Abstract
The corticotropin-releasing factor (CRF) ligand family has diverse effects on the CNS, including the modulation of the stress response. The ligands' effects are mediated by binding to CRF G protein-coupled receptors. We have determined the 3D NMR structure of the N-terminal extracellular domain (ECD1) of the mouse CRF receptor 2β, which is the major ligand recognition domain, and identified its ligand binding site by chemical-shift perturbation experiments. The fold is identified as a short consensus repeat (SCR), a common protein interaction module. Mutagenesis reveals the integrity of the hormone-binding site in the full-length receptor. This study proposes that the ECD1 captures the C-terminal segment of the ligand, whose N terminus then penetrates into the transmembrane region of the receptor to initiate signaling. Key residues of SCR in the ECD1 are conserved in the G protein-coupled receptor subfamily, suggesting the SCR fold in all of the ECD1s of this subfamily.
For more than a century, the ability of the body to adapt to stressful stimuli and the role of stress maladaptation in human diseases have been intensively investigated. However, in 1981 the isolation and characterization of corticotropin-releasing factor (CRF) (1) forged a major breakthrough in understanding the human stress response. Today, a considerable body of evidence suggests that peptides of the CRF family, i.e., CRF (1), (frog) sauvagine, (fish) urotensin, and the mammalian urocortins (Ucns) 1, 2, and 3 (2–5), play biologically diverse roles by activating CRF receptors (6, 7).
The CRF receptors, encoded by two distinct receptor genes, exist in multiple splice variant forms and display both species and tissue differential expression (6, 7). Studies with transgenic mice expressing functionally disabled receptors have underscored the importance of CRF-R1 in regulating the hypothalamic–pituitary–adrenal axis in its response to stress (8). CRF-R2 plays an important role in modulating the CNS response to stress (8) and has a unique role in cardiac function (9) and pancreatic hormone release (10).
The CRF receptors belong to the B1 subfamily of G protein-coupled receptors (GPCRs). The GPCRs consist of seven transmembrane helices but present a large variety of different conformations in their extracellular domains (ECDs) to cover their different functions (11, 12). Indeed, a major ligand binding site on both CRF receptors is the N-terminal ECD (ECD1) (13–19). The inhibitory binding constants for a bacterially expressed soluble protein, ECD1–CRF-R2β [comprising amino acids 39–133 of mouse CRF-R2β (mCRF-R2β)] are: 11.8 (7.4–18.9) nM, 53.7 (18.7–154) nM, and 21.1 (15.3–29.0) nM for Ucn1, Ucn2, and astressin, respectively. Here, we present the 3D structure of ECD1–CRF-R2β and its hormone peptide binding site to provide a structural rationale for characterizing the mechanism of activation of this family of GPCRs.
Materials and Methods
Mutagenesis. The myc–mCRF-R2β, in which a c-myc epitope is inserted between residues 29 and 30, and all of the point mutants were created by overlap extension PCR with mCRF-R2β as the template. The PCR products were subcloned into pCDNA3, and the complete sequences were confirmed by automated sequencing.
Protein Expression. A cDNA encoding amino acids 39–133 of mCRF-R2β was inserted into pET-32a(+) (Novagen) with KpnI and XhoI, and its integrity was confirmed by automated sequencing. The sequence of the protein is: GSGMKETAAAKFERQHMDSPDLGT [mCRF-R2β (39–133)], in which an S-tag sequence (used for purification) is underlined and the additional amino acids are part of the thrombin cleavage site and the KpnI cloning site. The N-terminal residues are highly flexible as determined by NMR (data not shown). The protein was expressed in minimal media containing 4 g/liter 13C-d-glucose and 1g/liter 15N-ammonium sulfate. Protein purification was carried out as described (16). Twenty liters of expression media yielded one sample of 13C,15N-labeled ECD1–CRF-R2β with a concentration of ≈0.2 mM.
Radioreceptor Assays. Mutant receptors or myc–mCRF-R2β were transiently transfected into COSM6 cells followed by binding to crude membrane preparations as described (20). Binding was performed in triplicate, as described (16).
NMR Experiments. All of the NMR spectra were recorded at 25°C on a Bruker 700-MHz spectrometer equipped with four radio-frequency channels and a triple-resonance cryo-probe with shielded z-gradient coil. The NMR samples contained 0.2 mM 13C,15N-labeled ECD1–CRF-R2β in 10 mM BisTris(HCl)/95% H2O/5% D2O at pH 7.4. Sequential assignment and structure determination were performed with the standard protocol for 13C,15N-labeled samples (21). 1H, 13C, and 15N backbone resonances were assigned by using the triple-resonance experiments HNCA and CBCA(CO)NH and 3D 15N-resolved [1H,1H]-NOESY experiments. The side-chain signals were assigned from HCCH-correlation spectroscopy (COSY) and 13C-resolved [1H,1H]-NOESY experiments. Aromatic side-chain assignments were obtained with 2D double quantum filtered-COSY, 2D [1H,1H]-NOESY in D2O, and 3D 1H-total correlation spectroscopy-relayed ct-[13C,1H]-heteronuclear multiple quantum correlation (HMQC) experiments. Distance constraints for the calculation of the 3D structure were derived from 3D 13C-,15N-resolved [1H,1H]-NOESY and 2D [1H,1H]-NOESY spectra recorded with a mixing time of 80 ms.
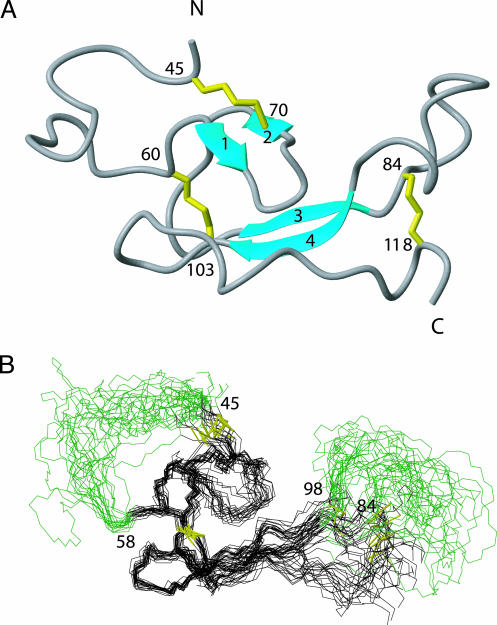
Structure Determination. A total of 3,881 nuclear Overhauser effects were observed in the NOESY spectra, leading to 1,089 meaningful distance restraints and 362 angle restraints (Table 1). These structural restraints were used as an input for the structure calculation with the program cyana (22) followed by restrained energy minimization using the program insight. A total of 100 conformers were initially generated by cyana, and the bundle of 20 conformers with the lowest target function was used to represent the 3D NMR structure. The small residual constraint violations in the 20 refined conformers and the good coincidence of experimental nuclear Overhauser effects and short interatomic distances (data not shown) show that the input data represent a self-consistent set and that the restraints are well satisfied in the calculated conformers (Table 1). The deviations from ideal geometry are minimal, and similar energy values were obtained for all 20 conformers. The quality of the structures determined is reflected by the small backbone rms deviation values relative to the mean coordinates of residues 58–83 and 99–113 of ≈0.8 Å (see Table 1 and Fig. 1B). The bundle of 20 conformers representing the NMR structure has been deposited in the Protein Data Bank database with ID codes 1U34.
Table 1. Parameters characterizing the NMR structure of ECD1–CRF-R2β.
| No. of distance constraints | 1,089 |
| No. of dihedral angle constraints | 362 |
| Average upper limit distance constraint violations, Å | 1.88 ± 0.94 |
| Average dihedral angle constraint violations | 11.8 ± 10.3 |
| Intraprotein energy after minimization, * kcal·mol-1 | -2,092.3 ± 49.8 |
| Coordinate precision, Å, residues 58-83, 99-113 | |
| rms deviation to the mean for N, C, and C′ | 0.81 ± 0.20 |
| rms deviation to the mean for all the heavy atoms | 1.30 ± 0.25 |
| Structural quality, Ramachandran plot,† % | |
| In most favored region | 54.9 ± 2.57 |
| In the allowed region | 31.9 ± 2.29 |
| In the additionally allowed region | 7.4 ± 1.62 |
| In the disallowed region | 4.8 ± 1.09 |
The parameters are given for an ensemble of 20 lowest-energy conformers (of 100 structures calculated). None of these final structures exhibit nuclear Overhauser effect-derived violations >0.2 Å or dihedral angle restraint violations >5°.
The cyana structures were parameterized with the cff91 force field. The minimizations were conducted in vacuum by using conjugate gradients to a maximum derivative of 1.0 kcal/molA2 with discover.
Structure quality was analyzed by using procheck. Most of the angles in the disallowed region are in the disordered region.
Fig. 1.
The 3D structure of ECD1–CRF-R2β.(A) A ribbon diagram of the lowest energy conformer highlighting the β-sheets in cyan and the disulfide bonds in yellow. (B) Superposition of 20 conformers representing the 3D NMR structure. Only amino acid residues 44–119 are shown. The bundle is obtained by superimposing the backbone Cα carbons of residues 58–83 and 99–113. The program molmol was used to generate the figures, and in Figs. 2, 3, 4 the conformer with the lowest cyana target function is used to represent the 3D structure of ECD1–CRF-R2β.
Chemical-Shift Perturbation Experiments. [15N,1H]-HMQC experiments of 0.05 mM ECD1–CRF-R2β in 10 mM BisTris(HCl)/95% H2O/5% D2O at pH 5 were measured in the absence and presence of an equimolar concentration of either astressin or CRF. Backbone assignment at pH 5 has been achieved after pH-dependent chemical-shift changes of the cross-peaks in a series of [15N,1H]-HMQC experiments measured at pH 7, 6.5, 5.5, and 5. The assignment was then verified by the measurement of a HNCA experiment at pH 5.
Results
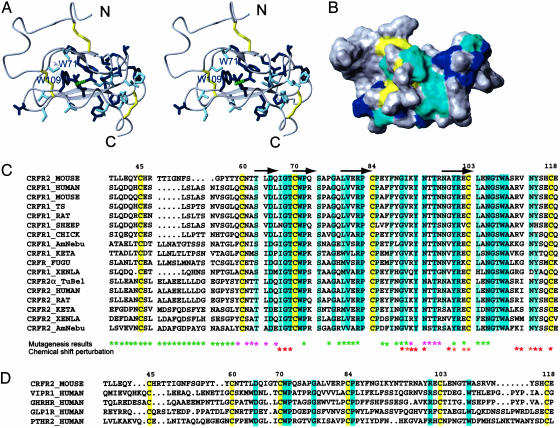
Description of the Structure. The NMR structure of 13C,15N-labeled ECD1–CRF-R2β has been determined by using triple-resonance experiments for the backbone assignment and NOESY experiments for the distance restraints (see Materials and Methods and Table 1). The NMR structure of ECD1–CRF-R2β contains two antiparallel β-sheet regions comprising residues 63–64 (β1 strand), 70–71 (β2 strand), 79–82 (β3 strand), and 99–102 (β4 strand) (Fig. 1 A). The polypeptide fold is stabilized by three disulfide bonds between residues Cys-45–Cys-70, Cys-60–Cys-103, and Cys-84–Cys-118 (16) and by a central core consisting of a salt bridge involving Asp-65–Arg-101, sandwiched between the aromatic rings of Trp-71 and Trp-109 (Fig. 2A). The two β-sheets, interconnected by this core, form the scaffold flanked by two disordered regions (residues 39–58 and 84–98). Furthermore, the core is surrounded by a second layer of highly conserved residues, Thr-69, Val-80, and Arg-82, and conservatively conserved residues, Thr-63, Ser-74, and Ile-67 (dark and light blue residues in Fig. 2C). The other conserved residues include Pro-72 and Pro-83, which are presumably important for ending the β-strands, as well as Gly-77, Asn-106, and Gly-107 located in the hinge regions of the two β-sheets, probably important for their relative orientation. Another cluster of conserved residues is present in the disordered loop between strands β3 and β4 (Gly-92, Phe-93, Asn-94, and Thr-96). In contrast, the disordered loop from residues 39–58 is highly variable in amino acid sequence.
Fig. 2.
Mapping the conserved amino acids onto the 3D structure of ECD1–CRF-R2β. Stereoview (A) and surface view (B) of the 3D structure with side chains of the conserved amino acids within the B1 family of GPCRs colored dark blue and similar residues colored light blue. The salt bridge between Asp-65 and Arg-101 is shown by the green dashed line. (C) Sequence alignment of the ECD1 of the CRF-R family. (D) Sequence alignment of the ECD1 of the B1 GPCR family. Only a representative set of sequences is shown. Highlighted in yellow are conserved cysteines, conserved amino acids throughout the whole B1 family are blue, and amino acids conserved >80% throughout the whole B1 family are light blue. Mutagenesis studies (references in Fig. 5) for the identification of receptor–ligand interaction are summarized here: Magenta stars represent amino acid segments proposed to be involved in hormone binding, and green stars represent amino acid segments that are less important for binding (note: these studies indicate that only some, but not necessarily all, of the indicated amino acid residues are involved in binding). Amino acids identified to be involved in hormone binding from chemical-shift perturbation studies are designated by red and orange (see Fig. 3 for more details). The β-sheet secondary structure elements are labeled by an arrow above the sequence. TS, tree shrew; AmNebu, Ameriurus Nebulosus; TuBel, Tupaia Belangeri; VIPRI, vasoactive intestinal peptide receptor 1; GHRH, growth hormone releasing hormone receptor; GLPIR, glucagon-like peptide 1 receptor; PTHR2, parathyroid hormone receptor2.
The structure of ECD1–CRF-R2β is identified as a short consensus repeat (SCR) commonly found in proteins of the complement system (23, 24), including the first SCR module of the human β2-glycoprotein (25) (PDB ID code 1C1Z), the closest structure found by the dali server (26). Among GPCRs, the SCR domain has been predicted to occur in the N-terminal domain of only the γ-aminobutyric acid receptor (27).
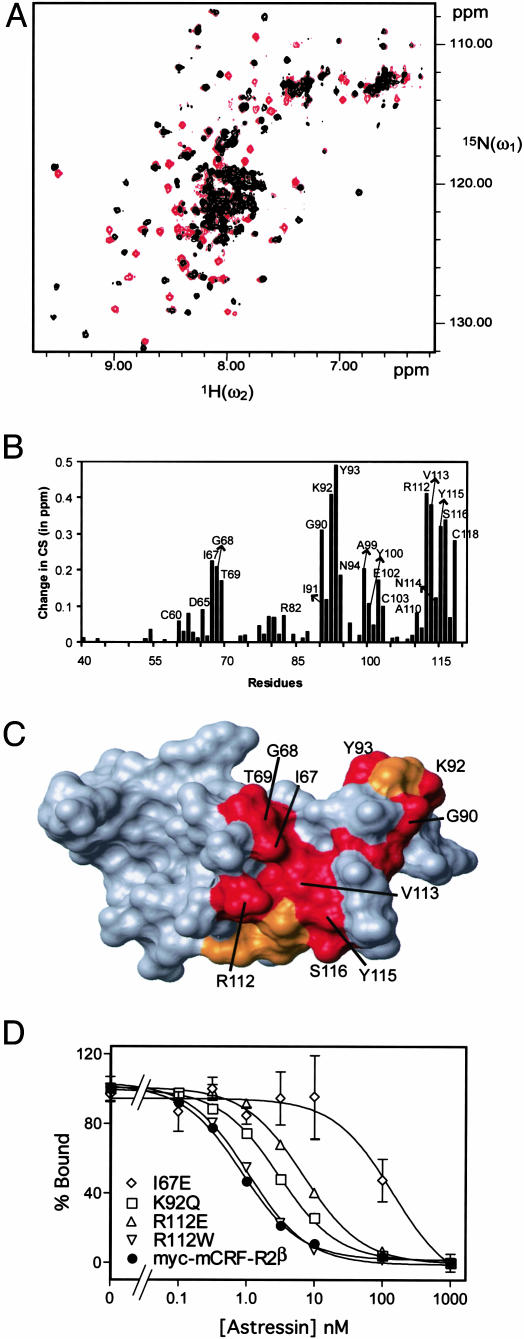
Peptide Hormone Binding Site. To obtain detailed structural insights about the binding interface, we studied the interaction between the potent peptide antagonist astressin (20) and ECD1–CRF-R2β by using NMR chemical-shift perturbation experiments (28). Fig. 3A shows the HMQC spectra of 15N-labeled ECD1–CRF-R2β in the absence and presence of equimolar astressin. The largest chemical-shift perturbations are observed in the segments comprising residues 67–69, 90–93, 102–103, and 112–116 (Fig. 3B and Fig. 5, which is published as supporting information on the PNAS web site). These residues are clustered in the cleft region between the tip of the first β-sheet and the edge of the “palm” of the second β-sheet (Fig. 3C). The observed changes in the chemical shifts in the disordered loop region 85–98 are indicative of a folding after ligand binding. This interpretation is supported by CD data that revealed a conformational change toward a more structured ECD1–CRF-R2β upon ligand binding (16). A structure–evolution approach that assumes the conservation of the ligand–receptor interface within the CRF-R family and that concomitantly screens the surface of ECD1 for patches with conserved and similar amino acids highlights the same surface region (Fig. 2B). Furthermore, studies of mutant CRF-R2β, which show reduced binding affinities, serve to confirm the integrity of the binding site in the full-length receptor. The mutation R112E in myc–mCRF-R2β results in a ≈7-fold decrease in the affinity for astressin. The inhibitory binding constants, Kis, are: 7.2 (6.3–8.3) nM for myc–mCRF-R2β(R112E) compared to 1.1 (0.8–1.5) nM for myc–mCRF-R2β. Introducing the mutation, I67E, results in a larger decrease in the affinity for astressin: Ki = 128 (85–191) nM (Fig. 3D). The I67E mutation also reduces (by ≈3-fold) the affinity for agonist sauvagine (data not shown). Mutations of residues T69 or N114, which show only small chemical-shift perturbations upon binding to astressin, do not significantly influence the binding affinities. These data suggest that these small chemical-shift perturbations are an indirect effect of binding. Mutagenesis studies of CRF receptors reported (18, 19, 29) are also consistent with the proposed interaction surface (Figs. 2C and 5).
Fig. 3.
Identification of the binding site of astressin on the 3D structure of ECD1–CRF-R2β.(A)2D[15N,1H]-HMQC spectra of ECD1–CRF-R2β in the absence (red contours) and presence (black contours) of astressin. (B) Plot of the normalized chemical-shift changes [Δ(δ(1H))2 +Δ(δ(15N))2/5]1/2 observed in the complex versus the amino acid sequence (28). (C) Surface representation of ECD1–CRF-R2β showing the amino acids involved in binding with astressin. Residues with large chemical-shift perturbations (>0.2 ppm) are colored red and residues with chemical-shift perturbations between 0.1 and 0.2 ppm are colored orange. (D) Competitive displacement (see Materials and Methods) by astressin of [125I-DTyr0]-astressin bound to membranes from COSM6 cells transiently expressing myc–mCRF-R2β (•); myc–mCRF-R2β(K92Q) (□); myc–mCRF-R2β(R112E) (▵); myc-mCRF-R2β(I67E) (♦); and myc-mCRF-R2β(R112W) (▿). One representative experiment, repeated at least twice, is shown.
The hormone binding site, identified in this study, also provides a structural basis for explaining the binding specificity of ligands. CRF-R2β binds with high affinity to Ucn 1, Ucn 2, Ucn 3, and the antagonist astressin, but with lower affinity to CRF. On the other hand, CRF binds to CRF-R1 with higher affinity than does Ucn 2 or Ucn 3. These different binding specificities of CRF receptors are explained by the presence of different amino acids in the binding pocket (Figs. 2 and 3). For example, the point mutations R112W and K92Q, replacing the residues R112 or K92 in mCRF-R2β with the residues found in xCRF-R1, result in a 2- to 3-fold lower binding affinity for astressin: Ki = 1.9 (1.0–3.4) nM for myc–mCRF-R2β(R112W) and Ki = 3.1 (2.4–3.9) nM for myc–mCRF-R2β(K92Q). To ensure the conservation of the proposed binding site for different ligands, the chemical-shift perturbation experiment was also performed with CRF. In the presence of CRF, the same cross-peaks of ECD1–CRF-R2β were affected as were influenced by astressin. However, instead of a chemical-shift change, the cross-peaks were broadened beyond detection, probably because of slow conformational exchange induced by the low binding affinity of CRF (data not shown).
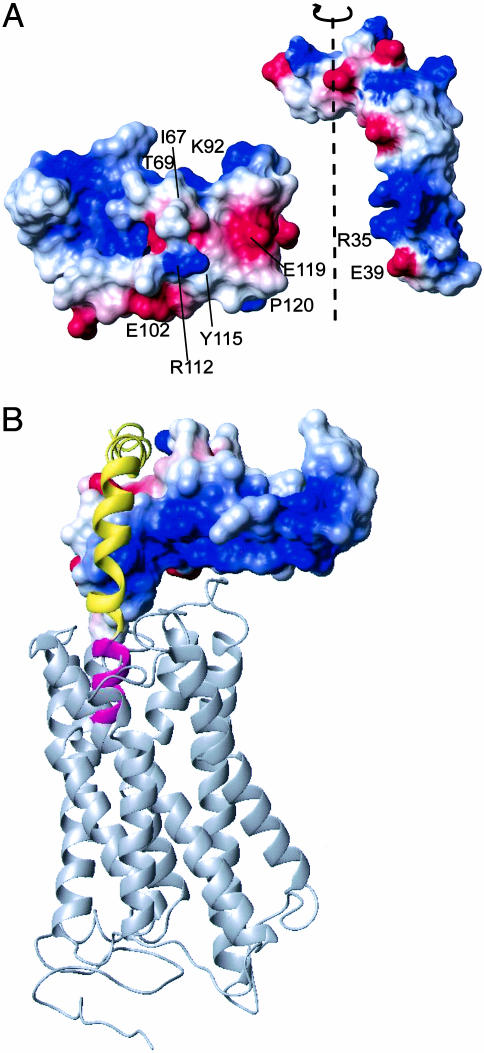
Model for Receptor Activation. The surface potential of the 3D structure provides an insight into receptor activation mechanism. An accumulated distribution of positive charges on the “back side” of the structure displayed in Figs. 1, 2, 3 (Arg-47, Arg-82, Arg-97) suggests its orientation toward the negatively charged ECDs 2–4 and the transmembrane segment of CRF-R2β (Fig. 4B). This orientation is further supported by the observation that amino acid replacements between CRF-R2 and CRF-R1 with negative charges on the back side of the ECD1 have their counterpart in replacements with positive charges in ECDs 2–4. Based on the relative orientation of ECD1 and the 3D structure of ECD1–CRF-R2β and astressin B (Fig. 4), we propose that hormone binding and receptor activation occur in two steps. First, the ligand binds with its C-terminal segment to the solvent exposed binding site of ECD1 (30). Second, for an agonist ligand, the N-terminal segment, known to be important for signaling, penetrates into the transmembrane segment of the receptor, producing activation of the receptor (Fig. 4B). In contrast, the peptide antagonist astressin lacks the first 11 N-terminal residues and, hence, is unable to penetrate the transmembrane and other ECDs of the receptor and thus, fails to activate it. An important prerequisite for this two-step mechanism is the observed kink in the peptide ligand astressin (Fig. 4) (C.R.R.G., J.E.R. and R.R., unpublished work).
Fig. 4.
The two-step model for hormone binding and receptor activation. (A) The surface presentation of ECD1–CRF-R2β and astressin B (unpublished data) with the electrostatic potential of both molecules. Color code is blue for positive charges, red for negative charges, and white for neutral surface. The proposed binding interface between ECD1–CRF-R2β and the ligand is indicated in an opened view. Proposed electrostatic interactions include Glu-39–Arg-112 (ECD1) and Arg-35–Glu-119 (ECD1), as well as hydrophobic interactions (Leu-37, Ile-41 of the ligand with Tyr-115 and Pro-120 of ECD1). (B) Schematic of the hormone binding in the full-length receptor. Shown as a yellow and pink helix is the peptide hormone structure containing a kink at residue ≈24. The N-terminal segment important for receptor activation and signaling (30) is shown in pink. The positively charged surface of the ECD1 is facing the transmembrane segment. The transmembrane segment of the receptor (gray) is modeled by using the rhodopsin structure (Protein Data Bank ID code 1HZX). Orientation is rotated relative to the standard orientation of A by 90° and 180° along the vertical axes and horizontal axes, respectively.
Conservation of SCR in B1 GPCR. The B1 receptors are encoded by 15 genes in humans; the ligands for these receptors are polypeptide hormones of 27-to 141-aa residues. A structure-based analysis of the amino acid sequences of this receptor subfamily suggests that the SCR fold of the ECD1 domain must be conserved in all of the B1 family receptors (Fig. 2D). This prediction is based on (i) the conserved disulfide bonds and their identical arrangement in the ECD1s of CRF-R1, CRF-R2β, PTHR, and GLP-1R (16, 31, 32), and (ii) the conserved salt bridge (Asp-65 and Arg-101) surrounded by the two conserved tryptophan residues (Trp-71 and Trp-109), which have been identified as the key residues in the core of ECD1. Additionally, two prolines, which have been proposed to be crucial for ending the β-sheet (Pro-72, Pro-84), and Gly-77, are also conserved in the receptor subfamily.
Initial analysis of the 3D structure of ECD1 provides an explanation for the profound effect of the Asp-60–Gly mutation (position 65 in CRF-R2β) in another member of this family, namely, the mouse growth hormone-releasing factor (GRF) receptor (33). This mutant GRF receptor is impaired in its ability to bind and transduce the GRF-induced cAMP response, with the physiological consequences of a hypoplastic pituitary and a dwarf (little) phenotype (33). This mutation in the SCR motif would prevent the formation of the structurally important core salt bridge, thereby hindering the correct folding of ECD1 and concomitantly high-affinity ligand binding.
Discussion
The structure for the ECD1 of the CRF-R2β that is presented provides a further structural basis for the concept of the modular nature of GPCRs. All GPCRs consist of seven transmembrane helices but differ extensively in their ECDs. ECD1 of the rhodopsins is very small and contains an antiparallel β-sheet as secondary structural motif (11). The Methuselah ECD1 consists of three β-sheet-rich domains that are interconnected by disulfide bonds (12); the two-domain structure of the extracellular ligand-binding region of the metabotropic glutamate receptor forms a homodimer covalently linked by disulfide bonds (34, 35). The ECD1 of the B1 subfamily of GPCRs is now found to be an SCR module and is further identified as the major site of peptide hormone interaction.
Because the SCR domains are often involved in protein–protein interactions, the structure also raises the possibility of receptor–receptor interactions between this family of peptide hormone receptors and other receptors. For example, in the complement system, the interaction of CD55 with CD97 involves the SCR and epidermal growth factor (EGF) modules of each of the proteins, respectively (24). This observation suggests possible receptor–receptor interactions between the B1 subfamily of GPCRs and EGF-like receptors.
The ECD1–CRF-R2β structure not only gives insight into the modular nature of ligand–receptor and receptor–receptor interactions, but also provides a framework for the proposed two-step activation mechanism of this family of receptors.
Supplementary Material
Acknowledgments
We thank R. Kaiser, J. Vaughan, and Drs. J. Gulyas, N. Justice, and S. Koerber for suggestions and assistance. R.R. is a Pew Scholar. This research was supported in part by the Foundation for Research, the Robert J. Jr. and Helen C. Kleberg Foundation, and National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Diseases Grant DK26741 W.W.V. is a Foundation for Research Senior Investigator.
Abbreviations: CRF, corticotropin-releasing factor; mCRF-R2β, mouse CRF receptor 2β; ECD, extracellular domain; SCR, short consensus repeat; GPCR, G protein-coupled receptor; Ucn, urocortin; HMQC, heteronuclear multiple quantum correlation.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 1U34).
References
- 1.Vale, W., Spiess, J., Rivier, C. & Rivier, J. (1981) Science 213, 1394–1397. [DOI] [PubMed] [Google Scholar]
- 2.Vaughan, J., Donaldson, C., Bittencourt, J., Perrin, M. H., Lewis, K., Sutton, S., Chan, R., Turnbull, A. V., Lovejoy, D., Rivier, C., et al. (1995) Nature 378, 287–292. [DOI] [PubMed] [Google Scholar]
- 3.Reyes, T. M., Lewis, K., Perrin, M. H., Kunitake, K. S., Vaughan, J., Arias, C. A., Hogenesch, J. B., Gulyas, J., Rivier, J., Vale, W. W. & Sawchenko, P. E. (2001) Proc. Natl. Acad. Sci. USA 98, 2843–2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lewis, K., Li, C., Perrin, M. H., Blount, A., Kunitake, K., Donaldson, C., Vaughan, J., Reyes, T. M., Gulyas, J., Fischer, W., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 7570–7575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hsu, S. Y. & Hsueh, A. J. (2001) Nat. Med. 7, 605–611. [DOI] [PubMed] [Google Scholar]
- 6.Perrin, M. H. & Vale, W. W. (2002) in Understanding G Protein-Coupled Receptors and Their Role in the CNS, eds. Pangalos, M. N. & Davies, C. H. (Oxford Univ. Press, New York), pp. 505–545.
- 7.Dautzenberg, F. M. & Hauger, R. L. (2002) Trends Pharmacol. Sci. 23, 71–77. [DOI] [PubMed] [Google Scholar]
- 8.Bale, T. L. & Vale, W. W. (2004) Annu. Rev. Pharmacol. Toxicol. 44, 525–557. [DOI] [PubMed] [Google Scholar]
- 9.Brar, B. K., Jonassen, A. K., Stephanou, A., Santilli, G., Railson, J., Knight, R. A., Yellon, D. M. & Latchman, D. S. (2000) J. Biol. Chem. 275, 8508–8514. [DOI] [PubMed] [Google Scholar]
- 10.Li, C., Chen, P., Vaughan, J., Blount, A., Chen, A., Jamieson, P. M., Rivier, J., Smith, M. S. & Vale, W. (2003) Endocrinology 144, 3216–3224. [DOI] [PubMed] [Google Scholar]
- 11.Palczewski, K., Kumasaka, T., Hori, T., Behnke, C. A., Motoshima, H., Fox, B. A., Le Trong, I., Teller, D. C., Okada, T., Stenkamp, R. E., et al. (2000) Science 289, 739–745. [DOI] [PubMed] [Google Scholar]
- 12.West, A. P., Jr., Llamas, L. L., Snow, P. M., Benzer, S. & Bjorkman, P. J. (2001) Proc. Natl. Acad. Sci. USA 98, 3744–3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perrin, M. H., Sutton, S., Bain, D. B., Berggren, T. W. & Vale, W. W. (1998) Endocrinology 139, 566–570. [DOI] [PubMed] [Google Scholar]
- 14.Perrin, M. H., Fischer, W. H., Kunitake, K. S., Craig, A. G., Koerber, S. C., Cervini, L. A., Rivier, J. E., Groppe, J. C., Greenwald, J., Moller Nielsen, S. & Vale, W. W. (2001) J. Biol. Chem. 276, 31528–31534. [DOI] [PubMed] [Google Scholar]
- 15.Hofmann, B. A., Sydow, S., Jahn, O., Van Werven, L., Liepold, T., Eckart, K. & Spiess, J. (2001) Protein Sci. 10, 2050–2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perrin, M. H., DiGruccio, M. R., Koerber, S. C., Rivier, J. E., Kunitake, K. S., Bain, D. L., Fischer, W. H. & Vale, W. W. (2003) J. Biol. Chem. 278, 15595–15600. [DOI] [PubMed] [Google Scholar]
- 17.Assil, I. Q., Qi, L. J., Arai, M., Shomali, M. & Abou-Samra, A. B. (2001) Biochemistry 40, 1187–1195. [DOI] [PubMed] [Google Scholar]
- 18.Dautzenberg, F. M., Higelin, J., Brauns, O., Butscha, B. & Hauger, R. L. (2002) Mol. Pharmacol. 61, 1132–1139. [DOI] [PubMed] [Google Scholar]
- 19.Dautzenberg, F. M. & Wille, S. (2004) Regul. Pept. 118, 165–173. [DOI] [PubMed] [Google Scholar]
- 20.Perrin, M. H., Sutton, S. W., Cervini, L. A., Rivier, J. E. & Vale, W. W. (1999) J. Pharmacol. Exp. Ther. 288, 729–734. [PubMed] [Google Scholar]
- 21.Cavanagh, J., Fairbrother, W. J., Palme, A. G. I. & Skelton, N. J. (1996) Protein NMR Spectroscopy, Principles, and Practice (Academic, New York).
- 22.Guntert, P., Mumenthaler, C. & Wuthrich, K. (1997) J. Mol. Biol. 273, 283–298. [DOI] [PubMed] [Google Scholar]
- 23.Bork, P., Downing, A. K., Kieffer, B. & Campbell, I. D. (1996) Q. Rev. Biophys. 29, 119–167. [DOI] [PubMed] [Google Scholar]
- 24.Lin, H. H., Stacey, M., Saxby, C., Knott, V., Chaudhry, Y., Evans, D., Gordon, S., McKnight, A. J., Handford, P. & Lea, S. (2001) J. Biol. Chem. 276, 24160–24169. [DOI] [PubMed] [Google Scholar]
- 25.Schwarzenbacher, R., Zeth, K., Diederichs, K., Gries, A., Kostner, G. M., Laggner, P. & Prassl, R. (1999) EMBO J. 18, 6228–6239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holm, L. & Sander, C. (1994) Proteins 19, 165–173. [DOI] [PubMed] [Google Scholar]
- 27.Hawrot, E., Xiao, Y., Shi, Q. L., Norman, D., Kirkitadze, M. & Barlow, P. N. (1998) FEBS Lett. 432, 103–108. [DOI] [PubMed] [Google Scholar]
- 28.Grzesiek, S., Bax, A., Clore, G. M., Gronenborn, A. M., Hu, J. S., Kaufman, J., Palmer, I., Stahl, S. J. & Wingfield, P. T. (1996) Nat. Struct. Biol. 3, 340–345. [DOI] [PubMed] [Google Scholar]
- 29.Wille, S., Sydow, S., Palchaudhuri, M. R., Spiess, J. & Dautzenberg, F. M. (1999) J. Neurochem. 72, 388–395. [DOI] [PubMed] [Google Scholar]
- 30.Nielsen, S. M., Nielsen, L. Z., Hjorth, S. A., Perrin, M. H. & Vale, W. W. (2000) Proc. Natl. Acad. Sci. USA 97, 10277–10281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grauschopf, U., Lilie, H., Honold, K., Wozny, M., Reusch, D., Esswein, A., Schafer, W., Rucknagel, K. P. & Rudolph, R. (2000) Biochemistry 39, 8878–8887. [DOI] [PubMed] [Google Scholar]
- 32.Bazarsuren, A., Grauschopf, U., Wozny, M., Reusch, D., Hoffmann, E., Schaefer, W., Panzner, S. & Rudolph, R. (2002) Biophys. Chem. 96, 305–318. [DOI] [PubMed] [Google Scholar]
- 33.Lin, S.-C., Lin, C. R., Gukovsky, I., Lusis, A. J., Sawchenko, P. E. & Rosenfeld, M. G. (1993) Nature 364, 208–213. [DOI] [PubMed] [Google Scholar]
- 34.Tsuchiya, D., Kunishima, N., Kamiya, N., Jingami, H. & Morikawa, K. (2002) Proc. Natl. Acad. Sci. USA 99, 2660–2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kunishima, N., Shimada, Y., Tsuji, Y., Sato, T., Yamamoto, M., Kumasaka, T., Nakanishi, S., Jingami, H. & Morikawa, K. (2000) Nature 407, 971–977. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.