The capacity to mount an immune response that eliminates infection of a host by a microbial pathogen is critical for species survival and propagation. Mammals have developed complex immune systems that integrate innate responses, such as phagocytosis and the production of antimicrobial peptides after Toll-like receptor activation, along with adaptive responses, such as antibody production and activation of helper and cytotoxic T cell populations. By contrast, simpler organisms have much less complex immune systems that are focused exclusively on generating innate immune defenses. Over the last few years, it has become apparent that mitogen-activated protein (MAP) kinase (MAPK)-signaling pathways play an essential and central role in the immune responses within each of these species.
Invertebrate Innate Immune Responses
In the absence of a functional immune system, it was long thought that invertebrate animals could not mount a meaningful defense against pathogens. However, this view changed with the realization that the Toll signaling pathway in flies is used to generate antimicrobial peptides that thwart bacterial invaders (1). Although Caenorhabditis elegans appear to lack a Toll receptor signaling-based defense mechanism, recent work has identified a role for a p38 MAPK-signaling pathway in defense toward infection and death caused by bacterial pathogens. Armed with the initial discovery that wild-type C. elegans is susceptible to infection and eventual death by growth on Psuedomonas aeruginosa, Ausubel and colleagues (2) asked which genes were required for resistance to this pathogen. Using genetics, they discovered several mutations that made otherwise wild-type animals succumb to the bacterium. Careful mapping and cloning studies revealed that two of the genes required for resistance were nsy-1 and sek-1, the C. elegans MAPK kinase kinase and MAPK kinase homologs (2). RNA interference studies demonstrated that the p38 MAPK encoded by the pmk-1 gene was the downstream target of these two enzymes (2). This pathway can also protect C. elegans from infection by Salmonella enterica by a mechanism involving the activation of a programmed cell death pathway (3).
In a recent issue of PNAS, two key articles (4, 5) provided compelling evidence that integrated signaling mechanisms involving components of both the PMK-1 and KGB-1 pathways are critical for the innate response of C. elegans to challenge either with whole bacteria or bacterial toxins. It appears that these responses are achieved by transcriptional and posttranslational regulation of MAPK pathway members. Interestingly, two new downstream genes that are regulated by MAPK have been identified by microarray analysis and were found to play an essential role in the innate immune response within this organism.
Genetics of Innate Immunity in C. elegans
Kim et al. (5) demonstrate that the function of PMK-1 in innate defense mechanisms is under the regulation of two components of c-Jun-NH2-terminal kinase (JNK)-signaling pathways (Fig. 1). Specifically, they show that the C. elegans MEK-1 MAPK kinase protein, which was known to be involved in activating the JNK MAPKs, JNK-1, is also required for the activation of PMK-1. This finding is surprising because the mammalian MEK-7 has not been found to interact with p38 kinase family members. Consistent with a role of MEK-1 in innate immunity, this group showed that mek-1 mutant animals are hypersensitive to pathogenic bacterial challenge. However, unlike pmk-1, neither jnk-1, kgb-1, nor kgb-2 mutant animals were found to be hypersensitive to bacterial pathogens. In contrast, kgb-1 mutant animals, but not pmk-1 mutant animals, were hypersensitive to heavy metal stress. Taken together, these data demonstrate that MEK-1 can regulate innate immunity by activating PMK-1 or, instead, can govern stress responses by activating KGB-1, or both. In addition to MEK-1 involvement, this group also showed that the function of PMK-1 in innate immune defenses was also subject to negative regulation imposed by the dual-specificity VHP-1 MAPK phosphatase. Thus, it would appear that crosstalk between MAPK signaling pathways can occur at several levels to regulate innate defense mechanisms in C. elegans, an observation that may have important implications for our understanding the roles played by these pathways in higher organisms, including mammals.
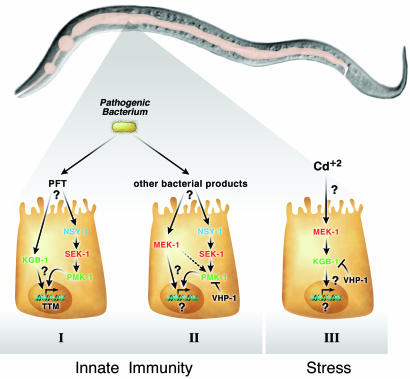
Fig. 1.
MAPK and JNK pathways regulate innate immunity and resistance to heavy metal toxicity. Depicted is the nematode C. elegans, highlighting the digestive tract of this worm. From left to right is the pharynx, intestine that spans almost the entire animal, and the posterior with the anus. (I) Once ingested, bacterium expressing a PFT interacts with the intestinal cells in an unknown manner, causing the up-regulation of the MAPK kinase kinase (MAPKKK) NSY-1, which activates the MAPK kinase SEK-1 to ultimately up-regulate the p38 MAPK PMK-1. Additionally, the PFT increases the activity of the JNK-related kinase KGB-1 by an unknown mechanism. The combined action of PMK-1 and KGB-1 results in the increased expression of ttm-1 and ttm-2, which function to increase resistance to PFT. (II) The ingestion of pathogenic bacterium, such as Pseudomonas aerugonisa, results in the interaction of bacterial products with the intestinal cells. This interaction, although unknown, results in the up-regulation of the MAPKKK pathway, although the impact of these events on the transcriptome is still not known. Unlike interaction with PFT (I), this interaction does not require the JNK kinase KGB-1. PMK-1 is negatively regulated by the VHP-1 dual specificity phosphatase (MKP7). (III) In response to heavy metal stress, the JNK-1 kinase KGB-1 becomes activated by MAPKKK pathway to regulate resistance to stress. This interaction does not require the p38 MAPK PMK-1, and again the influence on the transcriptome of activating this pathway is unknown. The activity of KGB-1 is negatively regulated by VHP-1.
The Transcriptome of C. elegans Innate Immunity
These observations raise an important question, namely, how do these signaling cascades elaborate host defense mechanisms? With the idea of identifying transcriptional targets of MAPK signaling pathways that play a role in the primordial immune response in C. elegans, Huffman et al. (4) embarked upon DNA microarray analysis to identify differentially regulated genes after exposure to bacteria expressing the Bacillus thuringensis Cry5B crystal pore-forming toxin (PFT). To distinguish these genes from others that lead to death of the organism, Huffman et al. included a control treatment involving exposure to the heavy metal cadmium. This treatment can also induce death in a manner that resembles that associated with toxin treatment, albeit by a distinct mechanism as described in ref. 4. In the analysis performed by Huffman et al., nearly 1,000 genes were altered in response to either cry5B or the heavy metal, although only a handful of these genes were specific to cry5B treatment. Intriguingly, the sek-1 and pmk-1 genes, as well as the kgb-1 gene, were among those that were upregulated specifically by toxin treatment. Therefore, it appears that, in response to a bacterial toxin, members of the MAPK signaling pathway are up-regulated at the transcriptional, and presumably also the posttranscriptional, level.
The importance of SEK-1 and PMK-1 for resistance to the bacterial toxin was confirmed by demonstrating that animals harboring mutations in these genes were hypersensitive to toxin treatment. Again, crosstalk between this p38 MAPK signaling pathway and a JNK MAPK signaling pathway is necessary for this host defense mechanism because animals that were deficient in kgb-1, but not in other JNK family kinases, were also hypersensitive to the toxin. However, because animals deficient in kgb-1 were still competent for mounting a protective response against challenge with pathogenic bacteria, the precise details of the molecular crosstalk mechanisms must also be different.
In other experiments, Huffman et al. (4) performed microarray studies to identify genes that are activated in response to toxin treatment specifically in animals that have an intact SEK-1 pathway. Presumably, these genes would include those that are candidate effectors of the host defense mechanism, and they could then be subsequently identified by the RNA interference method. Indeed, two such genes were identified when this approach was used, and one of these genes, ttm-1 (toxin-regulated targets of MAPK), appears to encode a cation efflux channel that may protect against cytotoxic cations that arise within the cytoplasm of cells after pore formation.
MAPK Target Genes Are Required for Innate Immunity
C. elegans has a remarkable ability to sense its environment, providing a method to help this soil-dwelling animal avoid potential toxic compounds. This is evident by the fact that this nematode can readily distinguish among different alcohols as either attractants or repellants (6). However, it is not clear how the worm can deal with substances, such as the bacterial PFT, which it presumably cannot readily sense until after it is taken up in the digestive system. From the studies by Huffman et al. (4) and Kim et al. (5), it is clear that one approach is to mount a cellular defense mechanism, i.e., to activate the MAPK pathway, to help deal with the stress of acute exposure and ensure fitness until the animal can move away from the toxic source. Huffman et al. directly tested this hypothesis and discovered that the MAPK pathway is required for survival to acute exposure to toxin.
Besides mounting a cellular defense mechanism, it is logical to think that the worm may have evolved other methods to help it survive and possibly move away from a potentially toxic environment. In this regard, perhaps the worm has linked responses within its intestine, where cry5B functions as a toxin, to the intricate behaviors of movement to help the worm exit a potentially harmful environment. Does the worm change its behavior (i.e., move faster or turn more) when treated with bacteria such as P. aerugonisa or cry5B toxin? Alternatively, the worm may program itself to cease ingestion of food (i.e., stop pharyngeal pumping) or increase its defecation rate in response to P. aerugonisa or to treatment with toxins like cry5B. It is noteworthy that an intact MAPK pathway is required for proper sensory neuron development (7, 8). It is possible that the same system used in the intestine to sense toxin uptake may also be used in the nervous system to coordinate behaviors to exit, or even stop eating, in a potentially harmful environment.
It is interesting to speculate that similar crosstalk mechanisms between p38 and JNK MAPK signaling pathways and related effector mechanisms may also operate to regulate innate immune responses in higher organisms, such as mammals. If so, one might expect bacteria that infect mammals to evolve elaborate measures to counteract these host defenses. Consistent with this idea, Bacillus anthracis, the causative agent of anthrax, produces a secreted exotoxin, lethal toxin, which acts to proteolytically cleave and inactivate the majority of the major MAPK kinases, including those involved in activating p38 MAPKs and the JNK MAPKs (9–11). Indeed, it is already known that anthrax lethal toxin can disable known elements of the innate and adaptive immune responses in mammals (12–14). Because Huffman et al. (4) demonstrated that the role of the p38 MAPK pathway in protecting cells from the action of bacterial PFTs also extends to a mammalian cell type, we consider it likely that critical effector functions that lie downstream of the integrated p38 and JNK MAPK signaling pathways in C. elegans are conserved in mammals. If so, B. anthracis would have evolved to express lethal toxin to inactivate these primordial defense mechanisms.
References
- 1.Anderson, K. V. (2000) Curr. Opin. Immunol. 12, 13–19. [DOI] [PubMed] [Google Scholar]
- 2.Kim, D. H., Feinbaum, R., Alloing, G., Emerson, F. E., Garsin, D. A., Inoue, H., Tanaka-Hino, M., Hisamoto, N., Matsumoto, K., Tan, M. W. & Ausubel, F. M. (2002) Science 297, 623–626. [DOI] [PubMed] [Google Scholar]
- 3.Aballay, A., Drenkard, E., Hilbun, L. R. & Ausubel, F. M. (2003) Curr. Biol. 13, 47–52. [DOI] [PubMed] [Google Scholar]
- 4.Huffman, D. L., Abrami, L., Sasik, R., Corbeil, J., van Der Goot, F. G. & Aroian, R. V. (2004) Proc. Natl. Acad. Sci. USA 101, 10995–11000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim, D. H., Liberati, N. T., Mizuno, T., Inoue, H., Hisamoto, N., Matsumoto, K. & Ausubel, F. M. (2004) Proc. Natl. Acad. Sci. USA 101, 10990–10994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sengupta, P., Colbert, H. A., Kimmel, B. E., Dwyer, N. & Bargmann, C. I. (1993) Ciba Found. Symp. 179, 235–244; discussion, 244–250. [DOI] [PubMed] [Google Scholar]
- 7.Sagasti, A., Hisamoto, N., Hyodo, J., Tanaka-Hino, M., Matsumoto, K. & Bargmann, C. I. (2001) Cell 105, 221–232. [DOI] [PubMed] [Google Scholar]
- 8.Tanaka-Hino, M., Sagasti, A., Hisamoto, N., Kawasaki, M., Nakano, S., Ninomiya-Tsuji, J., Bargmann, C. I. & Matsumoto, K. (2002) EMBO Rep. 3, 56–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vitale, G., Bernardi, L., Napolitani, G., Mock, M. & Montecucco, C. (2000) Biochem. J. 352, 739–745. [PMC free article] [PubMed] [Google Scholar]
- 10.Duesbery, N. S., Webb, C. P., Leppla, S. H., Gordon, V. M., Klimpel, K. R., Copeland, T. D., Ahn, N. G., Oskarsson, M. K., Fukasawa, K., Paull, K. D. & Vande Woude, G. F. (1998) Science 280, 734–737. [DOI] [PubMed] [Google Scholar]
- 11.Chopra, A. P., Boone, S. A., Liang, X. & Duesbery, N. S. (2003) J. Biol. Chem. 278, 9402–9406. [DOI] [PubMed] [Google Scholar]
- 12.Dang, O., Navarro, L., Anderson, K. & David, M. (2004) J. Immunol. 172, 747–751. [DOI] [PubMed] [Google Scholar]
- 13.Agrawal, A., Lingappa, J., Leppla, S. H., Agrawal, S., Jabbar, A., Quinn, C. & Pulendran, B. (2003) Nature 424, 329–334. [DOI] [PubMed] [Google Scholar]
- 14.Webster, J. I., Tonelli, L. H., Moayeri, M., Simons, S. S., Jr., Leppla, S. H. & Sternberg, E. M. (2003) Proc. Natl. Acad. Sci. USA 100, 5706–5711. [DOI] [PMC free article] [PubMed] [Google Scholar]