Abstract
In many extant animal and plant species in Europe and North America a correlation exists between the geographical location of individuals and the genetic relatedness of the mitochondrial (mt) DNA sequences that they carry. Here, we analyze mtDNA sequences from cave bears, brown bears, cave hyenas, and Neandertals in Europe before the last glacial maximum and fail to detect any phylogeographic patterns similar to those observed in extant species. We suggest that at the beginning of the last glacial maximum, little phylogeographic patterns existed in European mammals over most of their geographical ranges and that current phylogeographic patterns are transient relics of the last glaciation. Cycles of retreat of species in refugia during glacial periods followed by incomplete dispersal from one refugium into other refugia during interglacial periods is likely to be responsible for the deep genetic divergences between phylogeographic clusters of mtDNA seen today.
Keywords: ancient DNA, glacial refugia, mitochondrial DNA, Pleistocene, population structure
The mitochondrial (mt) DNA gene pools of animals and plants are often subdivided into clades of phylogenetically related mtDNA types that show no or little overlap in their geographical occurrence (1, 2). Similarly, subspecies or closely related species with nonoverlapping geographic distributions often hybridize in limited hybrid zones (3). These geographical separations of closely related populations are generally explained as a relic of the last glacial maximum (≈25,000–10,000 years B.P.) during which many species were reduced to refugia in nonglaciated areas (3). However, the genetic divergences between the clades of related mtDNA sequences often date far into the Pleistocene or even earlier (refs. 4 and 5 and references therein). Because many different species show a similar age for the divergence between the mitochondrial clades (6), this divergence has been interpreted as evidence for long separation of the respective populations (6). However, recent work where mtDNA sequences from late Pleistocene brown bears in Alaska were retrieved has shown that at least some mtDNA clades that are today geographically separated occurred together at a single location ≈35,000 years ago (7, 8). This suggests that phylogeographic patterns may have been less pronounced before the last glacial maximum than they are today. Although this might be a special case because brown bears immigrated to North America only 50,000–70,000 years ago and became widespread even later (9), these findings indicate that caution is warranted when historical inferences are drawn from modern population data.
A large number of European species show strong phylogeographic patterns concordant with isolation of populations in a few Pleistocene refugia in southern Europe (see, e.g., refs, 2, 4, and 6 for reviews). However, some species, such as European wolves (10), do not show any phylogeographic patterns. This absence has been suggested to be due to their high geographic mobility, resulting in the erosion of any signal of Pleistocene population separations (10). In agreement with this, autosomal microsatellites show little separation across the geographical boundaries of different mtDNA clades in brown bears (11, 12), probably as a result of the higher migration rate of males than females in this species.
Here, we analyze mtDNA sequences from late Pleistocene cave bears, brown bears, cave hyenas, and Neandertals and fail to find any evidence for phylogeographic patterns before the last glacial maximum. As the last glacial maximum ended only ≈10,000 years ago (13, 14), but the time between the second-to-last and the last glacial maximum allowed for ≈100,000 years of migration and population intermingling (13), we suggest that populations displayed considerably less phylogeographic patterns shortly before the last glacial maximum, whereas today phylogeographic patterns have vanished only for species with high migration rates. Thus, the phylogeographic patterns currently observed are transient relics of the last glacial maximum and do not represent long-term adaptations to different environments.
Materials and Methods
Bears. Cave bears from Germany, France, Italy, Austria, and Croatia and brown bears from Austria were extracted as described (15). Primers and PCR conditions used for cave bear (15) and brown bear (16) mtDNA amplifications have been described previously. Altogether, 134 bp of the mitochondrial control region were amplified in cave bears along with ≈270 bp in brown bears.
Carbon dating (Table 1, which is published as supporting information on the PNAS web site) was performed at Beta Analytic (Miami) and at the Oxford Radiocarbon Accelerator Unit (Oxford).
Hyenas. Cave hyenas from 15 different localities in Europe and Asia were extracted as described (15). A 366-bp piece of the cytochrome b gene was amplified in four overlapping pieces (see Supporting Text, which is published as supporting information on the PNAS web site).
Amplification, reamplification, cloning, and sequencing were done as described (15). Amplifications were carried out with a 3-min activation step at 94°C, followed by 60 cycles at 93°C for 30 s, 52°C for 60 s, and 72°C for 45 s. Carbon dating (Table 1) was performed at the Vienna Environmental Research Accelerator (VERA) (Vienna).
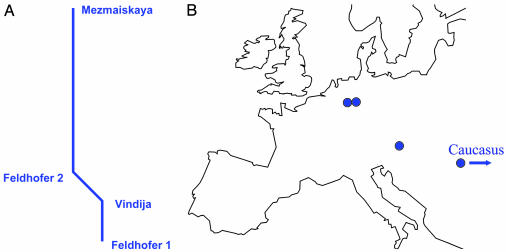
Neandertals. Published mtDNA sequences from four individuals from Feldhofer, Germany (17, 18), Mezmaiskaya, Russia (19), and Vindija, Croatia (20) were considered.
Authenticity of DNA Sequences. We applied criteria for the authenticity of the ancient DNA sequences as described (21). In particular, we only used DNA sequences for which each nucleotide position was determined from at least two independent amplifications and several clones have been sequenced from each amplification. These criteria are crucial to avoid misinterpretations due to miscoding lesions present in ancient DNA molecules. When discrepancies between two initial amplifications occurred, a third amplification was performed to identify the nucleotide sequence that is reproducible (22). Published DNA sequences where these criteria were not fulfilled were not analyzed.
Because it has recently been claimed that an uncharacterized mutagenic factor exists in ancient DNA extracts that may influence ancient DNA sequences (ref. 23, but see also ref. 24), we checked whether such a mutagenic factor exists in five Neandertal extracts (18, 25). In brief, following the procedure described in ref. 23, we amplified 274 bp of the mtDNA control region from 50 ng of chimpanzee DNA in the presence of either 1 μl of Neandertal extract, 1 μl of extraction blank, or 1 μl of water. After 25 cycles, PCR products were cloned and several clones were sequenced. The frequencies of nucleotide misincorporation did not differ between the amplifications done in the presence of Neandertal extracts and in the presence of water or extraction blanks (data not shown). Thus, no mutagenic factor of the described type (23) exists in these extracts.
Definition of Clades. Neighbor-joining trees were reconstructed by using the program mega2 (26). Phylogeography relies on the definition of the subunits or “clades” among the DNA sequences investigated. However, no generally applied definition of such clades exists (2). To avoid possible biases due to knowledge of the geographical origin of the sequences, we defined clades as monophyletic groups separated by the longest internal branch and required that such a branch should be supported by bootstrap values >90%. The most recent common ancestors of DNA sequences within caves were dated as in ref. 16.
Results
Cave Bears. The European cave bear (Ursus spelaeus) ranged from Spain to the Ural and the Caucasus during the Pleistocene (27). It was the sister species to the brown bear (Ursus arctos), with which it shared a common ancestor ≈1.5 million years ago (28, 29). In contrast to the omnivorous brown bear, cave bears were strict herbivores at least during the late Pleistocene (28, 30). They became extinct between 20,000 and 10,000 years B.P (31). We extracted DNA from 53 cave bears that range in age from 22,000 years B.P. to at least 72,000 years B.P. and stem from 15 different localities in and around the Alps. Of these, 40 yielded amplifiable DNA. In addition, published sequences from 43 cave bears (15, 16, 29) were included in the analyses. The 83 cave bear DNA sequences represent 26 different haplotypes (i.e., unique DNA sequences) from 27 localities. The longest internal branch in an unrooted neighbor-joining tree is supported by a bootstrap value of 94% and divides the tree into two clades of 5 and 21 haplotypes, respectively (Fig. 1A). When the geographical origin of the bears is taken into account, the two clades show extensive geographical overlap (Fig. 1B).
Fig. 1.
Phylogeography of Pleistocene cave bears. (A) Unrooted neighbor-joining tree for 26 cave bear mtDNA haplotypes. The longest branch divides the sequences into two clades present in 94% of bootstrap replicates. (B) Map of Europe showing the geographical distribution of the two cave bear mtDNA clades (in blue and red, respectively) before the last glacial maximum. Some dots represent more than one cave.
Brown Bears. Brown bears (U. arctos) exist today in Europe, Asia, and North America. We determined DNA sequences from two brown bears found in different caves in Austria, Winden and Ramesch. The bones were dated to 47,420 (Ramesch) and 39,940 years B.P. (Winden) (Beta-171310 and Beta-171311). From each brown bear we amplified ≈270 bp of the mitochondrial control region in three overlapping fragments. A neighbor-joining tree was estimated by using these two sequences and 18 DNA haplotypes (16) representing 117 contemporary European brown bears. The bootstrap support for the longest internal branch is 99% (Fig. 2A). These two clades have been described earlier for contemporary brown bears (32, 33). Today, they show a phylogeographic pattern where one of them occurs in Western Europe (“western” clade) and the other one occurs in Eastern Europe, Asia, and North America (“eastern” clade). They meet in Europe in Sweden and Romania (Fig. 2B) with a small zone of geographical overlap (12, 33). The Pleistocene bear that belongs to the “western” clade comes from Winden, a location 200 km east of Ramesch, where the bear that belongs to the “eastern” clade was found (Fig. 2). Moreover, Ramesch is located ≈500 km west of the area in Romania where the “eastern” and “western” clades meet today (33). Thus, although we cannot currently draw conclusions about the exact phylogeography of Pleistocene brown bears in Europe, it is clear that if any phylogeographic pattern existed, it must have been different from what is found today and must have involved considerably more geographic overlap between the two clades than today, because the alternative explanation, a complete inversion of the geographical distribution of the two clades, seems highly unlikely.
Fig. 2.
Phylogeography of Pleistocene brown bears. (A) Unrooted neighbor-joining tree for 18 modern and 2 ancient brown bear mtDNA haplotypes from Eurasia. The longest branch divides the sequences into two clades supported in 99% of bootstrap replicates. The “western” clade (blue) occurs today in western Europe and the “eastern” clade (red) in eastern Europe, Asia, and North America. The two Pleistocene mtDNA sequences are black. (B) Map of Europe showing the geographical location of the Pleistocene brown bears. R, Ramesch; W; Winden. The current areas of overlap between the two clades in Romania and Sweden are shown in black.
Cave Hyenas. During most of the Pleistocene cave hyenas lived in Eurasia, ranging from Spain to northeastern China (31). Most authors treat them as a subspecies (Crocuta crocuta spelaea) of the extant spotted hyena (Crocuta crocuta), although some authors give them species status (Crocuta spelaea). Their extinction is likely to have occurred between 20,000 and 10,000 years B.P (31). We determined 366 bp of the mitochondrial cytochrome b gene from 18 individuals originating from 15 different caves from western France to the Altai (Siberia). Eight samples were carbon-dated and their ages vary from 37,000 to >50,000 years. The 18 animals carry four mtDNA haplotypes. In an unrooted neighbor-joining tree, the longest internal branch separates two clades with 100% bootstrap value (Fig. 3A). One clade occurs across the whole range investigated, whereas the other clade is found geographically intermingled with the first one in the central part of the range (Fig. 3B). Thus, the easternmost sequence of the clade with the larger geographical distribution occurs in the Altai Mountains, ≈5,000 km east of the easternmost location of the other clade, whereas the westernmost sequence of the former occurs about 1,500 km west of the westernmost sequence of the latter.
Fig. 3.
Phylogeography of Pleistocene cave hyenas. (A) Unrooted neighbor-joining tree for four cave hyena mtDNA haplotypes. The longest branch divides the sequences into two clades supported in 100% of bootstrap replicates. (B) Map of Europe showing the distribution of cave hyena mtDNA clades (blue and red, respectively) before the last glacial maximum.
Neandertals. Neandertals were archaic humans living in Europe and western Asia during the Pleistocene. They were closely related to modern humans with whom they share a common mtDNA ancestor ≈500,000 years ago (34, 35). They appear in the fossil record ≈350,000 years B.P. and disappear ≈29,000 years B.P (19, 34, 36). Mitochondrial control region sequences of 333–357 bp from four Neandertal individuals ranging in age from 29,000 to >42,000 years B.P. have been published (17–20). The four mtDNA sequences differ at one to seven positions and thus exhibit a genetic diversity among themselves similar to that of current modern humans (18, 25). Because of the few differences between the Neandertal mtDNA sequences, it is not possible to define clades that fulfill our criteria (see Materials and Methods) (Fig. 4). However, it is worth noting that one Neandertal individual from Feldhofen in Germany carried a mtDNA sequence more similar to that of a Neandertal individual in Croatia than to another Neandertal individual from Feldhofen (18). This finding suggests that, at least in central and western Europe, no strong geographical clustering existed among the Neandertals before the last glacial maximum.
Fig. 4.
Neandertal mtDNA variation. (A) Unrooted neighbor-joining tree for four Neandertal mtDNA sequences. (B) Map showing the geographical provenience of the Neandertal individuals.
Discussion
Lack of Pleistocene Phylogeographic Patterns. The species investigated are all the Eurasian species for which enough ancient mtDNA sequences are available to draw conclusions about phylogeographic patterns during the Pleistocene. Remarkably, in none of these four species is any phylogeographic pattern observed. By contrast, many extant European species display western and eastern mtDNA clades that meet in central Europe (e.g., see refs. 4 and 6 for recent reviews). For example, this situation is present in extant European brown bears (32, 33). Thus, whereas no phylogeographic patterns are seen in the four late Pleistocene species studied, such patterns are commonly seen in current species in the same area.
Three of the four investigated species are now extinct in Europe. Thus, the possibility exists that, in the case of these species, different haplotypes never became separated in different refugia. For three reasons, we consider this unlikely. First, the divergence times between the two mtDNA clades in the cave bears and the cave hyenas, respectively, are similar to the divergence of mtDNA clades for the species in which extant phylogeographic patterns exist. If no separation of mtDNA haplotypes in glacial refugia would have existed in these species, we would expect the divergences to be more recent. Second, the brown bear, which is closely related to the cave bear, carries a phylogeographically structured mtDNA pool today. Third, the two mtDNA clades identified for the cave hyenas are present also in extant African hyenas and show strong phylogeographical structure there today (N.R. and M.H., unpublished data). To investigate whether these species carried phylogeographically structured gene pools in the past, it would be necessary to analyze Pleistocene samples from the refugial areas, where the separation of mtDNA haplotypes would be expected to persist over several glacial cycles (see below). Unfortunately, because the refugia are located in southern Europe (6) and DNA survival in palaeontological samples is strongly temperature-dependent (37), this is technically difficult.
It should be noted that all four species studied here are likely to have been highly mobile. This mobility would accelerate the decay of phylogeographic patterns over time, as has been suggested for wolves (10), and make the findings less likely to be of general relevance for other late Pleistocene mammals. However, for both brown bears (11) and spotted hyenas (38) female dispersal seems to be limited and phylogeographic patterns occur in spotted hyenas in Africa (39, N.R. and M.H., unpublished data). Furthermore, in addition to bears (32, 33), phylogeographic patterns have been found for several large mammals in Europe, such as red deer and roe deer (40, 41); and in species where no phylogeographic pattern could be detected, such as for pine martens (42) or otters (43), this lack has been attributed to survival and subsequent recolonization from single refugia rather than rapid dispersal. Therefore, we consider it unlikely that the lack of phylogeographic patterns would be due to high dispersal rates in all four species analyzed.
One possible and trivial explanation for the absence of phylogeographic patterns in the late Pleistocene species is that, at any particular point in time, phylogeographic patterns existed, but that these patterns were different at different times. As a consequence, no phylogeographic patterns would be detected when samples of different age are analyzed. Geographical changes of the boundaries of mtDNA clades could be caused, for example, by climatic changes during the middle and late Pleistocene (44–46), by random fluctuations of population ranges (47), or by movements of so-called “tension zones” (48, 49). For many reasons, we consider this unlikely. First, even if we restrict our analyses only to cave bears that differ by <4,000 years in age, mtDNA clades overlap geographically (Fig. 6, which is published as supporting information on the PNAS web site). Second, at least some of the populations studied have been stable over a long time. For example, similar or identical mtDNA sequences were recovered at Vindija (Croatia) from cave bears that differ in age by ≈30,000 years (15). Third, at two cave sites where many cave bear mtDNA sequences were retrieved (Vindija, Croatia, and the Ach Valley, Germany), each DNA sequence differs from the next closest sequence in the same cave by just one substitution, whereas the sequences differ by 8–11 substitutions between caves. Because all substitutions observed in one cave have likely occurred subsequent to each other, this finding indicates that none of the intermediate DNA sequences has been lost by drift. Thus, neither of the two populations is likely to have undergone severe population size fluctuations since the time of its most recent common mtDNA ancestor, which is estimated at 130,000 years.
The data therefore suggest that phylogeographic patterns in European animals during the late Pleistocene were much less pronounced than they are today.
Decay of Phylogeographic Patterns over Time. If the phylogeographic patterns observed today were caused by a restriction of animal population ranges to refugia during glacial maxima, it is reasonable to assume that the time between the “release” of the species from the refugia and the time point when the mtDNA sequences are sampled is crucial for the extent to which patterns are seen. The last glacial maximum ended ≈10,000 years ago (13, 14). By contrast, the penultimate glacial maximum ended 130,000 years ago (13). Because almost all Pleistocene samples studied are <70,000 years B.P., current populations had an approximately five times shorter period at their disposal for migrations from glacial refugia than the late Pleistocene species studied here. We suggest that this explains the common occurrence of phylogeographic pattern in the current mtDNA gene pool of many mammals in Europe and the absence of such patterns in the late Pleistocene.
This suggestion is supported by observations in current animals. For example, the absence of phylogeographic patterns in wolves has been explained by their high migration rate (10). Similarly, in brown bears, analyses of autosomal microsatellite loci (11, 50) show no phylogeographic pattern, whereas mtDNA sequences do show such a pattern. The fact that the former are transmitted through both males and females but the latter are transmitted by females, which are more sedentary, supports the notion that the existence of phylogeographical patterns is inversely correlated with dispersal rates.
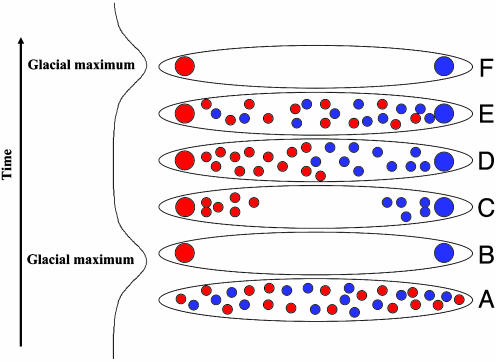
If migration indeed eroded phylogeographic structure between the last two glacial maxima, this raises the question of why the molecular divergences between mtDNA clades in many species are several hundreds of thousands of years old (for a review see refs. 5 and 6), i.e., much older than the last glacial maximum. Two factors probably contribute to this. First, the date of divergence of two genetic lineages found in two distinct populations includes polymorphism that existed in the ancestral population from which the two lineages emerged. It therefore gives only a maximum divergence time for a population separation that is generally much more recent (51). Dates of the divergences within mtDNA clades are therefore useful complementary estimates because they give a minimal age of the divergence between populations. Second, between glacial maxima, mtDNA sequences from one refugium often may not have reached the refugium in which another clade evolved, even if substantial mixing in other areas occurred. The reason for this is that the population densities in the refugia are likely to have been high and these high densities may have slowed down or prevented introgression of mtDNA into these regions (ref. 52 and references therein and ref. 53). As a result, the mtDNA clades might have evolved almost independently in the refugia despite extensive mixing in the rest of the geographic range of the species between glacial maxima (Fig. 5). The mtDNA divergence times may therefore not reflect the time when the current phylogeographic patterns were established but the times of the first population divergences. This hypothesis is compatible with the fact that identical mtDNA sequences in brown bears are found from Estonia to Alaska, yet little overlap between the different mtDNA clades is seen in western Europe (11). Thus, although the brown bears carrying the “eastern” mtDNA clade migrated thousands of kilometers to the east, they did not migrate a few hundred kilometers to the west into the area occupied by the “western” mtDNA clade. The most plausible explanation for this is that the presence of brown bears in the latter area slowed down migration so efficiently that the “eastern” mtDNA clade did not penetrate Western Europe during the past 10,000 years. This scenario is further supported by studies of dispersal of brown bears from growing populations into areas of low population density in Sweden (12).
Fig. 5.
Schematic figure showing the possible effects of glacial cycles on phylogeographic mtDNA patterns. (A) A population that lacks phylogeographic structure is shown shortly before a glacial maximum. (B) During a glacial maximum only individuals in the refugia survive. By drift, different mtDNA types become fixed in the two refugia. (C) After the glaciation, recolonization from the refugia occurs. (D) Individuals from the two clades meet to form a “hybrid zone.” (E) Migration eventually erases the phylogeographic pattern for most of the population's range, but the higher population density slows migration into the refugia. (F) During a subsequent glacial maximum the refugia are likely to remain distinct with respect to mtDNA clades.
Implications for Conservation Genetics. Conservation geneticists (54) are often torn between the wish to prevent inbreeding depression (55) and the desire to preserve populations that have been historically separated as distinct gene pools (often referred to as “evolutionary significant units,” e.g., ref. 56). Our results suggest that, when phylogeographic patterns of mtDNA variants in the absence of obvious physical barriers are seen in a species, this finding may often represent an intermediate state of a spontaneous diffusion process after the removal of a barrier, e.g., past glacial maxima. No inherent reason exists to assume that the mixing of such “populations” defined by mtDNA clades would have detrimental effects. This argument is all the more valid because nuclear DNA haplotypes, which are more likely to confer phenotypic effects that represent adaptations to different environments than mtDNA haplotypes, are less likely to show phylogeographic patterns due to their larger effective population size that makes the fixation of single haplotypes in small populations less likely. Conservation efforts might thus often be better directed toward preserving and restoring connections between suitable habitats to allow gene flow between populations (56).
Conclusions
We suggest that the decay of phylogeographic structure depends on three mutually nonexclusive factors: (i) the time since a geographical barrier disappeared, (ii) the migration rate of the species considered, and (iii) the population densities in the colonized areas. We furthermore suggest that the ancient separation of mtDNA clades in the current gene pool of many animals in Europe is the result of independent evolution of mtDNA sequences in the refugia over several glacial and interglacial periods, whereas extensive mixing occurred between populations in most of their geographical ranges outside the refugia. To test the generality of this scenario, it will be important to determine ancient DNA sequences from several species that currently show phylogeographic patterns to elucidate whether such patterns existed before the last glacial maximum.
Supplementary Material
Acknowledgments
We thank many collaborators from the Max Planck Institute for Evolutionary Anthropology, Sylvio Tüpke for help with figure design, and Joshua Pollack and Molly Przeworski for comments on earlier versions of this manuscript. This work was funded by the Max Planck Society and the Deutsche Forschungsgemeinschaft.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: mt, mitochondrial.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AJ809162–AJ809334).
References
- 1.Avise, J. C., Giblin-Davidson, C., Laerm, J., Patton, J. C. & Lansman, R. A. (1979) Proc. Natl. Acad. Sci. USA 76, 6694–6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avise, J. C. (2000) Phylogeography (Harvard Univ. Press, Cambridge, MA).
- 3.Remington, C. (1968) Evol. Biol., 321–428.
- 4.Taberlet, P., Fumagalli, L., Wust-Saucy, A. G. & Cosson, J. F. (1998) Mol. Ecol. 7, 453–464. [DOI] [PubMed] [Google Scholar]
- 5.Hewitt, G. (1996) Biol. J. Linn. Soc., 247–276.
- 6.Hewitt, G. (2000) Nature 405, 907–913. [DOI] [PubMed] [Google Scholar]
- 7.Leonard, J. A., Wayne, R. K. & Cooper, A. (2000) Proc. Natl. Acad. Sci. USA 97, 1651–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barnes, I., Matheus, P., Shapiro, B., Jensen, D. & Cooper, A. (2002) Science 295, 2267–2270. [DOI] [PubMed] [Google Scholar]
- 9.Kurten, B. & Anderson, E. (1980) Pleistocene Mammals of North America (Columbia Univ. Press, New York).
- 10.Vila, C., Amorim, I. R., Leonard, J. A., Posada, D., Castroviejo, J., Petrucci-Fonseca, F., Crandall, K. A., Ellegren, H. & Wayne, R. K. (1999) Mol. Ecol. 8, 2089–2103. [DOI] [PubMed] [Google Scholar]
- 11.Waits, L., Taberlet, P., Swenson, J. E., Sandegren, F. & Franzen, R. (2000) Mol. Ecol. 9, 421–431. [DOI] [PubMed] [Google Scholar]
- 12.Swenson, J., Sandegren, F. & Soderberg, A. (1998) J. Anim. Ecol., 819–826.
- 13.Petit, J. R., Jouzel, J., Raynaud, D., Barkov, N. I., Barnola, J.-M., Basile, I., Bender, M., Chapellaz, J., Davis, M., Delaygue, G., et al. (1999) Nature 399, 429–436. [Google Scholar]
- 14.Waelbroeck, C., Duplessy, J. C., Michel, E., Labeyrie, L., Paillard, D. & Duprat, J. (2001) Nature 412, 724–727. [DOI] [PubMed] [Google Scholar]
- 15.Hofreiter, M., Rabeder, G., Jaenicke-Despres, V., Withalm, G., Nagel, D., Paunovic, M., Jambresic, G. & Pääbo, S. (2004) Curr. Biol. 14, 40–43. [DOI] [PubMed] [Google Scholar]
- 16.Hofreiter, M., Capelli, C., Krings, M., Waits, L., Conard, N., Munzel, S., Rabeder, G., Nagel, D., Paunovic, M., Jambresic, G., et al. (2002) Mol. Biol. Evol. 19, 1244–1250. [DOI] [PubMed] [Google Scholar]
- 17.Krings, M., Stone, A., Schmitz, R. W., Krainitzki, H., Stoneking, M. & Pääbo, S. (1997) Cell 90, 19–30. [DOI] [PubMed] [Google Scholar]
- 18.Schmitz, R. W., Serre, D., Bonani, G., Feine, S., Hillgruber, F., Krainitzki, H., Pääbo, S. & Smith, F. H. (2002) Proc. Natl. Acad. Sci. USA 99, 13342–13347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ovchinnikov, I. V., Gotherstrom, A., Romanova, G. P., Kharitonov, V. M., Liden, K. & Goodwin, W. (2000) Nature 404, 490–493. [DOI] [PubMed] [Google Scholar]
- 20.Krings, M., Capelli, C., Tschentscher, F., Geisert, H., Meyer, S., von Haeseler, A., Grossschmidt, K., Possnert, G., Paunovic, M. & Pääbo, S. (2000) Nat. Genet. 26, 144–146. [DOI] [PubMed] [Google Scholar]
- 21.Hofreiter, M., Serre, D., Poinar, H. N., Kuch, M. & Pääbo, S. (2001) Nat. Rev. Genet. 2, 353–359. [DOI] [PubMed] [Google Scholar]
- 22.Hofreiter, M., Jaenicke, V., Serre, D., von Haeseler, A. & Pääbo, S. (2001) Nucleic Acids Res. 29, 4793–4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pusch, C. M. & Bachmann, L. (2004) Mol. Biol. Evol. 21, 957–964. [DOI] [PubMed] [Google Scholar]
- 24.Serre, D., Hofreiter, M. & Pääbo, S. (2004) Mol. Biol. Evol. 21, 1463–1467. [DOI] [PubMed] [Google Scholar]
- 25.Serre, D., Langaney, A., Chech, M., Teschler-Nicola, M., Paunovic, M., Mennecier, P., Hofreiter, M., Possnert, G. G. & Pääbo, S. (2004) PLoS Biol. 2, 0313–0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar, S., Tamura, K. & Nei, M. (1993) MEGA: Molecular Evolutionary Genetics Analysis (Pennsylvania State Univ., University Park, PA).
- 27.Musil, R. (1980) Ursus Speleus-Der Hoehlenbaer (Museum fuer Urund Fruehgeschichte, Thuerigens, Weimar, Germany).
- 28.Kurten, B. (1976) The Cave Bear Story (Columbia Univ. Press, New York).
- 29.Loreille, O., Orlando, L., Patou-Mathis, M., Philippe, M., Taberlet, P. & Hänni, C. (2001) Curr. Biol. 11, 200–203. [DOI] [PubMed] [Google Scholar]
- 30.Bocherens, H., Fizet, M. & Mariotti, A. (1994) Paleogeogr. Paleoclimatol. Paleoecol., 213–225.
- 31.Kurten, B. (1968) Pleistocene Mammals of Europe (Weidenfeld and Nicolson, London).
- 32.Taberlet, P. & Bouvet, J. (1994) Proc. R. Soc. London Ser. B. 255, 195–200. [DOI] [PubMed] [Google Scholar]
- 33.Kohn, M., Knauer, F., Stoffella, A., Schröder, W. & Pääbo, S. (1995) Mol. Ecol. 4, 95–103. [DOI] [PubMed] [Google Scholar]
- 34.Klein, R. G. (2003) Science 299, 1525–1527. [DOI] [PubMed] [Google Scholar]
- 35.Krings, M., Geisert, H., Schmitz, R. W., Krainitzki, H. & Pääbo, S. (1999) Proc. Natl. Acad. Sci. USA 96, 5581–5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bischoff, J., Shamp, D., Arambu, A., Arsuaga, J., Carbonell, E. & Bermudez de Castro, J. (2003) J. Archaeol. Sci. 30, 275–280. [Google Scholar]
- 37.Smith, C. I., Chamberlain, A. T., Riley, M. S., Cooper, A., Stringer, C. B. & Collins, M. J. (2001) Nature 410, 771–772. [DOI] [PubMed] [Google Scholar]
- 38.Holekamp, K., Ogutu, J., Frank, H., Dublin, H. & Smale, L. (1993) Ethology 93, 285–299. [Google Scholar]
- 39.Albert, R., Hofer, H., East, M. & Pitra, C. (2000) Zool. Garten 70, 1–10. [Google Scholar]
- 40.Ludt, C. J., Schroeder, W., Rottmann, O. & Kuehn, R. (2004) Mol. Phylogenet. Evol. 31, 1064–1083. [DOI] [PubMed] [Google Scholar]
- 41.Vernesi, C., Pecchioli, E., Caramelli, D., Tiedemann, R., Randi, E. & Bertorelle, G. (2002) Mol. Ecol. 11, 1285–1297. [DOI] [PubMed] [Google Scholar]
- 42.Davison, A., Birks, J. D., Brookes, R. C., Messenger, J. E. & Griffiths, H. I. (2001) Mol. Ecol. 10, 2479–2488. [DOI] [PubMed] [Google Scholar]
- 43.Cassens, I., Tiedemann, R., Suchentrunk, F. & Hartl, G. B. (2000) J. Hered. 91, 31–35. [DOI] [PubMed] [Google Scholar]
- 44.Alley, R. B., Marotzke, J., Nordhaus, W. D., Overpeck, J. T., Peteet, D. M., Pielke, R. A., Jr., Pierrehumbert, R. T., Rhines, P. B., Stocker, T. F., Talley, L. D. & Wallace, J. M. (2003) Science 299, 2005–2010. [DOI] [PubMed] [Google Scholar]
- 45.Prokopenko, A. A., Williams, D. F., Kuzmin, M. I., Karabanov, E. B., Khursevich, G. K. & Peck, J. A. (2002) Nature 418, 65–68. [DOI] [PubMed] [Google Scholar]
- 46.Genty, D., Blamart, D., Ouahdi, R., Gilmour, M., Baker, A., Jouzel, J. & Van-Exter, S. (2003) Nature 421, 833–837. [DOI] [PubMed] [Google Scholar]
- 47.Irwin, D. E. (2002) Evolution 56, 2383–2394. [DOI] [PubMed] [Google Scholar]
- 48.Key, K. H. L. (1968) Syst. Zool. 17, 14–22. [Google Scholar]
- 49.Barton, N. H. & Hewitt, G. (1985) Annu. Rev. Ecol. Syst. 16, 113–148. [Google Scholar]
- 50.Paetkau, D., Amstrup, S. C., Born, E. W., Calvert, W., Derocher, A. E., Garner, G. W., Messier, F., Stirling, I., Taylor, M. K., Wiig, O. & Strobeck, C. (1999) Mol. Ecol. 8, 1571–1584. [DOI] [PubMed] [Google Scholar]
- 51.Nei, M. (1987) Molecular Evolutionary Genetics (Columbia Univ. Press, New York).
- 52.Flagstad, O., Walker, C. W., Vila, C., Sundqvist, A. K., Fernholm, B., Hufthammer, A. K., Wiig, O., Koyola, I. & Ellegren, H. (2003) Mol. Ecol. 12, 869–880. [DOI] [PubMed] [Google Scholar]
- 53.Hutchinson, W. F., van Oosterhout, C., Rogers, S. I. & Carvalho, G. R. (2003) Proc. R. Soc. London Ser. B. 270, 2125–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Frankham, R. (2003) C. R. Biologies 326, Suppl. 1, S22–S29. [DOI] [PubMed] [Google Scholar]
- 55.Crnokrak, P. & Roff, D. A. (1999) Heredity 83, 260–270. [DOI] [PubMed] [Google Scholar]
- 56.Moritz, C. (2002) Syst. Biol. 51, 238–254. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.