Abstract
Microperoxidases were initially isolated as peptide fragments containing covalently bound heme and are derived from naturally occurring c-type cytochromes. They are not only used as model compounds but also have potential applications as biosensors, electron carriers, photoreceptors, microzymes, and drugs. In a systematic attempt to define the minimal requirements for covalent attachment of hemes to c-type cytochromes, we have succeeded to produce artificial microperoxidases with peptide sequences that do not occur naturally and can be manipulated. The in vivo production of these microperoxidases requires targeting of the peptide to the bacterial periplasm, proteolytic processing of the signal peptide, and covalent attachment of heme to the signature motif CXXCH by the cytochrome c maturation proteins CcmA-H. The peptides that bind heme carry a C-terminal histidine tag, presumably to stabilize the heme peptide. We present a heme cassette that is the basis for the de novo design of functional hemoproteins.
Hemes have a broad range of physicochemical characteristics and are involved in many cellular processes in pro- and eukaryotes such as electron transport, enzymatic catalysis, or oxygen transport. At physiological pH, heme is insoluble and is always associated with peptides, proteins, or lipids. Heme-binding peptides, also referred to as microperoxidases (1), are used as biosensors (2, 3), electron carriers (4–6), photoreceptors (7, 8), molecules with specific electrochemical or catalytic properties (6, 9, 10), drugs (11), in biofuel cells (12), and as model compounds for understanding biological redox systems (5). Microperoxidases have been used to better characterize the structural and functional aspects of cytochromes (13), to obtain new, low molecular weight compounds able to mimic biologically active systems (14), and as molecular markers (15). So far, they have been synthesized chemically or by proteolytic digestion of naturally existing c-type cytochromes. We have developed a technique for the in vivo synthesis of artificial heme peptides based on our knowledge of the biogenesis of c-type cytochromes. These cytochromes carry one or more heme molecules each bound covalently to the amino acid sequence CXXCH. The two cysteines are crosslinked to the heme vinyl groups, resulting in two thioether bonds, and the histidine residue serves as an axial ligand to the heme iron. Bacterial c-type cytochromes are synthesized as precursor polypeptides with an N-terminal signal sequence for export of the polypeptide to the periplasm by the general protein type II secretion system (Sec) (16). Soluble c-type cytochromes are processed by leader peptidase after translocation (16). The covalent ligation of heme to the c-type cytochromes takes place on the periplasmic side of the cytoplasmic membrane. In Escherichia coli, eight cytochrome c maturation proteins CcmA-H (Ccm) are required specifically for this process (17, 18). The corresponding genes ccmA-H are expressed only under anaerobic growth conditions (19), when c-type cytochromes are produced for anaerobic respiration. We have constructed plasmid pEC86 (20), which expresses the ccmA-H operon constitutively from a vector promoter. This plasmid is now widely used for the overproduction of c-type cytochromes in E. coli. We have pursued the idea to define the minimal sequence requirements within the apocytochrome polypeptide for holocytochrome c formation by the Ccm system. We expressed small peptides containing a CXXCH motif in the E. coli periplasm, and at the same time provided pEC86, enabling covalent heme attachment. As a result, we present a mini-c-type cytochrome that can be produced in vivo and has characteristics similar to commercially available microperoxidases. To our knowledge, this peptide is also the smallest described to be secreted to the periplasm with a Sec-dependent signal peptide.
Materials and Methods
Strains, Plasmids, and Growth Conditions. E. coli strain DH5α (21) was used for cloning. Strain EC06 (17), harboring pEC86 (20) and the construct to be analyzed, was used for expression of heme peptides. pISC2 is an arabinose-inducible expression plasmid (19) and was used as the vector for construction of peptide expressing plasmids. Plasmid pRJ3290 (22) was used for expression of hexahistidine (H6)-tagged Bradyrhizobium japonicum cytochrome c550. Final antibiotic concentrations were 200 μg/ml–1 for ampicillin and 10 μg/ml–1 for chloramphenicol. For expression of peptides, 0.9 l of Luria-Bertani broth was inoculated with 9 ml of an overnight culture of the appropriate strain at 30°C. Cultures were grown to an OD600 of 0.8 and were induced with 0.1% arabinose. Seventeen hours after induction, cells were harvested by centrifugation at 3,300 × g.
Plasmid Construction. PCR products were cloned into pISC2 for expression of peptides. Primers complementary to the 3′ end of DNA fragments encoding C-terminal deletions of B. japonicum cytochrome c550 were designed to match the C-terminal 6–7 codons, followed by the sequence encoding the H6 tag (or Ala5His1 tag), a stop codon, and an EcoRI site. The vector primer pING1araB (23) was used as 5′ primer. Plasmids pMP126 and pMP132 encoding deletions located N-terminal to the CXXCH motif were constructed by the ligation of PCR products amplified with outwardly directed primers. Plasmids pMP360 and pMP370, encoding the shortest N-terminal sequences, were constructed with the QuikChange method (Stratagene). Plasmids encoding cytochrome c550 derivatives (19) were used as templates for all PCRs. Primer sequences and specific templates used are summarized in Table 2, which is published as supporting information on the PNAS web site. All constructs were confirmed by DNA sequencing with primer pING1araB (23). Sequencing was performed by and primers were purchased from Microsynth (Balgach, Switzerland).
Protein Extraction and Purification. Periplasmic proteins were isolated by extraction of cell pellets in 5 ml of 1 mg/ml–1 polymyxin B sulfate/500 mM sucrose/300 mM NaCl/100 mM Tris·HCl, pH 8. The suspension was stirred for 60 min at 4°C and was centrifuged at 15,000 × g for 20 min. The supernatant contained the periplasmic proteins. Total proteins were extracted by resuspending cell pellets in 5 ml of 1 M urea/3 M guanidinium-HCl/500 mM NaCl/20 mM Tris·HCl, pH 8. The suspension was frozen at –80°C, thawed at room temperature, stirred for 30 min, and centrifuged at 15,000 × g for 20 min. The supernatant contained total protein extracts. Alternatively, heme peptides were extracted in 1% Triton X-100 or Tween 80 in 500 mM NaCl/20 mM Tris·HCl, pH 8. H6-tagged peptides were bound to 0.5 ml of bed volume of nickel nitrilotriacetic acid beads (Qiagen), washed once with 5 ml of the corresponding extraction buffer, a second time with 5 ml of 50 mM Tris·HCl, pH 8/300 mM NaCl/20 mM imidazole, and eluted with four times 0.4 ml of 50 mM Tris·HCl, pH 8/300 mM NaCl/300 mM imidazole. Fractions 2–4 containing the H6-tagged peptides were pooled and used for further characterization. Because imidazole may serve as an axial ligand to the heme iron, heme peptides were also eluted with low pH instead of imidazole, by washing the nickel nitrilotriacetic acid beads with the bound heme peptide with five bed volumes of 100 mM NaH2PO4/10 mM Tris·HCl, pH 6.3, then with five bed volumes of the same buffer, pH 5.9, and eluted with the same buffer, pH 4.5. The pH of the eluted factions was readjusted with NaOH to 7.
Analytical Methods. Peptides were separated by SDS/18% PAGE as described by Laemmli (24). Gels were supplemented with 8.7% glycerol. Heme-bound peptides were detected in gels by using their intrinsic peroxidase activity by immersion of the gels in a solution of 10 mg of 3,3′-dimethoxybenzidine (Sigma), dissolved in 10 ml of H2O and 0.7% H2O2. Molecular masses of purified peptides were determined by matrix-assisted laser desorption ionization/time-of-flight MS on an Applied Biosystems Voyager-DE Elite mass spectrometer by using 2,5-dihydroxybenzoic acid as matrix. Optical spectra were recorded either on a Molecular Devices SPECTRAmax plus or on a Hitachi model U-3300 spectrophotometer and exported to a Microsoft excel table for calculations of difference spectra and determination of absorption maxima. Relative concentrations of purified heme peptides were determined from the reduced minus oxidized absorption spectra shown in Fig. 3A. The absorption difference between 550 and 536 nm is proportional to the amount of peptide-bound heme (25). Absolute concentrations of heme peptide were determined by the pyridine hemochrome assay (26). Quantifications for MP86 and MP251 by either method correlated with an SD of 15%. Microperoxidase MP-11 and horse heart cytochrome c (Sigma) were used as standards.
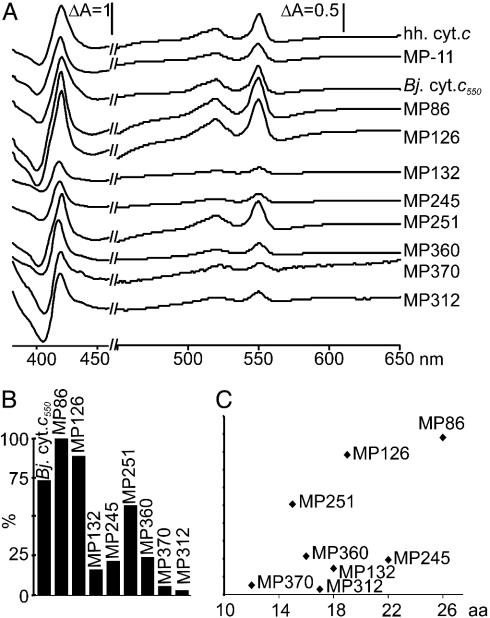
Fig. 3.
Length-yield correlation of purified microperoxidases. (A) Dithionite-reduced minus-air-oxidized difference spectra are shown. The y axis is expanded from 450 to 650 nm. For better resolution, the ΔA of MP370 and MP312 were magnified 3- and 10-fold, respectively. hh. cyt. c, horse heart cytochrome c. (B) Yields of heme peptides after purification over nickel nitrilotriacetic acid calculated from ΔA550–536 nm of the difference spectra. The value of MP86 was set to 100%. (C) Correlation of length and yield of heme peptides. The y axis corresponds to that of B.
Peroxidase Activity Assay. Peroxidase activity of heme peptides was determined essentially as described by Primus et al. (27). A total of 1 mM 3,3′-dimethoxybenzidine was used as substrate. Heme peptide was added at a final concentration of 0.2 μM. Reactions were started by the addition of 1 mM H2O2. Product formation was monitored by the change in absorbance at 460 nm (ε460 = 11.3 mM–1·cm–1) on a SPECTRAmax plus spectrophotometer. Initial reaction rates were calculated from the results of triplicate experiments.
Measurement of Oxidation-Reduction Potentials. Potentiometric titrations were performed as described by Dutton (28). Heme peptides were purified in the absence of imidazole, and titrations were performed under argon in 10 mM Tris·HCl, pH 7, with heme peptide concentrations of 3–10 μM. The following mediators from Sigma-Aldrich were used at a concentration of 10 μM: 2-hydroxy-1,4-naphthoquinone/9,10-anthraquinone-2,6-disulfonic acid disodium salt/anthraquinone-2-sulfonic acid sodium salt/phenazine ethosulfate. Optical spectra were recorded from 380 to 600 nm on a Hitachi model U-3300 spectrophotometer at various electrode potentials. A combination platinum and Ag/AgCl electrode (Willi Möller, Zürich) was used to measure the half-cell potential. The electrode was calibrated with a saturated solution of quinhydrone at pH 7. The degree of reduction of heme peptides was estimated from the ΔA500–536 nm. The potentials were adjusted by the addition of ≈1 μl of volumes of solutions, 10–50 mM Na2-dithionite (reductive titrations), and 3 mM K3Fe(CN)6 (oxidative titrations). Redox midpoint potentials were determined as a mean of at least four titrations.
Results
Common Features of Bacterial c-Type Cytochromes. The signal sequence that is required for Sec-dependent protein translocation across the cytoplasmic membrane and a CXXCH heme-binding motif are the only common signatures of c-type cytochromes, and are prerequisites for their posttranslational maturation. Because an α-helix is often found to precede the CXXCH motif of type I c-type cytochromes with the heme-binding site close to the N terminus of the mature protein (29–32), we wondered whether (partial) folding of the polypeptide or the establishment of secondary structures were important for the covalent attachment of heme to the polypeptide. We investigated the sequence requirements for heme ligation by stepwise truncation of a natural cytochrome c sequence on either side of the CXXCH signature motif and testing the products for heme binding.
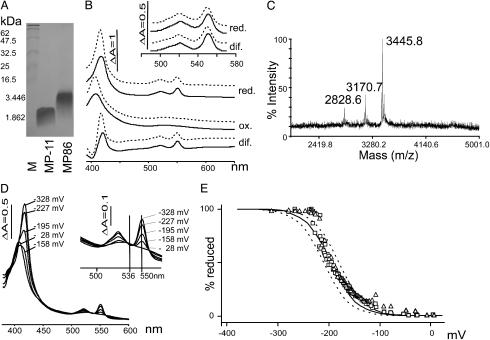
The N-Terminal 20 aa of B. japonicum Cytochrome c550 Are Sufficient for Heme Incorporation. Our model c-type cytochrome is the soluble B. japonicum cytochrome c550, encoded by the gene cycA. The mature protein contains 108 aa. Heme is bound covalently to the cysteines at positions 13 and 16 of the mature peptide. In B. japonicum, the protein is needed for anaerobic nitrate respiration (33). To express a truncated version of this cytochrome, plasmid pMP86 was constructed. It encodes the signal sequence plus the first 20 aa, including the CXXCH motif of this cytochrome, fused to a C-terminal H6 tag to facilitate protein purification (Table 1). Cells expressing this peptide in the presence of the Ccm proteins produced a red pigment. This pigment did not fractionate with the periplasmic fraction, as expected for a soluble c-type cytochrome. Fractionation of the heme-binding minicytochrome c with membranes rather than with the periplasm, suggests that unlike the full-length, globular cytochrome, this truncated peptide has more hydrophobic characteristics, most likely due to a relatively exposed heme, which may serve as a membrane anchor. This result is also supported by the finding that the heme peptides MP86 and MP251 could be solubilized with nonionic detergents Tween 80 and Triton X-100. Our standard protocol, solubilization in a buffer containing guanidinium-HCl and urea followed by purification on nickel nitrilotriacetic acid agarose, resulted in a soluble heme peptide, as revealed by SDS/PAGE and staining for covalently-bound heme (Fig. 1A). Coomassie staining of this gel revealed an additional protein in the range of 30 kDa that did not bind heme (data not shown). The spectral properties of the heme peptide in the presence and absence of imidazole were characteristic for a low spin c-type cytochrome (Fig. 1B). For air-oxidized MP86, an absorption maximum was observed at 408 nm in the absence of imidazole, and at 406 nm in the presence of imidazole. MP86 was reduced with dithionite but not with ascorbate, unlike its parent full-length cytochrome c550, which is also reduced with ascorbate. Dithionite-reduced spectra of MP86 had peak maxima at 417.5, 521.5, and 550 nm in the absence of imidazole, and at 416, 520, and 549.5 nm in the presence of 300 mM imidazole. Calculated reduced minus oxidized difference spectra showed an α-band of 550.5 nm, a β-band of 521 nm, and a γ-(Soret) band of 420.5 nm in the absence and of 549.5, 520, and 417.5 nm, respectively, in the presence of 300 mM imidazole (Fig. 1B). A yield of 202 μg MP86 heme peptide per liter of E. coli culture was calculated. MS analysis resulted in a main peak at 3445.8 m/z (Fig. 1C), which corresponded well with the calculated mass of the mature holopeptide (3,446 Da), confirming that the peptide contained heme and that its signal peptide had been proteolytically removed by leader peptidase. The mass of the hemeless, processed peptide was also found (m/z = 2,828.6), suggesting that the purified fraction contained both apo- and holo-MP86. An additional peak at m/z = 3,170.7 corresponded to the expected mass of the holopeptide missing two histidine residues. This finding is indicative of a partial C-terminal degradation. The midpoint potential of MP86 was determined to be –195 ± 13 mV (Fig. 1 D and E), which is in agreement with ascorbate not being able to reduce the heme peptide.
Table 1. Peptides and their properties.
| Molecular mass, Da
|
||||
|---|---|---|---|---|
| Peptide | Heme binding* | Peptide sequence | Theoretical, holo (apo) | Determined from purified peptides, holo (apo) |
| MP-11 | YES | VQKCAQCHTVE | 1,862.4 (1,245.4) | ND |
| cyt.c550-H6 | YES | B. japonicum cyt. c550-HHHHHH | 13,443 (12,826) | ND |
| MP86 | YES | QDAAAGKTSFNKCLACHAIGHHHHHH | 3,446 (2,829.1) | 3,445.8 (2,828.6) |
| MP105 | NO | VQKCAQCHTVE | (1,245.4) | ND |
| MP126 | YES | QDAAACLACHAIGHHHHHH | 2,683 (2,066.2) | 2,682.5 (2,064.5) |
| MP132 | YES | QDAACLACHAIGHHHHHH | 2,612 (1,995.1) | 2,612.5 |
| MP153 | NO | QDAAAGKTSFNKCLACHAIG | (2,006.2) | ND |
| MP204 | NO | QDAAAGKTSFNKCLACHAIGAAAAAH | (2,498.8) | ND |
| MP207 | NO | QDAAAGKTSFNKCLACHAIGAAAAHA | (2,498.8) | ND |
| MP210 | NO | QDAAAGKTSFNKCLACHAIGAAAHAA | (2,498.8) | ND |
| MP213 | NO | QDAAAGKTSFNKCLACHAIGAAHAAA | (2,498.8) | ND |
| MP216 | NO | QDAAAGKTSFNKCLACHAIGAHAAAA | (2,498.8) | ND |
| MP220 | NO | QDAAAGKTSFNKCLACHAIGHAAAAA | (2,498.8) | ND |
| MP222 | YES | QDAAAGKTSFNKCLACHHHHHHH | 3,204.8 (2,587.8) | 3,207.1 (2,588.8) |
| MP245 | YES | QDAAAGKTSFNKCLACHHHHHH | 3,067 (2,450.7) | 3,067.1 |
| MP251 | YES | QDAAACLACHHHHHH | 2,305 (1,687.6) | 2,308.7 |
| MP259 | YES | QDAAACLACHHHHHHH | 2,441.8 (1,824.9) | 2,445.8 |
| MP312 | YES | VQKCAQCHTVEHHHHHH | 2,685 (2,068.2) | 2,683.4 (2066.3) |
| MP360 | YES | QDCLACHAIGHHHHHH | 2,470 (1,853.0) | 2,469.5 |
| MP370 | YES | QDCLACHHHHHH | 2,091.6 (1,474.6) | 2,090.5 |
ND, not determined.
Detection by optical spectroscopy of total cell proteins.
Fig. 1.
Biochemical and biophysical characteristics of MP86. (A) Heme stain of samples containing 1 nmol of heme peptide. Molecular weights of marker proteins and of the peptides are indicated on the left. (B) Optical spectra of MP86. Air-oxidized (ox.), reduced (5 mM dithionite; red.), and difference (dif.) spectra are shown in the presence (solid lines) and absence (dashed lines) of 300 mM imidazole. (Inset) The α/β region at higher magnification. (C) Results of MS. (D) Optical spectra for potentiometric titrations. (E) Determination of mid point potential with the Nernst equation. Curves from reductive titrations (▵) and oxidative titrations (□) as well as theoretical curves for n = 1 (one electron transfer) and E0 =–195 (solid line), –180 (dashed line), and –210 mV (dashed line) are shown.
We also tried to purify the apopeptide from cells lacking the Ccm plasmid pEC86, which did not produce the red pigment. However, the peptide was neither detected in whole-cell extracts nor in periplasmic fractions by Western blot analysis using α-His6 antibodies, Coomassie staining, or MS (data not shown). This result indicates that the apopeptide is not stable, which is in agreement with findings (19) that the full-length apocytochrome c550 of B. japonicum is unstable.
Peptide QDCLACHHHHHH Binds Heme in Vivo. To further truncate MP86, sequences located N-terminal, C-terminal, and both N- and C-terminal of the CLACH motif were deleted, resulting in peptides MP222 and MP245 (C-terminal deletions), MP126, MP132, and MP360 (N-terminal deletions), and MP251, MP259, and MP370 (C- and N-terminal deletions; Table 1). All peptides were found to bind heme covalently (Table 1). MP370 with a length of 12 aa was the shortest peptide that could bind heme in vivo. It contained the first two amino acids QD of mature cytochrome c550 to ensure efficient posttranslocational processing, followed by the CLACH heme-binding motif and five histidine residues for a C-terminal H6 tag. The resulting sequence QDCLACHHHHHH clearly does not occur in any natural c-type cytochrome and thus represents an artificial microperoxidase. We did not truncate the peptide further because a negative charge is often found at position +2 after the signal peptidase cleavage site. In addition, a cysteine following the signal peptide might direct the peptide to the lipoprotein maturation pathway that involves cleavage of the signal peptide by lipoprotein signal peptidase and simultaneous attachment of a lipid anchor to the cysteine, which would block heme binding (34).
MP-11 Peptide Does Not Bind Heme in Vivo. We next addressed whether a peptide with the sequence of commercially available microperoxidase MP-11 fused to the signal sequence of cytochrome c550 from B. japonicum could be expressed in its heme-binding form. MP-11 is a fragment of horse heart cytochrome c digested with pepsin (35). The periplasmic MP105 was expressed in the presence of pEC86. Total protein extracts and periplasmic proteins were analyzed for the presence of c-type cytochromes by absorption difference spectroscopy. The spectra did not show any specific heme absorption of a c-type cytochrome (data not shown). Unlike the shortest of our peptides, MP370, MP105 did not contain a C-terminal H6 tag. We therefore tested the influence of a H6 tag on heme binding of the peptide.
The Entire H6 Tag Is Required for Heme Incorporation. An H6 tag was fused to the C terminus of MP105 (the original MP-11). The resulting purified MP312 peptide (Table 1) was hardly detectable by heme staining, which was most likely due to low levels of peptide in the gel. Yet, its optical difference spectrum was typical for a c-type cytochrome (Fig. 3), and the obtained molecular mass of 2,683.35 Da (Table 1) clearly showed that covalent heme attachment was possible in this case. Quantification of holo-MP312 revealed the concentration to be 1.5 μM, corresponding to 4.7 μg of heme peptide in 1.2 ml. Hence, a C-terminal H6 tag enables the short peptide to bind heme covalently.
To test whether the H6 tag was also an important parameter for heme binding of the truncated cytochrome c, it was removed in the construct pMP153, which now encoded a mature cytochrome c peptide of 20 aa. This peptide did not bind heme, although it was longer than some of the previously expressed H6-tagged heme peptides such as MP126. This finding indicated that not the length of the peptide, but rather, the presence of one or more histidine residues is critical, perhaps serving as a sixth ligand to the heme iron. Therefore, the H6 tag of MP86 was replaced by alanine tags containing a single histidine residue at either of the six positions, resulting in peptides MP204, MP207, MP210, MP213, MP216, and MP220 (Table 1). None of the protein extracts derived from E. coli cultures harboring these peptide expressing plasmids had spectra typical for c-type cytochromes. We conclude that, rather than a histidine residue at a specific position, the entire H6 tag facilitated heme incorporation.
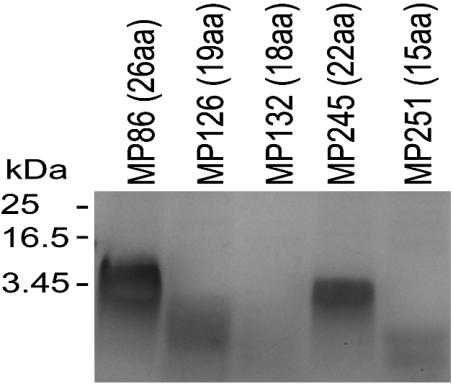
In Vivo-Expressed Heme Peptides Have Common Biochemical and Biophysical Characteristics but Sequence-Dependent Yields. The different heme peptides were isolated in parallel preparations of a comparative study where MP-11 and B. japonicum cytochrome c were also included. Equal volumes of purified peptide were separated on SDS/18% PAGE gels and stained for peroxidase activity (Fig. 2). All peptides showed heme binding, and their migration patterns correlated with the lengths of the mature peptides (Fig. 2). Staining intensities varied tremendously, despite the fact that all samples contained the same amount of the 30-kDa contaminating protein detected by Coomassie stain (data not shown). This variation was not only due to the different amounts of holopeptide loaded but also due to the length of the peptide (compare Fig. 2, lanes MP126, MP132, and MP245). Shorter peptides resulted in weaker stains probably due to diffusion during SDS/PAGE and staining procedures. Spectra of the new heme peptides were recorded in the air oxidized state and in the presence of ascorbate and dithionite, respectively. Only dithionite was found to reduce the heme peptides (data not shown). Dithionite-reduced minus-air-oxidized difference spectra of all heme peptides were compared with one another and no significant differences were found (Fig. 3A). The difference spectra were used to calculate the yields of purified holoheme peptides from 0.9 l of culture (Fig. 3B). Depending on the sequence, yields of holopeptide varied by up to a factor of 30. Neither the length of the entire peptide nor the N- or C-terminal extension from the CXXCH motif correlated with the yields (Fig. 3C). The biophysical properties of MP86 and MP251 were further characterized. Catalytic peroxidase activities were determined and compared with MP-11 in dependence of the pH. As described for MP-8 (27), the highest activity was found at alkaline pH >9 for MP86, MP251, and MP-11. At pH 6.5, all three samples had similar turnover numbers; MP86: kcat = 0.37 ± 0.03 s–1; MP251: 0.32 ± 0.03 s–1; and MP-11: 0.38 ± 0.03 s–1. The increase in activity at the respective pH optima was 4.4-fold for MP86, 12-fold for MP251, and 10-fold for MP-11, resulting in turnover numbers of 4.0 ± 0.44 s–1 for MP251, and 3.89 ± 0.59 s–1 for MP-11, which is twice that of MP86 (1.64 ± 0.077 s–1). The similar activities for MP86 and MP251 at pH 6.5 show that the lower intensity in heme stain is not due to lower specific activity, but rather to lower amounts of heme peptide in the gel. Midpoint potentials were determined by potentiometric titrations for MP86 to be –195 (± 13) mV (Fig. 1 D and E), and for MP251 to be –182 (± 8) mV in the absence of imidazole. In the presence of imidazole, no significant difference was found. The determined midpoint potentials explain why these heme peptides cannot be reduced by ascorbate (E0 = +58 mV).
Fig. 2.
Heme stains of purified microperoxidases. Equal volumes (10 μl) of purified peptides containing amounts of heme indicated in Fig. 3B were loaded in each lane. Molecular masses of marker proteins and of MP86 are indicated on the left.
Discussion
The posttranslational covalent insertion of heme into c-type cytochromes is a critical step in the maturation pathway of these proteins. In E. coli, it requires a multisubunit machinery with heme lyase activity encoded by the ccmABCDEFGH operon. The question of how the cytochrome polypeptide is selected to be a substrate for the heme lyase is based on its subcellular location and on its primary sequence. Heme attachment occurs in the periplasm and takes place exclusively in proteins that contain an N-terminal signal peptide for secretion into this compartment. In this work, we addressed the question of what is the minimal sequence requirement in the polypeptide that leads to covalent heme binding. Because the CXXCH heme-binding motif is the only known signature sequence common to c-type cytochromes, the theoretical minimal heme-binding peptide should be CXXCH fused to a signal sequence. However, a cysteine following the signal peptidase cleavage site is expected to direct the secreted polypeptide to the membrane after the covalent modification of the first cysteine with a lipid anchor and thus block heme ligation. Our system for the in vivo production of cytochrome c fragments is composed of two plasmids, one encoding a precursor peptide with a N-terminal signal sequence fused to a small peptide that contains the CXXCH motif, and pEC86 containing the ccmA-H genes that are expressed from the vector promoter for overproduction of the cytochrome c biosynthesis machinery.
We expected our system to allow the in vivo synthesis of commercially available microperoxidases, which are proteolytic fragments of naturally occurring cytochrome c. When the peptide VQKCAQCHTVE corresponding to MP-11 (Sigma) was fused to the signal peptide of our model cytochrome c550 of B. japonicum and was coexpressed with the ccm genes of pEC86, no heme binding could be observed. By contrast, our alternative approach to truncate a H6-tagged natural c-type cytochrome N- and C-terminally of its heme-binding motif resulted in the shortest known peptides translocated in a Sec-dependent way to the periplasm and binding heme. The H6 tag was introduced to allow detection of the apopeptides. Unexpectedly, we obtained small heme-binding peptides only when the tag was present. In line with this observation, we also achieved heme binding to MP-11 after adding a C-terminal H6 tag to the sequence. In a series of heme peptide constructs carrying five C-terminal alanine residues plus one histidine residue at either of the six C-terminal positions, we excluded the possibility that a histidine residue at a certain distance from the CXXCH motif was necessary. Hence, the C-terminal H6 tag as a whole seemed to facilitate heme binding. In the E. coli periplasm, apocytochromes c are rapidly degraded when heme binding is blocked (19). In the absence of the cytochrome c maturation system, we expected to produce the apoform of the H6-tagged microperoxidase. We failed to detect the apopeptide by immunoblots or MS, suggesting that the tag was not just contributing to the general stability of the peptide. The H6 tag either facilitated the recognition and selection of the peptide by the heme insertion machinery, or it stabilized the heme binding peptide by allowing one of the histidine residues to fold over the heme plane to form a sixth axial ligand to the iron. The presence of a strong sixth ligand to the heme iron seems likely from the sharp α- and β-bands of the difference spectra, as expected for low-spin heme and from the midpoint potentials. Although acetylated MP-11 had a significant difference in midpoint potentials, depending on the presence (–189 mV) or absence (–134 mV) of imidazole (14), no such difference was found for MP86 and MP251. Furthermore, the midpoint potential of the H6-tagged heme peptides in the absence of imidazole is close to the midpoint potential of acetylated MP-11 in the presence of the strong iron ligand imidazole. A requirement of the tag for heme insertion seems unlikely, because it is known that cytochromes c lacking a sixth heme ligand still have covalently bound heme (36, 37). We therefore suspect that the H6 tag protects the heme-binding peptide from degradation.
We could not establish a strict correlation between the length and the yield of the heme peptides. The latter depends on (i) efficiency of peptide translocation, (ii) signal sequence cleavage, and, (iii) recognition by the Ccm complex, and each of these steps may have certain, not necessarily overlapping sequence preferences. For example, cultures producing the shorter 15-mer MP251 resulted in ≈10 times more heme peptide than found in cultures producing the longer 17-mer MP312, but the latter had a positive charge close to the N terminus of the mature peptide, a feature that is known to inhibit Sec-dependent translocation (38). The shortest peptide that we found to bind heme, although with a low yield, was the 12-mer QDCLACHHHHHH. It suggests that recognition of the peptide by the Ccm apparatus is restricted to the CXXCH motif plus perhaps a few encompassing amino acids, and that a heme cassette consisting of CXXCH6 should be sufficient to allow covalent heme binding.
Recent attention has been devoted to the synthesis of both covalent and noncovalent heme-binding peptides, thereby enabling the solubilization of the hydrophobic porphyrins in aqueous solutions, which is required for many technological applications. They can be obtained after digestion of naturally occurring cytochrome c with trypsin or pepsin (1, 35) and subsequent chemical modifications for specific applications (39–41), or by de novo synthesis (42), which often is very complex. The source and variability of microperoxidases thus is limited to peptide sequences corresponding to the sequence encompassing the heme-binding site of naturally occurring c-type cytochromes or de novo chemical synthesis. We have created a number of artificial microperoxidases whose sequences do not correspond to any natural cytochrome c-derived fragment. Even though none of the heme peptides fractionated with the soluble periplasm, after solubilization with GuHCl/urea and subsequent Ni-affinity chromatography, all heme peptides remained soluble in aqueous solutions.
The rational design of functional metalloenzymes is highly desirable to develop new activities for tailored oxidations, bio-detoxification, engineering of new metabolites, and drugs. Our approach does enable the production of heme peptides with different properties, e.g., the specific activity of MP251 is more than double the specific activity of MP86 at alkaline pH. Additional tags such as the H6 tag enable specific immobilization of these microperoxidases to solid surfaces e.g., Ni2+-beads. The de novo design of heme proteins by peptide synthesis and assembly (4, 5, 7, 8) can now be expanded by the biotechnological introduction of a covalently bound heme group into a polypeptide. This procedure may open a new playground for the discovery of novel catalytic devices by combinatorial shuffling of naturally occurring redox active groups.
Supplementary Material
Acknowledgments
We thank David Knaff for technical advice on establishing potentiometric titrations, René Brunisholz and Helen Rechsteiner for support with MS, and Umesh Ahuja, Olaf Christensen, and Franz Narberhaus for helpful discussions. This work was supported by the Swiss National Foundation for Scientific Research.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Ccm, c-type cytochrome maturation proteins CcmA-H; Sec, bacterial type II protein secretion.
References
- 1.Feder, N. (1970) J. Histochem. Cytochem. 18, 911–1003. [DOI] [PubMed] [Google Scholar]
- 2.Tatsuma, T. & Watanabe, T. (1991) Anal. Chem. 63, 1580–1585. [DOI] [PubMed] [Google Scholar]
- 3.Huang, W., Jia, J., Zhang, Z., Han, X., Tang, J., Wang, J., Dong, S. & Wang, E. (2003) Biosens. Bioelectron. 18, 1225–1230. [DOI] [PubMed] [Google Scholar]
- 4.Sharp, R. E., Moser, C. C., Rabanal, F. & Dutton, P. L. (1998) Proc. Natl. Acad. Sci. USA 95, 10465–10470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robertson, D. E., Farid, R. S., Moser, C. C., Urbauer, J. L., Mulholland, S. E., Pidikiti, R., Lear, J. D., Wand, A. J., DeGrado, W. F. & Dutton, P. L. (1994) Nature 368, 425–432. [DOI] [PubMed] [Google Scholar]
- 6.Wilcox, S. K., Putnam, C. D., Sastry, M., Blankenship, J., Chazin, W. J., McRee, D. E. & Goodin, D. B. (1998) Biochemistry 37, 16853–16862. [DOI] [PubMed] [Google Scholar]
- 7.Rau, H. K., DeJonge, N. & Haehnel, W. (1998) Proc. Natl. Acad. Sci. USA 95, 11526–11531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takeda, S., Kamiya, N., Arai, R. & Nagamune, T. (2001) Biochem. Biophys. Res. Commun. 289, 299–304. [DOI] [PubMed] [Google Scholar]
- 9.Ricoux, R., Sauriat-Dorizon, H., Girgenti, E., Blanchard, D. & Mahy, J. P. (2002) J. Immunol. Methods 269, 39–57. [DOI] [PubMed] [Google Scholar]
- 10.Kadnikova, E. N. & Kostic, N. M. (2003) J. Org. Chem. 68, 2600–2608. [DOI] [PubMed] [Google Scholar]
- 11.Spector, A., Ma, W., Wang, R. R. & Kleiman, N. J. (1997) Exp. Eye Res. 65, 457–470. [DOI] [PubMed] [Google Scholar]
- 12.Willner, I., Katz, E., Patolsky, F. & Bückmann, A. F. (1998) J. Chem. Soc. Perkin. Trans. 2, 1817–1822.
- 13.Tezcan, F. A., Winkler, J. R. & Gray, H. B. (1998) J. Am. Chem. Soc. 120, 13383–13388. [Google Scholar]
- 14.Battistuzzi, G., Borsari, M., Cowan, J. A., Ranieri, A. & Sola, M. (2002) J. Am. Chem. Soc. 124, 5315–5324. [DOI] [PubMed] [Google Scholar]
- 15.Maurizi, G., Ciabini, A., Di Cioccio, V. & Ruggiero, P. (2000) Anal. Biochem. 286, 295–296. [DOI] [PubMed] [Google Scholar]
- 16.Thöny-Meyer, L. & Künzler, P. (1997) Eur. J. Biochem. 246, 794–799. [DOI] [PubMed] [Google Scholar]
- 17.Thöny-Meyer, L., Fischer, F., Künzler, P., Ritz, D. & Hennecke, H. (1995) J. Bacteriol. 177, 4321–4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grove, J., Tanapongpipat, S., Thomas, G., Griffiths, L., Crooke, H. & Cole, J. (1996) Mol. Microbiol. 19, 467–481. [DOI] [PubMed] [Google Scholar]
- 19.Thöny-Meyer, L., Künzler, P. & Hennecke, H. (1996) Eur. J. Biochem. 235, 754–761. [DOI] [PubMed] [Google Scholar]
- 20.Arslan, E., Schulz, H., Zufferey, R., Künzler, P. & Thöny-Meyer, L. (1998) Biochem. Biophys. Res. Commun. 251, 744–747. [DOI] [PubMed] [Google Scholar]
- 21.Hanahan, D. (1983) J. Mol. Biol. 166, 557–580. [DOI] [PubMed] [Google Scholar]
- 22.Ahuja, U. & Thöny-Meyer, L. (2003) J. Biol. Chem. 278, 52061–52070. [DOI] [PubMed] [Google Scholar]
- 23.Enggist, E., Schneider, M. J., Schulz, H. & Thöny-Meyer, L. (2003) J. Bacteriol. 185, 175–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laemmli, U. K. (1970) Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- 25.Appleby, C. A. (1969) Biochim. Biophys. Acta 172, 71–87. [DOI] [PubMed] [Google Scholar]
- 26.Fuhrhop, J.-H. & Smith, K. M. (1975) in Porphyrins and Metalloporphyrins, ed. Smith, K. M. (Elsevier Science, Amsterdam), pp. 757–869.
- 27.Primus, J. L., Boersma, M. G., Mandon, D., Boeren, S., Veeger, C., Weiss, R. & Rietjens, I. M. (1999) J. Biol. Inorg. Chem. 4, 274–283. [DOI] [PubMed] [Google Scholar]
- 28.Dutton, P. L. (1978) Methods Enzymol. 54, 411–435. [DOI] [PubMed] [Google Scholar]
- 29.Sogabe, S. & Miki, K. (1995) J. Mol. Biol. 252, 235–247. [DOI] [PubMed] [Google Scholar]
- 30.Bersch, B., Blackledge, M. J., Meyer, T. E. & Marion, D. (1996) J. Mol. Biol. 264, 567–584. [DOI] [PubMed] [Google Scholar]
- 31.Bartalesi, I., Bertini, I., Hajieva, P., Rosato, A. & Vasos, P. R. (2002) Biochemistry 41, 5112–5119. [DOI] [PubMed] [Google Scholar]
- 32.Ptitsyn, O. B. (1998) J. Mol. Biol. 278, 655–666. [DOI] [PubMed] [Google Scholar]
- 33.Bott, M., Thöny-Meyer, L., Loferer, H., Rossbach, S., Tully, R. E., Keister, D., Appleby, C. A. & Hennecke, H. (1995) J. Bacteriol. 177, 2214–2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Inouye, S., Franceschini, T., Sato, M., Itakura, K. & Inouye, M. (1983) EMBO J. 2, 87–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsou, C. L. (1951) Biochem. J. 49, 362–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Allen, J. W. & Ferguson, S. J. (2003) Biochem. J. 375, 721–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brandner, J. P., Stabb, E. V., Temme, R. & Donohue, T. J. (1991) J. Bacteriol. 173, 3958–3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kajava, A. V., Zolov, S. N., Kalinin, A. E. & Nesmeyanova, M. A. (2000) J. Bacteriol. 182, 2163–2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ippoliti, R., Picciau, A., Santucci, R., Antonini, G., Brunori, M. & Ranghino, G. (1997) Biochem. J. 328, 833–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheek, J., Low, D. W., Gray, H. B. & Dawson, J. H. (1998) Biochem. Biophys. Res. Commun. 253, 195–198. [DOI] [PubMed] [Google Scholar]
- 41.Low, D. W., Yang, G., Winkler, J. R. & Gray, H. B. (1997) J. Am. Chem. Soc. 119, 4094–4095. [Google Scholar]
- 42.De Luca, S., Bruno, G., Fattorusso, R., Isernia, C., Pedone, C. & Morelli, G. (1998) Lett. Pept. Sci. 5, 269–276. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.