Abstract
We completed a large insertional mutagenesis screen in zebrafish to identify genes essential for embryonic and early larval development. We isolated 525 mutants, representing lesions in ≈390 different genes, and we cloned the majority of these. Here we describe 315 mutants and the corresponding genes. Our data suggest that there are roughly 1,400 embryonic-essential genes in the fish. Thus, we have mutations in ≈25% of these genes and have cloned ≈22% of them. Re-screens of our collection to identify mutants with specific developmental defects suggest that ≈50 genes are essential for the development of some individual organs or cell types. Seventy-two percent of the embryonic-essential fish genes have homologues in yeast, 93% have homologues in invertebrates (fly or worm), and 99% have homologues in human. Yeast and worm orthologues of genes that are essential for early zebrafish development have a strong tendency to be essential for viability in yeast and for embryonic development in the worm. Thus, the trait of being a genetically essential gene is conserved in evolution. This mutant collection should be a valuable resource for diverse studies of cell and developmental biology.
To identify a significant fraction of the genes essential for early vertebrate development, we developed a method of insertional mutagenesis for the zebrafish using mouse retroviral vectors (1, 2) and applied the method in a large-scale screen. We identified mutants by visual inspection of embryos at 1, 2, and 5 days postfertilization (dpf), by which time they have developed into free-swimming and feeding larvae. Mutants that result in a visible defect by 5 dpf are almost invariably lethal. Here, we describe 315 mutants and their genes and present an analysis of the evolutionary conservation of these genes. The results argue that the mutant collection contains mutations in at least 25% of the genes essential for the development of many different embryonic organs and structures. Re-screens of the collection in our lab to identify mutations in specific developmental processes support this conclusion. This collection should be a valuable resource for diverse studies of cell and developmental biology in this vertebrate.
Materials and Methods
Mutagenesis and Gene Cloning. Retroviral-mediated insertional mutagenesis and the cloning of the mutated genes were carried out as described (1–3).
Comparative Genomic Analysis. The amino acid sequence of each fish gene was compared by blastp (4) to the reference genomes of Saccharomyces cerevisiae, Schizosaccharomyces pombe, Caenorhabditis elegans, Drosophila melanogaster, and Homo sapiens, as well as the nonredundant database of all organisms to find genes absent in the two yeast species but present in other unicellular eukaryotes. Comparative orthologous group (COG) analysis (5) to determine whether homologues were 1:1 orthologues or had other orthology relationships was done by iteratively blasting the top hits from each organism against the genomes of the others. We consider two genes from different species to be 1:1 orthologues if they are not only each other's reciprocal best blastp hits, but also if no other gene in either species' genome finds the gene as its top hit. A worm or yeast gene was defined as an “ancestor” of a human/fish gene if several human genes found the worm or yeast gene as their top hit, but no other worm or yeast gene found any of those human genes as their top hit.
Results
Identification of 25% of the Genes Essential for Early Zebrafish Development. We isolated 525 insertional mutants that have visible phenotypes by 5 dpf, a time when the embryo has developed from a fertilized egg to a free swimming larva that has begun to feed. Most of these mutations result in lethality, and many mutants die by 5 dpf. Almost all mutants that have not died by 5 days fail to inflate their swim bladder, a phenotype associated with certain death by 2 weeks of age. Thus, we refer to the mutants and mutations as “embryonic lethal(s)” and the mutated genes as “embryonic-essential” genes.
The insertional mutagenesis procedure and the methodology for identifying mutagenic inserts and cloning their flanking DNA have been described (1–3). A summary of the numbers of mutants isolated and flanking sequences and genes cloned is shown in Table 1. The 486 mutants for which we have obtained DNA sequence at the site of the mutagenic insertion probably represent 362 different loci; we have identified the mutated gene for 315 of these, 86 of which we have reported (1, 2). As discussed next, our findings suggest that these 362 loci represent ≈25% of the genes whose mutation leads to an embryonic lethal phenotype. Thus, there are only ≈1,400 such genes. The following evidence supports this conclusion.
Table 1. Insertional mutants recovered.
| Mutants | Loci | |
|---|---|---|
| Insertional mutants | 525 | ≈390 |
| Mutants with cloned junction DNA | 486 | 362 |
| Mutants with mutated gene identified | 438 | 315 |
Our collection includes mutations in 5 of 20 (25%) tRNA synthase genes and 26 of the 79 (33%) ribosomal protein genes in the zebrafish genome. We also have mutations in 23 of 97 genes (24%) for which a chemically induced mutant has also been identified and the mutated gene cloned by positional or candidate gene cloning (as of February 2004). Most of the latter genes encode transcription factors, receptors, and ligands. These data argue that we have screened ≈25% of the genes in the fish genome, whether extrapolated from housekeeping-type genes or genes with more specific developmental functions. The data in Table 2 also show that viral integrations do not occur preferentially into housekeeping-type genes because the allele frequency for housekeeping genes is no greater than that for other types of genes.
Table 2. Allele frequencies.
| Loci with one allele | 275 |
| Loci with two alleles | 62 |
| Loci with three allele | 17 |
| Loci with four alleles | 6 |
| Loci with five allele | 1 |
| Loci with seven alleles | 1 |
| Average allele frequency | 1.34 |
| Average allele freq, RP and tRS | 1.10 |
| Average allele freq, ENU cloned | 1.95 |
RP, ribosomal protein genes; tRS, tRNA synthase genes; ENU cloned, genes identified as the cause of ENU-induced mutants.
In theory, a second way to estimate the number of embryonicessential genes from these data is to apply the Poisson distribution to the numbers of single and multiple hits in Table 2 to calculate the number of loci that have not yet been mutated and hence the total number of loci that can give rise to an embryonic-visible phenotype. However, as is almost always seen in mutagenesis screens, the data do not fit the Poisson distribution precisely, making this approach unreliable. Attempting to apply a Poisson distribution under these circumstances usually results in an underestimate of the number of mutable loci (6, 7). For example, by using the number of loci hit twice in our study (62) and keeping either the number of loci hit (362) or the number of mutants obtained (486) fixed, solving the Poisson equation predicts that the total number of mutable loci is either 1,035 or 1,310 mutable loci. This finding is somewhat lower than (but not inconsistent with) the estimate of 1,400 genes obtained above.
The identities of the 315 mutated genes we have identified to date are provided in Fig. 1 (additional details about these genes are described in Table 3). A preliminary phenotypic description of the corresponding 315 mutants is provided in Table 3 and images are available at http://web.mit.edu/ccr/pnas_zebrafish_mutant_images/index.html. More detailed phenotypic descriptions of the defects in particular organs and structures will come from careful re-screens of the collection (see below).
Fig. 1.
Genes essential for zebrafish embryonic development identified by insertional mutagenesis. Genes are listed by mutant number and sorted by evolutionary conservation and gene function. Phenotypic descriptions are available in Table 3, and images are available at http://web.mit.edu/ccr/pnas_zebrafish_mutant_images/index.html.
If we take 1,400 as the number of zygotic genes whose mutation can lead to an embryonic visible/lethal phenotype in the zebrafish, the 315 genes listed here represent 22% of the total. Because mutated genes were cloned without regard to phenotype, these genes should be representative of the entire set of protein-coding genes that are genetically essential for early zebrafish development.
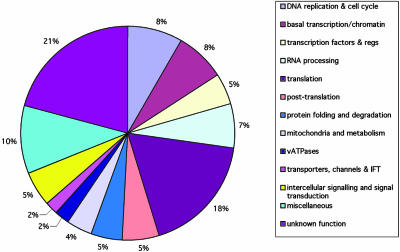
Fig. 2 provides a summary of the types of proteins encoded by the 315 embryonic essential genes based on their biochemical function. Many of the genes are probably essential for cell viability whereas others are likely to be required for more specific developmental processes. Mutations in cell-essential genes can survive for 1 to at least several days due to maternal supplies in the egg (8). Mutation of some cell-essential genes may not have been detected in our screen because maternal supplies of some genes are sufficient to sustain the embryo beyond 5 dpf, e.g., Dicer1 (9). About 20% of all of the genes we identified encode proteins of unknown biochemical function.
Fig. 2.
Types of genes whose mutation in zebrafish leads to an embryonic visible phenotype. The genes are assigned to the same categories as in Fig. 1 although some categories have been combined.
Evolutionary Conservation of Embryonic-Essential Zebrafish Genes. To better understand the genetic basis of animal development and its evolution, we asked whether the embryonic-essential fish genes have homologues in yeast, other single-celled eukaryotes (SCEs) (e.g., Giardia, Plasmodium, Trypanosoma, Chlamydomonas), worm, fly, and human genomes. Analysis of the completed human and mouse genomes has already indicated that 50% of their genes are present in SCEs and ≈80% in flies and/or worms (10, 11), (with homology defined as a blastp E value of <10–5), and our analysis of 50 fish genes selected at random from the July 2003 annotation of the zebrafish genome found a similar proportion of conserved genes (data not shown). Applying the same homology definition to our list of 315 embryonic-essential zebrafish genes, however, we found that a higher fraction, 74% of them, have homologues in yeast or other SCEs, and 93% are present in fly and/or worm. By using a more conservative cutoff requiring homology over 80% of the length of the protein and E < 10–20, 64% of the fish genes have homologues in yeast or some other SCE whereas 91% are present in flies or worms. Ninety-nine percent of the genes (all but 2 of the 315) have homologues in mammals. Thus, as found in other species (12, 13), the fish genes with essential functions are more likely than randomly selected genes to be conserved across species. The conservation of the 315 embryonic-essential fish genes in yeast and other SCEs, in worm, in fly, and in human is represented in Fig. 3 by using the more liberal definition of homology. The identity of the human, fly, worm, and yeast homologues of each essential fish gene can be found in Table 3.
Fig. 3.
Evolutionary conservation of essential zebrafish genes. Horizonal lines on the Left represent 315 different genes. The genes are listed in the same order as in Fig. 1. In the first four colored columns, the presence of a colored box indicates the presence of one or more homologous genes (blastp E value of <10–5) in yeast (either S. cerevisiae or S. pombe), C. elegans, D. melanogaster, or H. sapiens. Green boxes indicate genes without homologues in yeast but with homologues in other SCEs such as Giardia, Plasmodium, Trypanosoma, and/or Chlamydomonas. Thus, yellow boxes represent genes conserved through yeast, green are those found in SCEs other than yeast, red are those found in invertebrates but not SCEs, and blue are vertebrate-specific. The last two genes (with no colored boxes) seem to be fish-specific. The black boxes in the last three columns indicate genes whose mutation leads to one of three phenotypes: cystic kidney, chondrogenesis defects, or reduction or lack of melanocyte pigmentation.
Mutant Phenotypes and the Evolutionary Conservation of Genes Required for Specific Developmental Processes. About 30% of embryonic zebrafish mutants identified in gross morphological screens have developmentally “specific” and unique phenotypes whereas 70% display relatively “nonspecific” or common syndromes (7, 14). The latter more frequently result from mutations in cell-essential genes. Many mutants can be placed into either of the two broad phenotypic categories by superficial visual inspection. However, to identify mutants with phenotypic defects in specific organs or processes with certainty requires considerable effort. To accomplish this goal, we are re-screening the insertional mutant collection, a process called “shelf-screening.” Summaries of three such screens from our lab are shown in Fig. 3 Right.
Black boxes in the three columns in Fig. 3 Right identify genes specifically required (i) to prevent cystic kidney, probably because these genes are required for the normal development of kidney epithelial tubes (15), (ii) to form cartilage that appears normal after staining with Alcian blue (R.M.N., A.A., and N.H., unpublished observations) or (iii) for melanocyte pigmentation (E. Maldonado, A.A., and N.H., unpublished observations). Twelve genes when mutated resulted in kidney cysts, 8 in abnormal cartilage condensation as revealed by appearance after Alcian staining, and 11 affect melanocyte pigmentation. Because we have cloned ≈22% of the genes essential for early fish development, these results predict that mutations in ≈55 genes can give rise to cystic kidney, 36 to abnormal cartilage, and 50 to defects in melanocyte pigmentation in the zebrafish. As discussed later, many genes identified in each screen are components of a common pathway, with different pathways or processes emerging for each screen.
Fig. 3 shows the conservation of the genes identified in each of the three screens in the genomes of yeast, other SCEs, invertebrates (fly or worm), and human. Of the 12 genes that can give rise to cystic kidney when mutated, 6 are shared with SCEs but not with yeast. Three of these are homologues of genes identified in Chlamydomonas that encode intraflagellar transport (IFT) proteins required for flagellum formation (16) (see Discussion).
Formation of cartilaginous structures requires the deposition of proteoglycans in the extracellular matrix (17). Among the eight cartilage mutants identified, four have lesions in genes required for proteoglycan synthesis, predicting that ≈20 such genes would be found in a saturation screen in the fish. Three of these four genes are animal specific. In C. elegans, mutations in homologues of these four genes led to a “squashed vulva” phenotype (18, 19). Thus, whereas the genes are presumably required for proteoglycan synthesis in both worm and fish, they are involved in the formation and structural integrity of very different body parts in the two organisms.
Extending our earlier observations (2), 9 of 11 genes we identified as required for normal melanocyte pigmentation in fish encode v-ATPase subunits or associated proteins or proteins otherwise involved in intracellular vesicles. Whereas the main v-ATPase subunits are found in yeast, zebrafish required several animal-specific v-ATPase-associated proteins in addition.
The Trait of Being a Genetically Essential Gene Is Conserved in Evolution. As Fig. 3 above reveals, and has been observed in other species (12, 13), genes that are essential in zebrafish are highly conserved in evolution. We next asked whether genes that are genetically essential in zebrafish are also genetically essential in other species. It is convenient to ask this question of genes that have counterparts in yeast and worm because a large fraction of the annotated genes in these species have been deleted [yeast (12, 20)] or knocked down by using RNA interference (RNAi) [worm (13, 21)]. To make functional comparisons of genes across these species, however, it is necessary first to identify the yeast and worm genes that are not merely homologues of the essential fish genes, but that have orthologous relationships to them, because orthologues are more likely to perform the same function in their respective species. We determined which zebrafish genes have clear orthologous relationships with their homologues in S. cerevisiae and C. elegans by comparative orthologous group (COG) analysis (5) (see Materials and Methods). Because the zebrafish genome is not yet fully sequenced and annotated, it cannot be used for this purpose, so we used the human genome as a “surrogate vertebrate” genome. We considered both genes that were 1:1 orthologues and genes for which a single yeast or worm gene is an “ancestor” of two or more human paralogues. The results of the analysis are shown in Fig. 4 for yeast (Left) and for worm (Right).
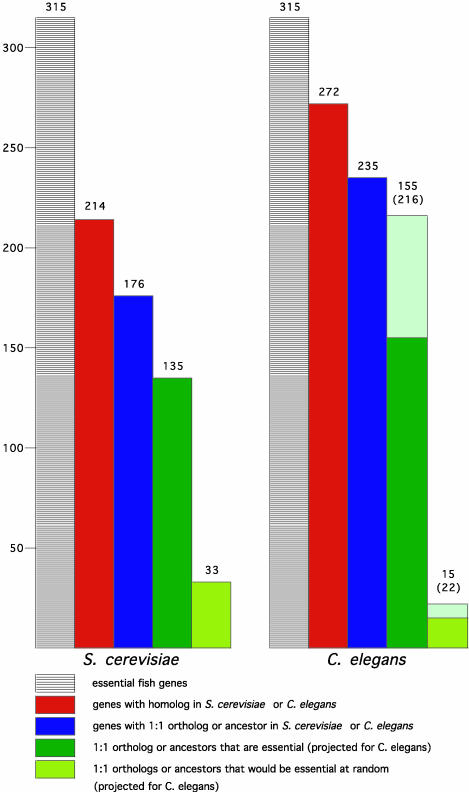
Fig. 4.
Essentialness of genes is evolutionarily conserved. Shown is an analysis of S. cerevisiae (Left) and C. elegans (Right) genes that are homologous to the essential fish genes. In each case, the leftmost columns represent 315 essential fish genes, the red columns show how many of these have homologues in yeast (214) or worm (272), and the blue columns show which have a 1:1 orthologue or “ancestor” gene in yeast (176) or worm (235). The dark green columns represent the number of these yeast or worm orthologues that are essential in their respective species, 135 of the 176 yeast genes, and 155 of the 235 worm genes. The pea green columns show the number that would be predicted to be essential at random, 33 of the 176 yeast genes, and 15 of the 235 worm genes. Thus, the difference between the dark green and pea green columns is the enrichment. In the case of the worm, the pale green extensions to the two green columns represent projections of how many of the orthologues would be found to be essential if 100% of the worm genes had been successfully knocked down by RNAi; this estimate prorates both for the reported failure rate of RNAi and the number of genes for which no RNAi data have been reported. This calculation estimates that 216 of the 235 worm genes are likely to be essential whereas only 22 would be expected to be at random. Note that the percentage of worm orthologues that are essential that is stated in the text does not include the genes for which no RNAi data have been reported.
The left-most bars in Fig. 4 Left and Right represent the list of 315 essential fish genes, the red columns next to them show the number of these genes that have homologues in yeast or worm, and the blue columns show the fraction of homologues that have an orthologous relationship to the vertebrate genes. Having identified the yeast and worm orthologues, we then asked which of these genes are essential in their respective species. The results are shown by the dark green columns in Fig. 4.
In yeast, only 19% of protein-coding genes are essential for cell viability under optimal growth conditions, despite the fact that most “nonessential” yeast genes seem to be single-copy (12). In contrast, among the yeast genes that are 1:1 orthologues or ancestors of the vertebrate genes in our study, 77% are essential in yeast. This is a 4-fold enrichment relative to the yeast genome at large (compare the number observed in dark green with the number predicted if random in pea green).
In C. elegans, RNAi analysis of protein-coding genes reveals that only 7% are required for embryo viability, an additional 1.5% for other developmental and/or physiological processes, and another 1.5% for wild-type growth (21). Yet among the worm genes that are orthologous to the vertebrate genes we identified and for which RNAi data have been reported, 72% are required for embryonic viability and a further 6% are associated with postembryonic developmental phenotypes, a total of 78%. This is an enrichment of nearly 10-fold over the worm genome at large. In the large RNAi screens in C. elegans that have been published to date, it was estimated that only 78% of embryonic-essential genes were detected, largely due to inefficiencies in the RNAi technology (21). Correcting for this failure rate, we conclude that nearly all of the worm orthologues of essential fish genes might in fact be essential for embryonic development (see lighter colored extensions on the green columns in Fig. 4 Right). In summary, this analysis reveals that genetically essential genes have a strong tendency to retain this special status through evolution from yeast to vertebrates.
Discussion
We have described the completion of a large genetic screen in zebrafish, the isolation of insertional mutations in ≈25% of the embryonic-essential genes of the fish, and the molecular cloning of ≈22% of all such genes. This collection of mutants will be a valuable resource for the study of many cellular and developmental processes in a vertebrate. Many of the genes we identified are probably required for cell viability, others for more specific developmental processes including patterning, differentiation or physiology. Twenty percent of the genes encode proteins that have no known or sufficiently clearly identifiable biochemical function. About three quarters of the genes we cloned have homologues in yeast or other single-celled organisms, and ≈25% are animal-specific with 7% overall being vertebrate-specific.
Our results imply that there are only ≈1,400 genes that when mutated result in a visible, usually lethal phenotype in the zebrafish embryo and 5-day-old larva. This number is fewer than the 2,400 such genes proposed by Haffter et al. (7) from data obtained in a large chemical mutagenesis screen by using ENU as the mutagen. We do not think the discrepancy is due to the inability of retroviral vectors to target genes than can be mutated by ENU because genes mutated by ENU and cloned by positional or candidate gene cloning were mutated at the same efficiency as genes encoding ribosomal proteins or tRNA synthetases in our screen. Rather, it seems likely that the discrepancy reflects inaccuracies in both calculations. The failure to achieve saturation in either screen and the fact that the data for single and multiple hits do not fit the Poisson distribution precisely in either screen make highly accurate calculations impossible.
The phenotypic descriptions of most of our mutants remain preliminary, and re-screens of the collection are needed to identify the specific defects in most mutants. The first three such re-screens to be completed, which we summarized here, are revealing in this respect. The screen for cystic kidney in particular supports our conclusion that our mutant collection contains 25% of the genes essential for diverse developmental processes in the embryo. Of 12 genes identified in the kidney cyst screen, 7 seemed to be novel when first cloned, whereas another is PKD2, a gene known to be mutated in human polycystic kidney disease (22). In humans, cystic kidney disease results from a failure of epithelial cells in kidney tubes and ducts to differentiate properly and to cease dividing. This defect can result from defects in primary cilia located on the epithelial cells of kidney tubules and ducts. When we gained access to the mostly unpublished sequences of 13 genes that encode IFT proteins in Chlamydomonas, and which are required for flagellum formation or function in that species, we found that 3 of our 7 “novel” genes were in fact fish homologues of 3 of the 13 IFT genes. It is possible that others of the “novel” genes identified in the fish kidney cyst screen will prove to be yet unidentified IFT genes, or other genes involved in a pathway linking cilia to cell differentiation and cell division in vertebrates. The finding that we have mutations in 3 of 13 IFT genes whose sequences are available is consistent with the notion that our mutant collection includes 25% of the genes essential for processes involved in embryonic development. The results of the kidney cyst screen also suggest that genetic screens in vertebrate animals can reveal many of the genes in a pathway and argue that redundancy may be no more of a problem in genetic screens in vertebrates than it has been in genetic screens in invertebrate animals.
The data from the three shelf screens described here, along with the data showing the conservation of the fish genes in the genomes of single-celled eukaryotes and invertebrate animals, show how many of the same genes used in SCEs and invertebrates are used for different biological tasks in vertebrate animals although their cellular functions are similar in the different species. Thus, IFT genes used by Chlamydomonas to make motile flagella are used to make cilia with different sensory roles in different organisms, for example, on chemosensory neurons sensing salt concentrations in the worm (23) and on kidney epithelial cells in vertebrates (24). Similarly, the same proteoglycan synthesis enzymes that the worm uses to make a properly shaped vulva (18, 19) are used in the fish to make a vertebrate-specific structure, cartilage. And the production of melanin pigment in the fish, which takes place in acidic subcellular compartments (25), requires the same genes to acidify these compartments that yeast cells use to acidify vacuoles (26).
The fact that there is such a small number of embryonic-essential genes and that they include genes that comprise coherent genetic pathways of development suggests that the genetically essential genes have a unique status in biological processes. Consistent with this possibility, we found that the yeast or worm orthologues of genes that are essential in fish have a high probability of also being essential in these species. Thus genes that can be detected in genetic screens, and in particular those that are essential for early viability, seem to have retained this special status through evolution. The implications of this observation are not known, but we suggest that these genes may be all or most of the genes that are absolutely required for many biological processes whereas most other genes may serve to assist these critical genes in making biological processes more robust. Evolution may have required that the number of genetically essential genes be small and that they remain the same genes.
By identifying about a quarter of the embryonic lethal genes in the zebrafish, our studies provide strong evidence that genetic screens in vertebrate animals, as in invertebrates, can reveal molecular pathways of development. Whereas redundancy may hide some genes from this analysis, the fact that the genes that we do find for particular phenotypes can define a biochemical complex or pathway suggests that forward genetic screens can indeed identify many if not most of the genes that are key players in specifying complex developmental processes in vertebrates.
Supplementary Material
Acknowledgments
We thank E. Maldonado and J. Davenport for assistance in screening for mutants, G. Golling and K. Anderson for help with gene identification, Maryann Haldi for phenotypic documentation, M. Cunningham for maintenance of the mutant lines of fish, and T. Angelini, C. Doller, and S. Farrington for maintenance of our zebrafish colony. We thank Bob Bosselman for his interest and support. This work was supported by a National Institutes of Health grant from the National Center for Research Resources (to N.H. and A.A.) and a grant from Amgen (to N.H.). Additional funding came from the Ford Foundation (to N.H.). R.M.N was supported by a postdoctoral fellowship from the National Institutes of Health.
This contribution is part of the special series of Inaugural Articles by members of the National Academy of Sciences elected on April 20, 2004.
Abbreviations: SCE, single-celled eukaryotes; IFT, intraflagellar transport; RNAi, RNA interference; dpf, days postfertilization.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession numbers can be found in Table 3, which is published as supporting information on the PNAS web site).
See accompanying Biography on page 12789.
References
- 1.Amsterdam, A., Burgess, S., Golling, G., Chen, W., Sun, Z., Townsend, K., Farrington, S., Haldi, M. & Hopkins, N. (1999) Genes Dev. 13, 2713–2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Golling, G., Amsterdam, A., Sun, Z., Antonelli, M., Maldonado, E., Chen, W., Burgess, S., Haldi, M., Artzt, K, Farrington, S., et al. (2002) Nat. Genet. 31, 135–140. [DOI] [PubMed] [Google Scholar]
- 3.Amsterdam, A. (2003) Dev. Dyn. 228, 523–534. [DOI] [PubMed] [Google Scholar]
- 4.Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tatusov, R. L., Koonin, E. V. & Lipman D. J. (1997) Science 278, 631–637. [DOI] [PubMed] [Google Scholar]
- 6.Meneely, P. M. & Herman R. K. (1979) Genetics 92, 99–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haffter, P., Granato, M., Brand, M., Mullins, M. C., Hammerschmidt, M., Kane, D. A., Odenthal, J., van Eeden, F. J. M., Jiang, Y.-J., Heisenberg, C.-P., et al. (1996) Development (Cambridge, U.K.) 123, 1–36. [DOI] [PubMed] [Google Scholar]
- 8.Kane, D. A., Hammerschmidt, M., Mullins, M. C., Maischein, H.-M., Brand, M., van Eeden, F. J. M., Furutani-Seiki, M., Granato, M., Haffter, P., Heisenberg, C.-P., et al. (1996) Development (Cambridge, U.K.) 123, 57–66. [DOI] [PubMed] [Google Scholar]
- 9.Wienholds, E., Koudijs, M. J., van Eeden, F. J., Cuppen, E. & Plasterk R. H. (2003) Nat. Genet. 35, 217–218. [DOI] [PubMed] [Google Scholar]
- 10.International Human Genome Sequencing Consortium (2001) Nature 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 11.Mouse Genome Sequencing Consortium (2002) Nature 420, 520–562. [DOI] [PubMed] [Google Scholar]
- 12.Giaever, G., Chu, A. M., Ni, L., Connelly, C., Riles, L., Veronneau, S., Dow, S., Lucau-Danila, A., Anderson, K., Andre, B., et al. (2002) Nature 418, 387–391. [DOI] [PubMed] [Google Scholar]
- 13.Fraser, A. G., Kamath, R. S., Zipperlen, P., Martinez-Campos, M., Sohrmann, M. & Ahringer, J. (2000) Nature 408, 325–330. [DOI] [PubMed] [Google Scholar]
- 14.Driever, W., Solnica-Krezel, L., Schier, A. F., Neuhauss, S. C. F., Malicki, J., Stemple, D. L., Stainier, D. Y. R., Zwartkruis, F., Abdelilah, S., Rangini, Z., et al. (1996) Development (Cambridge, U.K.) 123, 37–46. [DOI] [PubMed] [Google Scholar]
- 15.Sun, Z., Amsterdam, A., Pazour, G. J., Cole, D. G., Miller, M. S. & Hopkins, N. (2004) Development (Cambridge, U.K.), in press. [DOI] [PubMed]
- 16.Rosenbaum, J. L. & Witman, G. B. (2002) Nat. Rev. Mol. Cell Biol. 3, 815–825. [DOI] [PubMed] [Google Scholar]
- 17.Schwartz, N. B. & Domosicz, M. (2002) Glycobiology 12, 57R–68R. [DOI] [PubMed] [Google Scholar]
- 18.Herman, T. & Horvitz, H. R. (1999) Proc. Natl. Acad. Sci. USA 96, 974–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hwang, H.-Y., Olson, S. K., Esko, J. D. & Horvitz H. R. (2003) Nature 423, 439–443. [DOI] [PubMed] [Google Scholar]
- 20.Winzeler, E. A., Shoemaker, D. D., Astromoff, A., Liang, H., Anderson, K., Andre, B., Bangham, R., Benito, R., Boeke, J. D., Bussey, H., et al. (1999) Science 285, 901–906. [DOI] [PubMed] [Google Scholar]
- 21.Kamath, R. S., Fraser, A. G., Dong, Y., Poulin, G., Durbin, R., Gotta, M., Kanapin, A., Le Bot, N., Moreno, S., Sohrmann, M., et al. (2003) Nature 421, 231–237. [DOI] [PubMed] [Google Scholar]
- 22.Mochizuki, T., Wu, G., Hayashi, T., Xenophontos, S. L., Veldhuisen, B., Saris, J. J., Reynolds, D. M., Cai, Y., Gabow P. A., Pierides, A., et al. (1996) Science 272, 1339–1342. [DOI] [PubMed] [Google Scholar]
- 23.Cole, D. G., Diener, D. R., Himelblau, A. L., Beech, P. L., Fuster, J. C. & Rosenbaum, J. L. (1998) J. Cell Biol. 141, 993–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nauli, S. M., Alenghat, F. J., Luo, Y., Williams, E., Vassilev, P., Li, X., Elia, A. E., Lu, W., Brown, E. M., Quinn, S. J., et al. (2003) Nat. Genet. 33, 129–137. [DOI] [PubMed] [Google Scholar]
- 25.Bhatnagar. V., Anjaiah, S., Puri, N., Darshanam, B. N. & Ramaiah, A. (1993) Arch. Biochem. Biophys. 307, 183–192. [DOI] [PubMed] [Google Scholar]
- 26.Stevens, T. H. & Forgac M. (1997) Annu. Rev. Cell Dev. Biol. 13, 779–808. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.