a result, the junction of each deleted region contained an insertion of 2 aa (Gln and Leu). For the two largest deletions, ΔGP SUΔ103-352 and ΔGP SUΔ103-402, epitope-tagged versions were prepared, in which the Env coding sequences were fused at the C terminus with the influenza HA tag (ΔGP SUΔ103-352HA and ΔGP SUΔ103-402HA) (Fig. 1A). These epitope-tagged constructs were generated to facilitate detection of the deleted proteins, since an antibody to JSRV Env that reliably detects proteins in Western blots or immunoprecipitations is not currently available.
Transformation of NIH 3T3 cells by JSRV Env SU deletion constructs.
To test if domains in JSRV SU are important for cell transformation, the SU deletion plasmids were tested for transformation of NIH 3T3 cells in focus formation assays as described previously (15) (Table 1). As expected, the full-length JSRV Env expression construct, ΔGP, efficiently induced foci of transformed NIH 3T3 cells. However, none of the SU Env deletion constructs was able to induce NIH 3T3 cell transformation (Table 1). Similar results were obtained in three independent NIH 3T3 transformation assays; at least two different plasmid DNA preparations were tested for each construct. The HA-tagged full-length JSRV Env construct, ΔGPHA, also induced focus formation of NIH 3T3 cells but with a somewhat lower efficiency, as described previously (14) (Table 1). The two HA-tagged large SU deletion constructs, ΔGP SUΔ103-352HA and ΔGP SUΔ103-402HA, also failed to transform NIH 3T3 cells (Table 1).
TABLE 1.
Transformation by JSRV Env SU deletion constructs in NIH 3T3 cells
| JSRV Env protein | No. of foci observed
|
||
|---|---|---|---|
| Expt 1 | Expt 2 | Expt 3 | |
| PCDNA 3.1 | 0 | 0 | 0 |
| ΔGP | 27 | 24 | 28 |
| ΔGP SU102/103PstI | 0 | 0 | 0 |
| ΔGP SUΔ103-152 | 0 | 0 | 0 |
| ΔGP SUΔ103-202 | 0 | 0 | 0 |
| ΔGP SUΔ103-252 | 0 | 0 | 0 |
| ΔGP SUΔ103-302 | 0 | 0 | 0 |
| ΔGP SUΔ103-352 | 0 | 0 | 0 |
| ΔGP SUΔ103-402 | 0 | 0 | 0 |
| ΔGPHA | 16 | 14 | 10 |
| ΔGP SUΔ103-352HA | 0 | 0 | 0 |
| ΔGP SUΔ103-402HA | 0 | 0 | 0 |
In light of the inability of any of the nested deletions to show transformation, an additional set of SU constructs containing sequential smaller (50-aa) deletions were generated (Fig. 1B). In addition, the PCR-based cloning strategy also yielded SU insertion mutants containing the PstI restriction site linker at various positions in SU; these insertions would add 2 aa (Gln and Leu) to the mature protein (Fig. 1B). This set of constructs was then tested in the NIH 3T3 focus formation assay (Fig. 1C). However, all of these constructs also failed to transform NIH 3T3 cells, even the PstI linker insertions.
Expression of the Env deletion constructs.
The inability of all of the SU deletion constructs to transform NIH 3T3 cells suggested that domains of JSRV SU downstream of position 102 in the N terminus of Env are important for transformation. However, several alternative reasons could explain why some or all of the mutants failed to transform: differences in expression and/or stability of env mRNA or Env protein; failure of the deleted envelope proteins to localize to the plasma membrane; and improper folding of the mutated protein, leading to instability and/or lack of transforming activity.
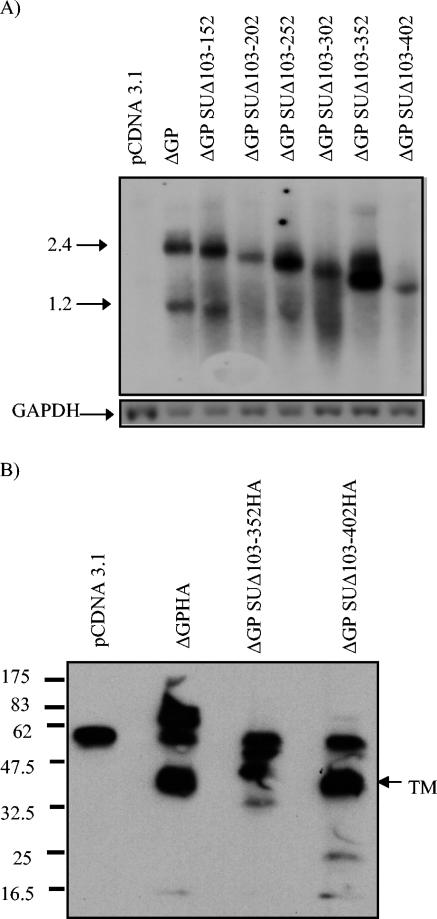
To determine if deletion of SU RNA sequences affected mRNA expression, we carried out Northern blot analysis for JSRV Env RNA from 293T cells transiently transfected with the nested deletion constructs (Fig. 2A). In general the levels of Env mRNA were comparable to those expressed by the full-length Env expression plasmid ΔGP. As expected, the nested deletions through the JSRV envelope resulted in correspondingly smaller Env mRNAs. For ΔGP-transfected cells, two sizes of JSRV-specific envelope mRNA were evident: a 2.4-kb RNA corresponding to full-length spliced env mRNA, and a 1.2-kb RNA corresponding to a premature cleavage/polyadenylation event near the SU-TM border (15, 23). The nested deletion constructs showed decreases in size for both of these RNAs, as expected. The sequential deletion constructs also expressed similar amounts of JSRV Env mRNA (data not shown).
FIG. 2.
RNA and protein encoded by SU deletion constructs. (A) 293T cells were transiently transfected with the plasmids indicated, and 48 h after transfection, total RNA was analyzed by agarose gel electrophoresis and Northern blot hybridization with a JSRV env probe. Bands corresponding to the spliced 2.4-kb env mRNA and the 1.2-kb prematurely polyadenylated env transcript (22, 23) are indicated for the ΔGP lane. The deletion constructs show a corresponding decrease in the size of these bands, as expected. Rehybridization of the same blot with a glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probe is also shown. (B) Western blot analysis of 293T cells analyzed 48 h after transfection with the HA-tagged full-length ΔGPHA or the Env SU deletion construct ΔGP-SUΔ103-352HA or ΔGP SUΔ103-402HA is shown. Cell lysates were first immunoprecipitated with anti-HA-antibody prior to SDS-PAGE and Western analysis with an anti-HA antibody. The 37-kDa TM protein expressed in ΔGPHA-transfected cells is indicated. A nonspecific band of ca. 55 kDa (likely immunoglobulin heavy chain) was detected in all lanes.
To test whether elimination of SU protein sequences affected the stability of the resulting Env proteins, the HA epitope-tagged versions of the two largest SU deletions, ΔGP SUΔ103-352HA and ΔGP SUΔ103-402HA, were studied. Transiently transfected 293T cells were analyzed by immunoprecipitation with an anti-HA antibody, followed by SDS-PAGE and Western blot analysis for the HA epitope (Fig. 2B). For comparison, cells transfected with HA-tagged full-length Env protein expression construct ΔGPHA were analyzed. For the ΔGPHA-transfected cells, a protein of ca. 37 kDa was observed corresponding to the predicted size of the cleaved TM protein, as described previously (14), as well as a higher-molecular-mass protein of ca. 80 kDa, presumably corresponding to the uncleaved envelope polyprotein precursor. (A nonspecific band of ca. 55 kDa was evident in all lanes, likely representing immunoglobulin heavy chain carried over from the immunoprecipitation step.) The ΔGP SUΔ103-352HA-transfected cells showed a reactive protein slightly larger than the 37-kDa TM protein. This presumably was due to inefficient cleavage by the cellular protease that normally cleaves between SU and TM. The deletion endpoint for this construct (residue 352) was quite close to the putative SU/TM cleavage site (predicted to be at residue 378). The SUΔ103-402HA-transfected cells showed a protein that had an electrophoretic mobility similar to that of the wild-type 37-kDa TM protein. In fact, this deletion would have removed the putative SU/TM cleavage site as well as 26 downstream amino acids, although this difference in size would not be resolved under the conditions of the SDS-PAGE. Most importantly, the relative amounts of envelope protein detected were similar for the wild-type and deletion constructs. Thus, for these two deletions, elimination of the SU domain did not lead to instability of the resulting protein.
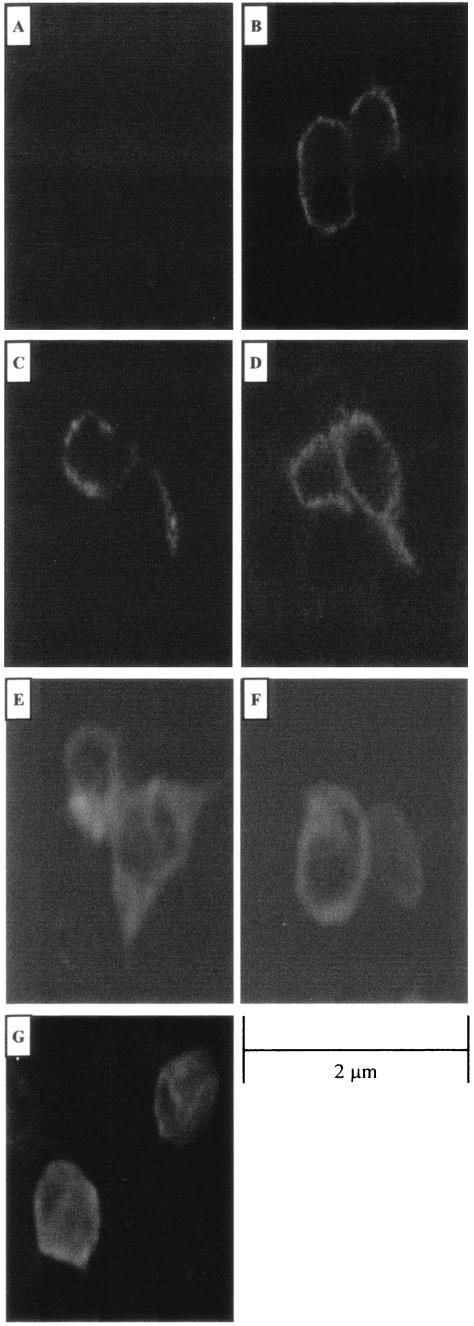
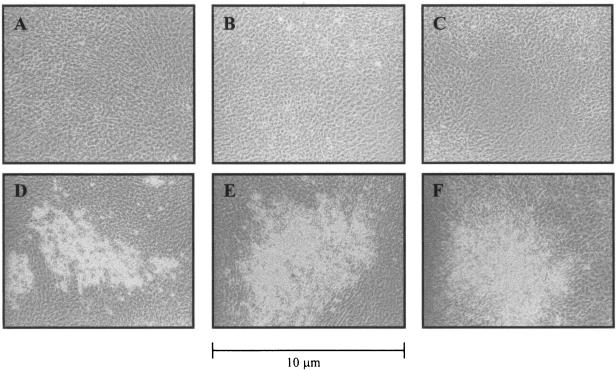
To address the possibility that deletions of the SU domain interfered with Env protein localization to the plasma membrane and thus influenced the transforming activity of the truncated SU Env proteins, we used confocal microscopy to visualize the location of the deleted Env proteins (Fig. 3). 293T cells were transiently transfected with the HA-tagged full-length Env expression plasmid, ΔGPHA, or the HA-tagged deletion constructs, ΔGP SUΔ103-352HA and ΔGP SUΔ103-402HA, and confocal immunofluorescence microscopy with an anti-HA antibody was performed. The envelope protein expressed from both deletion constructs showed the same ring-like plasma membrane staining pattern as for ΔGPHA (Fig. 3).
FIG.3.
Intracellular localization of JSRV Env proteins. Confocal microscopy of 293T cells transiently transfected with the empty expression vector pCDNA 3.1 (A), the HA-tagged full-length expression plasmid ΔGPHA (B), the HA-tagged SU deletion construct ΔGP SUΔ103-352HA, ΔGP SUΔ103-402HA, ΔGP SUΔ253-302HA, or ΔGP SUΔ353-402HA (C, D, E, and F, respectively), or the HA-tagged SU Env PstI insertion construct ΔGP SU102/103PstIHA (G) is shown. HA-tagged Env proteins were detected by staining with a fluorescein isothiocyanate-conjugated monoclonal antibody to HA. Bar, 2 μm. Magnification, ×600.
We also epitope tagged versions of three additional SU mutants: two 50-aa deletions (ΔGP SUΔ252-302HA and ΔGP SUΔ353-402HA) and a linker insertion (ΔGP SU102/103 PstI HA). All three mutants expressed high levels of the Env protein in Western blots, and the patterns indicated that they were glycosylated. However, these proteins were deficient for cleavage into SU and TM, which might reflect defective transport to the cell surface (data not shown). Confocal immunofluorescence microscopy was performed on 293T cells transiently transfected with these three mutants, as also shown in Fig. 3. ΔGP SUΔ252-302HA and ΔGP SUΔ353-402HA showed the same plasma membrane distribution as for ΔGPHA. ΔGP SU102/103PstIHA showed protein at the plasma membrane, but some was also retained intracellularly. In total, four of the five epitope-tagged SU mutations showed plasma membrane localization similar to that of wild-type JSRV Env protein, while one showed a partial defect in transport to the plasma membrane.
Transformation in rat 208F cells.
It seemed important to carry out transformation assays with another cell line, since cells of different tissue types or species can respond differently to a given oncogene (14). Rat 208F fibroblasts have also been used in JSRV transformation assays by other investigators (3, 13). These cells have several advantages compared to NIH 3T3 cells: (i) transformation occurs more rapidly (foci evident beginning at 2 weeks); (ii) the transformed cells are typically more distinct from the nontransformed cells; and (iii) the background monolayers remain flat longer, allowing for extended assays that might detected weakly transforming mutants. Thus, we transfected the minimal SU deletions and insertions into rat 208F cells and assayed their ability to induce foci of transformation. Table 2 gives a summary of three similar experiments performed with this set of constructs. As expected, the wild-type envelope expression construct ΔGP was able to transform rat 208F cells. It was noteworthy that two additional constructs gave efficient focus formation in these cells: ΔGP ΔSU14-52, with a deletion in SU sequences upstream of the predicted ER localization signal (signal peptide), and ΔGP SU52/53PstI, encoding two additional amino acids in the reading frame of JSRV Env, also transformed 208F cells (Table 2). For both of these mutants, the resulting SU proteins would likely be identical to wild-type SU, since the mutated regions presumably would be cleaved off during removal of the signal peptide in the ER. In contrast, all the other minimal deletion and insertion constructs, in which disruption of the SU coding sequences lies downstream of the ER signal sequence, were repeatedly unable to transform 208F cells, consistent with the results obtained in the NIH 3T3 transformation assays. It was interesting that the ΔGP SU13/14PstI insertion mutant did not induce foci in 208F cells; one possible explanation is that this insertion interfered with transport of the protein into the ER.
TABLE 2.
Transformation of JSRV Env mutant constructs in 208F cells
| JSRV Env protein | No. of transformed foci at wka:
|
No. of colonies picked at wkb:
|
Foci re-formationc | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 3 | 4 | 5 | 6 | 7 | −8 | 4 | 7 | ||
| Mock | 0 | 0 | 0 | 1 | 1 | 1 | ND | 1 | 0/1 | |
| pCDNA 3.1 | 0 | 0 | 0 | 0 | 0 | 1 | ND | 1 | 0/1 | |
| ΔGP | 36 ± 3 | 68 ± 5 | 149 ± 13 | TMTC | 3 | ND | 3/3 | |||
| ΔGP SUΔ14-52 | 13 ± 9 | 27 ± 6 | 69 ± 21 | TMTC | 3 | ND | 3/3 | |||
| ΔGP SUΔ103-152 | 0 | 0 | 0 | 1 | 1 | 1 | ND | 1 | 0/1 | |
| ΔGP SUΔ203-252 | 0 | 0 | 0 | 0 | 0 | 2 | ND | 2 | 0/2 | |
| ΔGP SUΔ253-302 | 0 | 0 | 0 | 0 | 1 | 1 | ND | 1 | 0/1 | |
| ΔGP SUΔ353-402 | 0 | 0 | 0 | 1 | 1 | 2 | ND | 1 | 0/1 | |
| ΔGP SU13/14PstI | 0 | 0 | 0 | 0 | 0 | 0 | ND | ND | ND | |
| ΔGP SU52/53PstI | 21 ± 3 | 34 ± 5 | 59 ± 7 | 76 ± 22 | 86 ± 18 | TMTC | 3 | ND | 3/3 | |
| ΔGP SU102/103PstI | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ND | ND | ND |
| ΔGP SU202/203PstI | 0 | 0 | 0 | 1 | 1 | 1 | ND | 1 | 0/1 | |
| ΔGP Y590F | 0 | 0 | 0 | 1 | 1 | 4 ± 1 | ND | 3 | 0/3 | |
| ΔGP M593T | 0 | 13 ± 6 | 19 ± 5 | 30 ± 3 | 38 ± 1 | 54 ± 4 | 70 ± 5 | 3 | ND | 3/3 |
All transfections were performed three times, and the average number of foci at each time is indicated along with the average deviation from the mean. Numbers where no errors are given represent the total foci observed in the three experiments. TMTC, too many to count.
Foci were isolated from initial transfectants at either 4 weeks or 7 weeks posttransfection, as indicated. ND, not done.
Isolated colonies were reseeded in a Six-well plate and assayed for their ability to re-form foci 2 weeks posttransfer. Results are expressed as number of colonies re-forming transformed foci/number of colonies picked.
As shown in Table 2, after prolonged incubation in the 208F transformation assays, several of the mutant-transfected cultures showed small numbers (1-2) of foci. While these numbers were not substantially higher than the number of foci observed in control 208F cultures (mock transfected or transfected with pCDNA3.1), it remained possible that they resulted from expression of the mutant Env proteins. Therefore, these foci were picked and recultured in an attempt to isolate the transformants. However, as also shown in Table 2, none of these picked foci showed re-formation of transformed foci. In contrast, when foci were picked at 4 weeks from cultures transfected with wild-type ΔGP DNA or from the deletion and insertion constructs with high transformation activity, secondary transformed foci appeared in every case. Thus, it seems likely that the small numbers of foci appearing after prolonged passage of the transformation-negative construct transfections did not represent foci induced by the mutant Env proteins. However, low-efficiency transformation that is not immediately apparent could not be ruled out.
As described in the introduction, we previously have reported that the cytoplasmic tail of the Env TM domain is necessary for JSRV transformation of NIH 3T3 cells (22). In particular, a YXXM motif was found to be important, since mutation of either of the critical residues (Y590F and M593T) abolished transformation (22) (Fig. 1B). Recently, Liu et al. have reported that the YXXM motif is not necessary for transformation of 208F rat cells (12), so we tested the ΔGP Y590F and ΔGP M593T mutants for 208F transformation as well (Table 2). In the conditions used here, ΔGP Y590F did not transform 208F cells, and the small number of foci appearing after 7 weeks did not re-form when picked and regrown. We also performed 208F transformation assays under the culture conditions described by Liu et al. (plating at lower density; culture in 1 μM dexamethasone), and the results were the same as shown in Table 2. On the other hand, consistent with the results of Liu et al., the ΔGP M593T mutant induced transformed foci in 208F cells, although with reduced efficiency. Thus, the tyrosine residue at position 590 in the JSRV Env cytoplasmic tail is essential for transformation by JSRV Env in both rat and mouse fibroblasts, whereas the methionine residue at position 593 appears only essential to JSRV Env transformation in NIH 3T3 cells.
Complementation between SU and TM domains in JSRV transformation.
The experiments with the SU deletions and insertions presented above suggested the necessity of an SU domain in JSRV transformation. On the other hand, previously described studies involving mutants in the TM cytoplasmic tail (e.g., ΔGP Y590F) have implicated this region of JSRV Env in transformation as well. If the SU and TM domains act independently in facilitating transformation, we hypothesized that cotransfection of two nontransforming env constructs that are mutated in the TM and SU domains, respectively, might complement and induce formation of transformed foci. In the cotransfected cells, the TM mutant would supply wild-type SU protein, and the SU mutant would supply wild-type TM protein. Thus, 208F cells were cotransfected with the ΔGP SUΔ103-352 mutant and the ΔGP Y590F TM mutant (Table 3, experiment 1; Fig. 4A). While neither of the mutants individually induced transformed foci, cotransfection resulted in efficient focus formation (ca. 20 to 25% as efficient as wild-type ΔGP). The resulting foci were morphologically identical to foci resulting from ΔGP transfection (Fig. 5). Transformed colonies picked at 4 weeks re-formed foci upon reculture. Thus, complementation between the SU and TM mutants was observed. We also tested epitope-tagged versions of these two constructs in the cotransfection assay, as also shown in Table 3, experiment 1 (ΔGP-SUΔ103-352HA and ΔGP Y590FHA). Again, complementation for focus formation was observed, although the efficiency was somewhat less. The lower level of efficiency was expected, since addition of the HA epitope to the C terminus of the TM protein reduces the efficiency of transformation (Table 1).
TABLE 3.
Cotransfection of transformation-defective SU and TM mutants in 208F cellsa
| JSRV Env protein | No. of transformed foci at weekb:
|
No. of colonies picked at weekb 4c | No. of re-formed focid | |||||
|---|---|---|---|---|---|---|---|---|
| 2 | 3 | 4 | 5 | 6 | 7 | |||
| Expt I | ||||||||
| Mock | 0 | 0 | 0 | 0 | 0 | 0 | ND | ND |
| PCDNA 3.1 | 0 | 0 | 0 | 0 | 0 | 0 | ND | ND |
| ΔGP | 23 ± 5.5 | 101 ± 20 | 128 ± 17 | TMTC | 3 | 3/3 | ||
| ΔGP SUΔ103-352 | 0 | 0 | 0 | 0 | 0 | 0 | ND | ND |
| ΔGP Y590F | 0 | 0 | 0 | 0 | 0 | 0 | ND | ND |
| ΔGP Y590F + ΔGP SUΔ103-352 | 9 ± 1 | 21 ± 2 | 28 ± 8 | 46 ± 6.5 | 61 ± 6 | 69 ± 4.5 | 6 | 6/6 |
| ΔGP Y590FHA | 0 | 0 | 0 | 0 | 0 | 0 | ND | ND |
| ΔGP SUΔ103-352HA | 0 | 0 | 0 | 0 | 0 | 0 | ND | ND |
| ΔGP Y590FHA + ΔGP SUΔ103-352HA | 0 | 1 ± 1 | 5 ± 2 | 13 ± 2 | 22 ± 3 | 27 ± 1 | 3 | 3/3 |
| Expt 2 | ||||||||
| Mock | 0 | 0 | 0 | 0 | 0 | 0 | ND | ND |
| PCDNA 3.1 | 0 | 0 | 0 | 0 | 0 | 0 | ND | ND |
| ΔGP | 36 ± 4 | 70 ± 13 | 140 ± 26 | TMTC | 3 | 3/3 | ||
| ΔGP SUΔ103-352 | 0 | 0 | 0 | 0 | 0 | 0 | ND | ND |
| ΔGP SuxTMen | 0 | 0 | 0 | 0 | 0 | 0 | ND | ND |
| ΔGP SuxTMen + ΔGP SUΔ103-352 | 0 | 15 ± 2 | 16 ± 2 | 17 ± 1 | 23 ± 4 | 26 ± 4 | 3 | 3/3 |
| Expt 3e | ||||||||
| Mock | 0 | 0 | 0 | 0 | ND | ND | ||
| PCDNA 3.1 | 0 | 0 | 0 | 0 | ND | ND | ||
| ΔGP | 29 | 116 | TMTC | ND | ND | |||
| ΔGP Y590F + ΔGP SUΔ103-352 | 3 | 6 | 7 | 13 | ND | ND | ||
| ΔGP Y590F + ΔGP SUΔ253-302HA | 0 | 2 | 4 | 9 | ND | ND | ||
Diagrams of the two constructs used in the cotransfection/transformation assay are in Fig. 4.
All transfections were performed three times, and the average number of foci at each time is indicated along with the average deviation from the mean. Numbers where no errors are given represent the total foci observed in the three experiments. TMTC, too many to count.
Number of foci isolated from initial transfectants at 4 weeks posttransfection. ND, not done.
Isolated colonies were reseeded in a six-well plate and assayed for their ability to reform foci 2 weeks posttransfer. Results are expressed as number of colonies re-forming transformed foci/number of colonies picked.
Experiment 3 values represent transformed foci from a single assay.
FIG. 5.
Complementation between separate domains of JSRV Env in transformation. 208F cells transfected with no DNA (A), the nontransforming SU deletion mutant ΔGP SUΔ103-352 (B), and the nontransforming TM cytoplasmic tail tyrosine mutant ΔGP Y590F (C) are shown at 4 weeks posttransfection. A typical focus induced by the wild-type ΔGP (D) is shown. Foci arising in cells cotransfected with ΔGP SUΔ103-352 plus Y590F (E) or ΔGP SUΔ103-352 plus SuxTMen (F) appear identical to those induced by ΔGP. Bar, 10 μm. Magnification, ×100.
Complementation for transformation with another TM mutant was also tested. We previously described molecular cloning of endogenous JSRV-related proviruses from the sheep genome (22); these endogenous viruses are quite related to exogenous JSRV (>90% sequence homology), although one region of difference is the membrane-spanning domain and cytoplasmic tail of the TM protein. Since endogenous JSRVs are expressed in several normal sheep tissues (e.g., the female reproductive tract) (7, 20, 28), they presumably do not induce oncogenic transformation. Indeed, an envelope chimera consisting of the SU of JSRV but the TM of an endogenous JSRV-related virus (SUxTMen) does not transform NIH 3T3 cells (22). Cotransfection of SUxTMen and SUΔ103-352 is shown in Table 3, experiment 2, and in Fig. 4B and 5. Complementation for focus formation between the two mutants was observed, and when the resulting colonies were picked, they re-formed foci upon replating.
We also tested the SU deletion mutant ΔGP SUΔ252-302HA for complementation with ΔGP Y590F, as shown in Table 3, experiment 3. Transformed foci appeared with efficiency and time course similar to those for ΔGP 103-352HA cotransfected with ΔGP Y590F. As discussed above, the presence of the HA epitope on the C terminus of the ΔGP SUΔ252-302HA TM protein likely resulted in reduced transformation. These results indicated that complementation for transformation could be achieved with another SU deletion mutant. This SU deletion mutant showed plasma membrane localization, although it was not cleaved into SU and TM (see above). Thus, cleavage per se was not necessary for the ability of the TM domain to participate in transformation.
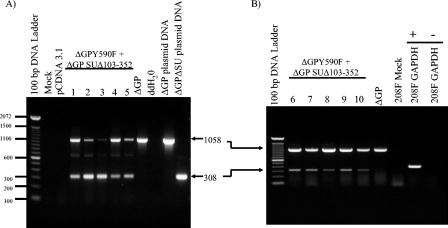
If the transformed foci described above resulted from genuine complementation, the transformants would all be expected to contain both mutant construct DNAs and to express them as RNA. To detect JSRV DNA, we used an env-specific PCR on genomic DNA recovered from five independent transformants (Fig. 6A). PCR with an upstream SU primer and a downstream TM primer yielded a product of 308 bp from ΔGP SUΔ103-352, while wild-type (or ΔGP Y590F) DNA gave a product of 1,058 bp (lanes on the right). PCR of DNA from five different foci resulting from cotransfection yielded fragments characteristic of ΔGP SUΔ103-352 and ΔGP Y590F in all cases. As expected, when this primer set was used to analyze transformed foci resulting from transfection with wild-type ΔGP, only the 1,058-bp band was amplified.
FIG. 6.
JSRV DNA and RNA in transformed 208F cells. (A) PCR was used to detect JSRV Env DNA in transformed 208F colonies picked at 4 weeks posttransfection. The primer set used for JSRV Env DNA detection spans the deleted region of SU and results in amplification of a 308-bp PCR product from ΔGP SUΔ103-352 (lane 12) or a 1,058-bp PCR product from full-length ΔGP (lane 11) plasmid DNA. PCR amplifications on DNA from five different transformed colonies picked from 208F cells cotransfected with ΔGPY590F plus ΔGP SUΔ103-352 (lanes 4 to 8) and for 208F cells transformed by ΔGP (lane 9) are shown. Mock-transfected or pCDNA 3.1-transfected 208F cells (lane 2 and 3, respectively) showed no signal, as expected. (B) RT-PCR was used to detect JSRV Env RNA in five transformed 208F cell colonies resulting from cotransfection of the same two plasmids as in panel A (lanes 2 to 7). The colonies analyzed in panels A and B were different. Both 308-bp and 1,058-bp RT-PCR products are apparent in lanes 2 to 7, whereas only the 1,058-bp PCR product is visible in 293T cells transformed by ΔGP (lane 8). There was no amplification from mock-transfected 208F cells (lane 9), and RT-PCR for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) RNA was also reverse transcriptase dependent (lanes 10 to 11: +, reverse transcriptase added to the PCR; −, no reverse transcriptase added).
To test for expression of the mutant constructs in the foci arising from cotransfection, RT-PCR with the same primer set was carried out on an additional five transformants (Fig. 6B). RT-PCR products characteristic of both ΔGP SUΔ103-352 and ΔGP Y590F mRNAs were detected in all foci analyzed. Taken together, the presence of both mutant DNAs in each transformant, as well as the corresponding mRNAs, supported the conclusion that the transformation arose from complementation between different domains of env. In addition, they did not support an alternative explanation of DNA recombination between the two mutant plasmids in which a wild-type env gene would be generated in the coprecipitated cells. If this were the predominant mechanism, not all transformants would be expected to retain and express the mutant plasmids. While the PCR and RT-PCR analyses could not distinguish between a recombinant wild-type DNA and ΔGP Y590F DNA, the 308-bp fragment was characteristic of only ΔGP SUΔ103-352, and it was readily detected in all transformants.
DISCUSSION
In previous studies, we have shown that expression of the JSRV envelope protein is sufficient to transform immortalized rodent fibroblasts cells in culture and that mutation of the YXXM motif in the cytoplasmic tail of the TM domain abolishes transformation (15, 22). Here, we examined the role of SU in JSRV Env-induced transformation by systematically deleting regions in SU. Progressive truncations in SU downstream of the ER signal sequence were unable to induce focus formation in NIH 3T3 fibroblasts, which suggested that domains of SU also may be important for transformation. The inability of these SU mutants to transform rodent fibroblasts was not due to low expression or stability of the deleted env mRNAs (Fig. 2A). For at least two of the larger deletion constructs (ΔGP SUΔ103-352HA and ΔGP SUΔ103-402HA), relatively normal levels of the truncated Env proteins also were expressed (Fig. 2B), and the resulting proteins were transported to the plasma membrane properly (Fig. 3).
We also tested the nested SU deletion constructs for transformation in another rodent fibroblast cell line, rat 208F (13). These cells have less background than NIH 3T3 cells in focus formation assays, and the assays can be carried for longer times (6 to 7 weeks). This increased the possibility of detecting mutants with low (but genuine) transformation activity. However, no transformation was observed with this same set of SU deletion constructs in 208F cells either. When a series of smaller deletions (50 aa) spaced at ca. 50-aa intervals throughout SU was tested, we only observed 208F cell transformation with a deletion upstream of the putative ER signal sequence (ΔGP SUΔ14-52) that would presumably express wild-type processed SU and TM proteins. This transformation was reassuring. A series of PstI linker insertion mutations in SU also did not transform NIH 3T3 or 208F cells, with the exception of a PstI insertion upstream of the ER signal sequence (ΔGP SU52/53PstI) that was able to transform 208F cells. While some of the deletion or insertion constructs may have had defects in expression of the mutant Env proteins, when five selected mutants were epitope tagged, all five expressed protein at normal levels. Four out of the five showed plasma membrane localization, while the fifth showed localization both at the plasma membrane and intracellularly. This supports the notion that a domain(s) of SU may be important for JSRV transformation.
If SU and TM act independently in transformation, then genetic complementation between a transformation-negative SU mutant (expressing wild-type TM) and a transformation-negative TM mutant (expressing wild-type SU) would be expected. As shown in Table 3, cotransfection with three different pairs of such mutants led to quite efficient transformation of 208F cells, while the individual SU or TM mutants were completely negative for transformation in the same assay. Similar results were also obtained when the cotransfections were carried out in NIH 3T3 cells and scored at 4 weeks (data not shown). These experiments strongly support an independent role for SU in JSRV transformation.
Potential alternate explanations for transformation in the cotransfection experiments did not seem likely. One possibility could be that recombination between the two transfected plasmids occurred, yielding a recombinant expressing wild-type Env protein, as mentioned in Results. However, such recombination would not be expected to occur extremely frequently, and the plasmids in the transfected cells do not replicate autonomously since they lack mammalian origins of replication. Thus, rare recombination events would not be amplified. At the same time, transformation for the most efficient combination (ΔGP Y590F and ΔGP SUΔ103-352; Table 3, experiment 1) was within 4.5-fold of the frequency observed for wild-type env. Moreover, as shown in the PCR and RT-PCR studies of Fig. 6, all transformants picked from the cotransfection plates (a total of 10) showed evidence for the presence and/or expression of the ΔGP SUΔ103-352 plasmid (as well as a plasmid that could have been either ΔGP Y590F or a wild-type recombinant). If DNA recombinants were responsible for the transformation, it seems unlikely that all 10 independent transformants would retain the ΔGP SUΔ103-352 plasmid during the focus assay and purification of the transformants (>8 weeks and many cell divisions).
The mechanism(s) of transformation by JSRV Env protein are the subject of considerable interest. It seems likely that different domains of SU and TM may interact with cellular cytoplasmic and/or cell surface proteins, leading to activation of different signal transduction pathways. Previous experiments have indicated that the cytoplasmic tail of the TM protein may activate the PI3-kinase/Akt pathway, since phosphorylated (activated) Akt can be detected in the transformed cells (1, 3, 13, 22, 31). However PI3-kinase itself is not required for JSRV transformation (14), which suggests that other proteins may also bind to the cytoplasmic tail of TM and initiate signal transduction. We favor the model that a cell surface protein binds to the SU domain and that this initiates signal transduction that collaborates with signals initiated from TM protein. One possible candidate for the cell surface protein could be the Toll-like receptor 4 (TLR-4) or TLR-2 protein. Several retroviral envelope proteins have been shown to bind to TLR-4 and/or TLR-2 and to initiate downstream signaling—most notably mouse mammary tumor virus (another betaretrovirus) (27). Preliminary evidence indicates that JSRV Env protein indeed binds TLR-4 (J. Rassa, N. Maeda, S. Ross, and H. Fan, unpublished data). It is interesting that for mouse mammary tumor virus, the region of the envelope protein that binds to TLR-4 is the C-terminal portion of SU (S. Ross, personal communication)—the same region of JSRV SU likely to be important for transformation. An alternate model for the complementation between SU and TM in transformation could be that the SU domain is important for multimerization of the TM domain, which then is responsible for activating signal transduction. The most likely complex for JSRV envelope protein in transformed cells would seem to be a trimer; other retroviral envelope proteins are generally believed to exist as trimers in virions (9). However, the region of Env protein involved in trimer formation is the extracellular domain of TM, not SU.
Many other viral oncogenes activate multiple signal transduction pathways by binding signaling molecules through different domains of the oncogene protein (e.g., SV40 large T-antigen and Rous sarcoma virus v-src protein (4, 11). Thus, the JSRV envelope protein appears to resemble other viral oncogene proteins in signaling through different protein domains. One consequence of this is that transformation of different cell types or cell lines may be more or less dependent on a particular pathway for transformation or on a particular binding motif in the oncogene protein. Indeed, it has been shown that while the YXXM motif in the cytoplasmic tail of TM is absolutely required for transformation of NIH 3T3 cells (22), mutation of the methionine residue (e.g., M593T) does not completely abolish transformation in rat 208F cells (13) (Table 2), and mutations in either the tyrosine or methionine residues do not completely abolish transformation in avian DF-1 cells (2).
Recently, Chow et al. (3) have also studied domains of env necessary for JSRV transformation. They reported that expression of only the TM domain was sufficient for transformation of 208F cells, if it was anchored to the plasma membrane by myristoylation. They also reported that deletion of the amino-terminal receptor-binding domain (RBD) of SU did not abolish transformation. This might seem to be inconsistent with the lack of transformation of the SU insertion and deletion mutants described here. As discussed above, some of the mutants in this study may have been improperly folded. In contrast, Chow et al. (3) employed computer modeling to predict a structure for the JSRV RBD, and their RBD deletion was designed to excise the RBD without disrupting the rest of the SU protein structure. It should also be noted that the RBD deletion of Chow et al. (3) still contained the C-terminal portion of SU protein. The large deletions of SU described here (ΔGP SUΔ103-352 and ΔGP SUΔ103-402) lacked essentially all of SU. Moreover, the complementation experiments (see below) indicate that the TM protein encoded by ΔGP SUΔ103-352 is functional for transformation. Taken together, this suggests that the C-terminal part of JSRV SU may be important for transformation.
Recently Danilkovitch-Miagkova et al. (5) have reported that JSRV SU protein is important for transformation of a human lung epithelial line (BAES-2). In these cells, they concluded that SU binds to the JSRV receptor, hyaluronidase-2 (Hyal-2), preventing appearance of Hyal-2 at the cell surface. In the absence of JSRV Env, Hyal-2 is normally complexed with the Stk/RON receptor tyrosine kinase; when Stk/RON is complexed with Hyal-2, it is not phosphorylated. They also showed that expression of JSRV Env in cells leads to loss of the Hyal-2/RON complex, and phosphorylation (activation) of Stk/RON. These results may be important for oncogenic transformation of lung epithelial cells (the normal target of JSRV tumorigenesis in sheep), but they are probably not involved in the transformation assays described here. The rodent Hyal-2 proteins do not normally function as JSRV receptors; in particular, murine Hyal-2 does not bind JSRV envelope (12). Moreover, Stk/RON is not expressed at high levels in rodent fibroblasts, such as NIH 3T3 or 208F. Thus, in ovine lung epithelial cells, two domains of SU may be important for transformation—the receptor binding domain (activating Stk/RON) and a C-terminal domain indicated in the experiments described here. In rodent fibroblasts, only the C-terminal domain may be important, and study of these cells may facilitate elucidation of the mechanism of its contribution to oncogenic transformation.
Acknowledgments
This work was supported by NIH grant R01CA094188.
The advice of Naoyoshi Maeda and Yasuo Inoshima is greatly appreciated. Support of the UCI Cancer Research Institute and the DNA sequencing core of the Chao Family Comprehensive Cancer Center is acknowledged.
REFERENCES
- 1.Alberti, A., C. Murgia, S. L. Liu, M. Mura, C. Cousens, M. Sharp, A. D. Miller, and M. Palmarini. 2002. Envelope-induced cell transformation by ovine betaretroviruses. J. Virol. 76:5387-5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen, T. E., K. J. Sherrill, S. M. Crispell, M. R. Perrott, J. O. Carlson, and J. C. DeMartini. 2002. The jaagsiekte sheep retrovirus envelope gene induces transformation of the avian fibroblast cell line DF-1 but does not require a conserved SH2 binding domain. J. Gen. Virol. 83:2733-2742. [DOI] [PubMed] [Google Scholar]
- 3.Chow, Y. H., A. Alberti, M. Mura, C. Pretto, P. Murcia, L. M. Albritton, and M. Palmarini. 2003. Transformation of rodent fibroblasts by the jaagsiekte sheep retrovirus envelope is receptor independent and does not require the surface domain. J. Virol. 77:6341-6350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Courtneidge, S. A. 1994. Protein tyrosine kinases, with emphasis on the Src family. Semin. Cancer Biol. 5:239-246. [PubMed] [Google Scholar]
- 5.Danilkovitch-Miagkova, A., F. M. Duh, I. Kuzmin, D. Angeloni, S. L. Liu, A. D. Miller, and M. I. Lerman. 2003. Hyaluronidase 2 negatively regulates RON receptor tyrosine kinase and mediates transformation of epithelial cells by jaagsiekte sheep retrovirus. Proc. Natl. Acad. Sci. USA 100:4580-4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeMartini, J. C., J. V. Bishop, T. E. Allen, F. A. Jassim, J. M. Sharp, M. de las Heras, D. R. Voelker, and J. O. Carlson. 2001. Jaagsiekte sheep retrovirus proviral clone JSRV(JS7), derived from the JS7 lung tumor cell line, induces ovine pulmonary carcinoma and is integrated into the surfactant protein A gene. J. Virol. 75:4239-4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeMartini, J. C., J. O. Carlson, C. Leroux, T. Spencer, and M. Palmarini. 2003. Endogenous retroviruses related to jaagsiekte sheep retrovirus. Curr. Top. Microbiol. Immunol. 275:117-137. [DOI] [PubMed] [Google Scholar]
- 8.Dirks, C., F. M. Duh, S. K. Rai, M. I. Lerman, and A. D. Miller. 2002. Mechanism of cell entry and transformation by enzootic nasal tumor virus. J. Virol. 76:2141-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Einfeld, D., and E. Hunter. 1988. Oligomeric structure of a prototype retrovirus glycoprotein. Proc. Natl. Acad. Sci. USA 85:8688-8692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan, H., M. Palmarini, and J. C. DeMartini. 2003. Transformation and oncogenesis by jaagsiekte sheep retrovirus. Curr. Top. Microbiol. Immunol. 275:139-177. [DOI] [PubMed] [Google Scholar]
- 11.Lavia, P., A. M. Mileo, A. Giordano, and M. G. Paggi. 2003. Emerging roles of DNA tumor viruses in cell proliferation: new insights into genomic instability. Oncogene 22:6508-6516. [DOI] [PubMed] [Google Scholar]
- 12.Liu, S. L., F. M. Duh, M. I. Lerman, and A. D. Miller. 2003. Role of virus receptor Hyal2 in oncogenic transformation of rodent fibroblasts by sheep betaretrovirus Env proteins. J. Virol. 77:2850-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu, S. L., M. I. Lerman, and A. D. Miller. 2003. Putative phosphatidylinositol 3-kinase (PI3K) binding motifs in ovine betaretrovirus Env proteins are not essential for rodent fibroblast transformation and PI3K/Akt activation. J. Virol. 77:7924-7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maeda, N., Y. Inoshima, D. A. Fruman, S. M. Brachmann, and H. Fan. 2003. Transformation of mouse fibroblasts by Jaagsiekte sheep retrovirus envelope does not require phosphatidylinositol 3-kinase. J. Virol. 77:9951-9959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maeda, N., M. Palmarini, C. Murgia, and H. Fan. 2001. Direct transformation of rodent fibroblasts by jaagsiekte sheep retrovirus DNA. Proc. Natl. Acad. Sci. USA 98:4449-4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller, A. D. 2003. Identification of Hyal2 as the cell-surface receptor for jaagsiekte sheep retrovirus and ovine nasal adenocarcinoma virus. Curr. Top. Microbiol. Immunol. 275:179-199. [DOI] [PubMed] [Google Scholar]
- 17.Miller, A. D., T. Curran, and I. M. Verma. 1984. c-fos protein can induce cellular transformation: a novel mechanism of activation of a cellular oncogene. Cell 36:51-60. [DOI] [PubMed] [Google Scholar]
- 18.Palmarini, M., and H. Fan. 2003. Molecular biology of jaagsiekte sheep retrovirus. Curr. Top. Microbiol. Immunol. 275:81-115. [DOI] [PubMed] [Google Scholar]
- 19.Palmarini, M., and H. Fan. 2001. Retrovirus-induced ovine pulmonary adenocarcinoma, an animal model for lung cancer. J. Natl. Cancer Inst. 93:1603-1614. [DOI] [PubMed] [Google Scholar]
- 20.Palmarini, M., C. A. Gray, K. Carpenter, H. Fan, F. W. Bazer, and T. E. Spencer. 2001. Expression of endogenous betaretroviruses in the ovine uterus: effects of neonatal age, estrous cycle, pregnancy, and progesterone. J. Virol. 75:11319-11327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palmarini, M., M. J. Holland, C. Cousens, R. G. Dalziel, and J. M. Sharp. 1996. Jaagsiekte retrovirus establishes a disseminated infection of the lymphoid tissues of sheep affected by pulmonary adenomatosis. J. Gen. Virol. 77:2991-2998. [DOI] [PubMed] [Google Scholar]
- 22.Palmarini, M., N. Maeda, C. Murgia, C. De-Fraja, A. Hofacre, and H. Fan. 2001. A phosphatidylinositol 3-kinase docking site in the cytoplasmic tail of the Jaagsiekte sheep retrovirus transmembrane protein is essential for envelope-induced transformation of NIH 3T3 cells. J. Virol. 75:11002-11009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palmarini, M., C. Murgia, and H. Fan. 2002. Spliced and prematurely polyadenylated Jaagsiekte sheep retrovirus-specific RNAs from infected or transfected cells. Virology 294:180-188. [DOI] [PubMed] [Google Scholar]
- 24.Palmarini, M., J. M. Sharp, M. de las Heras, and H. Fan. 1999. Jaagsiekte sheep retrovirus is necessary and sufficient to induce a contagious lung cancer in sheep. J. Virol. 73:6964-6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rai, S. K., J. C. DeMartini, and A. D. Miller. 2000. Retrovirus vectors bearing jaagsiekte sheep retrovirus Env transduce human cells by using a new receptor localized to chromosome 3p21.3. J. Virol. 74:4698-4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rai, S. K., F. M. Duh, V. Vigdorovich, A. Danilkovitch-Miagkova, M. I. Lerman, and A. D. Miller. 2001. Candidate tumor suppressor HYAL2 is a glycosylphosphatidylinositol (GPI)-anchored cell-surface receptor for jaagsiekte sheep retrovirus, the envelope protein of which mediates oncogenic transformation. Proc. Natl. Acad. Sci. USA 98:4443-4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rassa, J. C., J. L. Meyers, Y. Zhang, R. Kudaravalli, and S. R. Ross. 2002. Murine retroviruses activate B cells via interaction with toll-like receptor 4. Proc. Natl. Acad. Sci. USA 99:2281-2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanna, E., M. P. Sanna, C. Loddo, L. Sanna, M. Mura, T. Cadelano, and A. Leoni. 2002. Endogenous jaagsiekte sheep retrovirus RNA is expressed by different cell types in ovine foetus and placenta. Eur. J. Histochem. 46:273-280. [DOI] [PubMed] [Google Scholar]
- 29.Sharp, J. M., K. W. Angus, E. W. Gray, and F. M. Scott. 1983. Rapid transmission of sheep pulmonary adenomatosis (jaagsiekte) in young lambs. Brief report. Arch. Virol. 78:89-95. [DOI] [PubMed] [Google Scholar]
- 30.Verwoerd, D. W., E. M. De Villiers, and R. C. Tustin. 1980. Aetiology of jaagsiekte: experimental transmission to lambs by means of cultured cells and cell homogenates. Onderstepoort J. Vet. Res. 47:13-18. [PubMed] [Google Scholar]
- 31.Zavala, G., C. Pretto, Y. H. Chow, L. Jones, A. Alberti, E. Grego, M. De las Heras, and M. Palmarini. 2003. Relevance of Akt phosphorylation in cell transformation induced by Jaagsiekte sheep retrovirus. Virology 312:95-105. [DOI] [PubMed] [Google Scholar]