Abstract
A pH-dependent transition between delocalized and trapped mixed valence states of an engineered CuA center in azurin has been investigated by UV-visible absorption and electron paramagnetic resonance spectroscopic techniques. At pH 7.0, the CuA azurin displays a typical delocalized mixed valence dinuclear [Cu(1.5)····Cu(1.5)] spectra with optical absorptions at 485, 530, and 760 nm, and with a seven-line EPR hyperfine. Upon lowering of the pH from 7.0 to 4.0, the absorption at 760 nm shifted to lower energy toward 810 nm, and a four-line EPR hyperfine, typical of a trapped valence, was observed. The pH-dependent transition is reversible because increasing the pH restores all delocalized spectral features. Lowering the pH resulted in not only a trapped valence state, but also a dramatically increased reduction potential of the Cu center (from 160 mV to 340 mV). Mutation of the titratable residues around the metal-binding site ruled out Glu-114 and identified the C-terminal histidine ligand (His-120) as a site of protonation, because the His120Ala mutation abolished the above pH-dependent transition. The corresponding histidine in cytochrome c oxidases is along a major electron transfer pathway from CuA center to heme a. Because the protonation of this histidine can result in an increased reduction potential that will prevent electron flow from the CuA to heme a, the CuA and the histidine may play an important role in regulating proton-coupled electron transfer.
Dioxygen reduction catalyzed by cytochrome c oxidase (CcO) is one of the most important biological reactions to sustain life in aerobic organisms (1–3). The reduction results in a proton gradient, which in turns drives the synthesis of ATP, the energy source for many cellular processes. Three redox active metal centers, CuA, heme a, and heme a3-CuB, are involved in the reaction. Coupling electron transfer with proton transfer is the most critical step in this reaction.
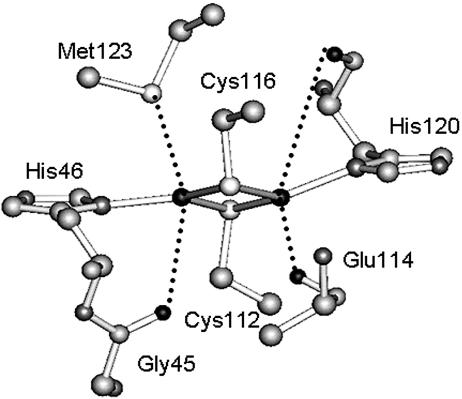
As an electron entry site for CcO, the CuA center contains a fully delocalized (class III) mixed valence (4) dinuclear [Cu(1.5)····Cu(1.5)] center bridged by two μ2 SCys thiolates (Fig. 1) (5–9). The distance between the two copper ions is short (≈2.4 Å) (5, 10–14), suggesting a weak Cu—Cu bond (6). Each copper is also coordinated by a Nδ(His), with weak axial ligand interactions approximately perpendicular to the plane defined by the Cu2(SCys)2 core.
Fig. 1.
The crystal structure of the CuA site in engineered CuA azurin (12).
Although dinuclear or multinuclear mixed valence copper complexes with the unpaired electron completely localized (class I) or partially delocalized (class II) were known for many years (15, 16), copper complexes with fully delocalized class III mixed valence states, such as in CuA center, are rare (17–24). Even rarer is the reversible conversion between different classes of compounds with delocalized and trapped valence states (25, 26). Studying these compounds and their valence state conversions can contribute significantly to the field of inorganic chemistry.
Herein, we report the first example of a reversible transition between delocalized and trapped mixed valence states of a dinuclear CuA center in azurin, triggered solely by pH. We have further identified the C-terminal histidine ligand to the CuA center as a site of protonation through site-directed mutagenesis. Because this histidine is directly in the pathway of electron transfer between the CuA center and the heme a center, and because the reduction potentials of the two states are dramatically different, this histidine may play a significant role in regulating proton-coupled electron transfer and thus in controlling CcO activity.
Materials and Methods
CuA azurin and its His-120 mutants were expressed in BLR(DE3) Escherichia coli and purified as reported (27, 28). A Glu114Gln mutant was made on the CuA azurin template by using the QuikChange site-directed mutagenesis procedure (Stratagene).
UV-visible (UV-vis) absorption spectra were obtained at 4°C on a Varian Cary 3E spectrophotometer. A mixed buffer (50 mM sodium acetate/50 mM Mes/50 mM Mops/50 mM Tris) was used for pH titration.
X-band (9.05 GHz) EPR spectra were collected on a Bruker (Billerica, MA) ESP 300. The protein samples (1 mM) were prepared in mixed buffer of desired pH with 50% glycerol as a glassing agent. Conditions are listed in the figure legends.
Electrochemical experiments were performed as described (29) using a CH Instruments (Austin, TX) Model 620A Electrochemical Analyzer. The pyrolytic graphite edge (PGE) electrode (typical electrode area = 3.6 mm2) was assembled according to the procedure described in the literature (30). The PGE electrode was treated with 10 μl of 100 mM chloroform solution of didodecyldimethylammonium bromide (DDAB) as described (29). Cyclic voltammetric experiments were carried out at 4°C with a normal three-electrode configuration consisting of a DDAB film-PGE electrode as a working electrode, an Ag/AgCl (Bioanalytical Systems, West Lafayette, IN; 3M NaCl) electrode as a reference electrode, and a platinum-wire auxiliary electrode (CH Instruments). The electrode voltage was cycled between –0.5 and +0.6 V vs. Ag/AgCl electrode with a scan rate of 500 mV/sina0.1–0.2 mM protein solution until the signal appeared. The electrode was then rinsed and transferred to a buffer solution of desired pH and allowed to equilibrate for 2 min before measurement. The reduction potential of the Ag/AgCl (3M NaCl) electrode was calibrated against the standard potential of a saturated calomel electrode (E = 0.253 V at 4°C) (31). The resulting potential for Ag/AgCl (3 M NaCl) of 221 mV (4°C) was added to obtain the reduction potential vs. normal hydrogen electrode (NHE). All reduction potentials in the text are reported as corrected values vs. NHE.
Results
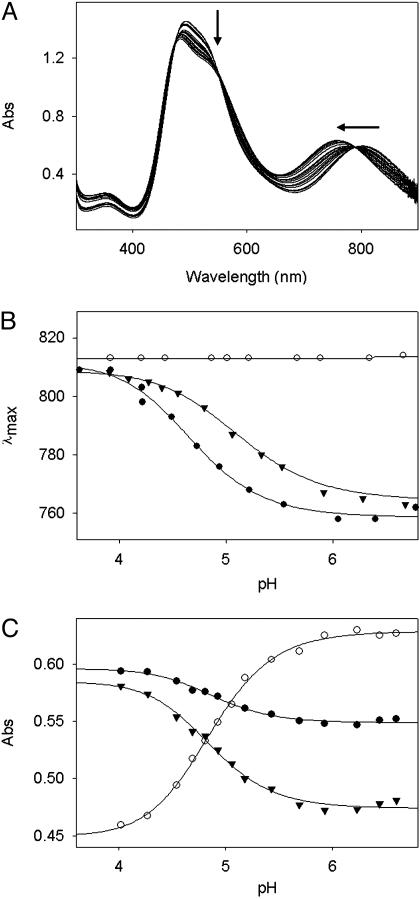
The UV-vis spectral changes of the CuA azurin with respect to pH are shown in Fig. 2A. The peak wavelength of the optical band around 800 nm was found to be shifted by 50 nm toward a shorter wavelength with increasing the pH from 4 to 7 (Fig. 2B). An isosbestic point at 789 nm suggests that two species are involved in this transition. A simple one-site protonation/deprotonation equilibrium model gives a good fit with the experimental data with a pKa value of 4.8 ± 0.1 (Fig. 2C). This pH-dependent transition is reversible because decreasing pH from 7 to 4 restores the original spectrum observed at pH 4.
Fig. 2.
pH-dependent UV-vis studies of CuA azurin and its variants. (A) Changes in the UV-vis spectrum of CuA azurin in the pH range of 4.1–6.8. Arrows indicate the direction of increasing pH. (B) Changes in the near-IR peak maxima of CuA azurin (•), Glu114Gln (▾), and His120Ala variants (○). (C) Changes in the peak intensities at 750 nm (○), 800 nm ((•), and 820 nm (▾). The solid lines represent the best fit of the experimental data.
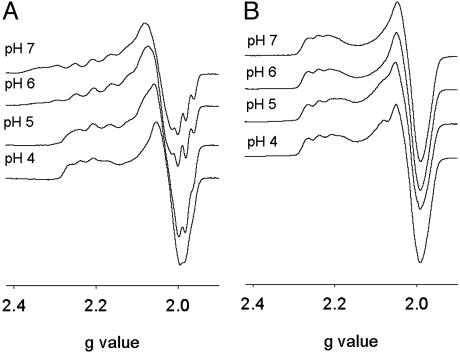
To obtain further insight into the states of the two species in the pH titration, EPR spectra at various pH were collected (Fig. 3A). At pH 7, a seven-line hyperfine pattern, with a hyperfine coupling constant of 60 × 10–4 cm–1, was observed in the g∥ region. As demonstrated and independently verified by multi-frequency EPR (28), the seven-line hyperfine is indicative of delocalization of one unpaired electron over two equivalent copper atoms, typical of a class III, delocalized mixed valence species. However, as the pH is lowered from pH 7, the seven-line structure is slowly merged into narrowly spaced lines. At pH 4, only a four-line hyperfine structure, with a hyperfine coupling constant of 42 × 10–4 cm–1, typical for type 1 copper centers, was observed. These results strongly suggest that the delocalized mixed valence dinuclear CuA center can be transformed to a trapped valence state by lowering the pH, and this transition is reversible. Increasing the pH from 7 to 10 resulted in a type 2 copper center (see Fig. 7, which is published as supporting information on the PNAS web site).
Fig. 3.
X-band EPR spectra of CuA azurin (A) and His120Ala mutant (B) at various pH (protein concentration 1 mM, microwave power 2 mW, modulation amplitude 4 G, and temperature 15 K).
Close inspection of the primary and secondary coordination spheres of the CuA site suggests that only Glu-114 and two His residues (His-46 and His-120) can be ionized in the pH range examined. Mutation of Glu-114 to Gln resulted in a similar pH dependence UV-vis absorption profile as the CuA azurin (Fig. 2B). However, mutations of His-120 to Asn, Asp, and Ala resulted in a totally different profile because the peak positions stay the same in the pH range of 4–7 (Fig. 2B). These results indicate that His-120 is the ionizable group responsible for the spectral changes observed. Further support of the above conclusion comes from EPR studies. The EPR spectra of His120Ala show the same four-line hyperfine feature through the pH range of 4–7 (Fig. 3B).
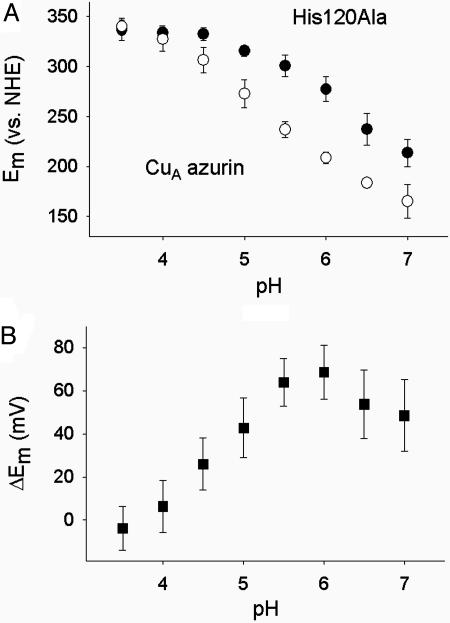
The reduction potential of CuA azurin and the His120Ala mutant were measured by cyclic voltammetry as a function of pH employing the didodecyldimethylammonium bromide film-pyrolytic graphite edge electrode method (Fig. 4) (29). At pH 5, the reduction potential of CuA azurin (270 mV) is very similar to those of native CuA centers reported previously (32, 33). However, the reduction potential increased dramatically with lowering pH. The reduction potential at pH 4 with a trapped valence state is ≈180 mV higher than that at pH 7 with its delocalized mixed valence state. The reduction potential of His120Ala mutant is similar to that of CuA azurin at low pH. As the pH increases, a decrease in the reduction potential was observed as in CuA azurin. However, the total decrease amounts to ≈110 mV in the case of His120Ala.
Fig. 4.
pH-dependent cyclic voltammetry studies of CuA azurin and its His120Ala variant. (A) Changes in the reduction potentials of CuA azurin (○) and His120Ala mutant ((•). (B) Changes in the differences of the reduction potentials (▾). ΔEm = Em (His120Ala) – Em (CuA azurin).
Discussion
Mixed valence compounds are an important class of inorganic complexes with promising applications such as in electron transfer (ET), magnetism, and catalysis. Mixed valence compounds have been classified into three groups, depending on whether an unpaired electron is completely localized (class I), partially delocalized (class II), or completely delocalized (class III) (4). The CuA center is one of a few examples of class III dinuclear copper complexes.
One of the most exciting phenomena in mixed valence coordination chemistry is the observation of reversible transitions between different classes of valence species. For example, one classic example is the temperature-dependent transition of mixed valence and trapped valence states in a dinuclear Cu compound (25). Another example is the regulation of the degree of electron delocalization in a dinuclear copper center in half-met state of hemocyanin by addition of halide ions (34). Now we have shown an example of reversible transition of mixed valence states in the CuA center in azurin triggered solely by pH. Before this work, several pH-dependent spectral changes of the CuA centers have been reported, most of which examine pH ranges from 6–10. For example, the pH-dependent spectral changes of soluble CuA domain from the Paracoccus denitrificans CcO have been examined in the pH range of 6–10 (32, 35). Significant changes in UV-vis absorption and circular dichroism were interpreted as the presence of two conformers: high-pH type 2 copper and low-pH delocalized mixed valence CuA center as evidenced by independent EPR studies (32). NMR studies on the CuA center from soluble domain of CcO from Paracoccus versutus indicate a decrease in the intensities of all paramagnetic signals in the pH range between 6.5 and 10. It was interpreted that the formation of an electron-localized species and slow electronic relaxation make the observation of the paramagnetic signal impossible (36).
The nature of valence localization observed in CuA azurin reported here at lower pH (from 4 to 6) is different from those described above at higher pH (from 6 to 10). Despite the significant change from delocalized mixed valence to trapped valence state as indicated by EPR (Fig. 3), the CuA center in azurin displays a very similar UV-vis spectrum in the two states, with only subtle differences (Fig. 2 A). Transformation of the CuA site into a type 2 copper at higher pH requires rearrangement of the Cu ligand set, and, as a result, it becomes a class I, localized mixed valence species, losing the optical feature of the purple CuA site. In contrast, protonation of His-120 in CuA azurin at lower pH does not entirely disturb the CuA site, and the major structure remains the same for the protonated species. Therefore, the valence-localized species in case of CuA azurin can be considered to be class II rather than class I mixed valence species, and the lower energy near IR peak observed at low pH can be assigned as an intervalence charge transfer band. Similar bands have been observed for Cu(I)Cu(II) class II mixed valence compounds in the visible/near IR region (37, 38). Although detailed spectroscopic analysis and assignments of the CuA center have been reported (39, 40), further detailed spectroscopic analysis is needed in the light of our finding that both delocalized and localized CuA centers display very similar UV-vis spectra.
The observation of a reversible pH-dependent transition between the delocalized and trapped valence states of the CuA center may be important not only in the fundamental understanding of coordination chemistry, but also in shedding new light on the biological function of the CuA center in CcO. Since the three-dimensional structure of the CcO was first elucidated by x-ray crystallography in 1995 (10, 41), the main research focus has been on the roles of the structural features in CcO activity, particularly on how the electrons and protons are transferred across this large complex and on how protons are coupled to ET. Located in subunit II of CcO, the CuA center mediates inter- and intramolecular ET by accepting electron from cytochrome c and passing it on to the heme a center. The electron is then further relayed to the catalytic heme a3-CuB site where 4 e– reduction of O2 to H2O occurs. As a terminal oxidase in the respiration chain, the driving forces for the ET reactions in CcO are not very large. For example, the driving force for ET is only ≈20 mV between cytochrome c and the CuA center (32, 33, 42), and 90 and 10 mV between the CuA center and heme a for oxidized and half-reduced enzyme, respectively (43, 44). Therefore any subtle structural changes that can influence the reduction potential of the CuA center could have a dramatic effect on the ET chains and thus the CcO function.
In this article, we have demonstrated that pH can exert not only subtle influences on the valence state of the CuA center, but also dramatic changes in reduction potentials. It is well known that the reduction potential of azurin is pH dependent due to the protonation/deprotonation equilibrium of the uncoordinated His-35 and His-83 residues (45, 46). Because no significant perturbation was observed in the position of His-35 and His-83 in CuA azurin upon introduction of the new metal-binding site by using loop-directed mutagenesis (12), protonation/deprotonation of His-35 and His-83 in CuA azurin should affect the reduction potential to a similar extent as in WT azurin. However, the difference in reduction potential (70 mV) of CuA azurin and His120Ala mutant at high pH (> 5.5) should purely represent the difference in reduction potential of the CuA center in the totally delocalized state (CuA with His-120 ligation) and in the valence-trapped state (His120Ala) in the same overall protein environment. This 70-mV increase is relatively large considering the small driving force of electron transfer from CuA center to heme a. Therefore, electron transfer from the CuA center with its disturbed electronic structure to heme a could be significantly hampered due to the unfavorable thermodynamic driving force.
Another important finding of this work is the identification of the His-120 ligand of CuA as the site of protonation that triggers the valence state change. As shown in Results, three titratable residues in the primary and secondary coordination spheres of the CuA site (Glu-114, His-46, and His-120) were investigated as possible candidates for a site of protonation. Glu-114 was ruled out because Glu114Gln mutation resulted in a similar pH profile. We cannot completely rule out His-46 as a possible site of protonation because proteins with mutation of His-46 cannot be expressed, probably because of loss of protein stability. However, our findings that mutations of His-120 to Asn, Asp, and Ala completely abolish the pH-dependent behavior strongly suggest that pH-dependent behavior is originated from His-120 protonation. This result is expected because this C-terminal histidine is closer to the surface of the protein in CuA azurin, and closer to the interface of subunit II and subunit I in the case of native CcO.
It seems that protonation of His-120 has disturbed the delicate balance between the two equivalent copper atoms and possibly makes the copper right next to the His-120 favor the reduced state as in other blue copper proteins. In blue copper proteins such as WT azurin, the C-terminal His is also at a similar location (47). Protonation of the C-terminal His ligand has been shown to occur in the reduced state of selected blue copper proteins such as plastocyanin, amicyanin, and pseudoazurin (48–52). However, no evidence for protonation has been found in the reduced state of other blue copper proteins such as azurin (53). Neither has observation been made on protonation of the oxidized states of any blue copper proteins. Therefore, the CuA site in azurin must have altered the properties of the C-terminal His ligand, making it titratable. Further structural analysis is necessary to find out the exact changes from the protonation.
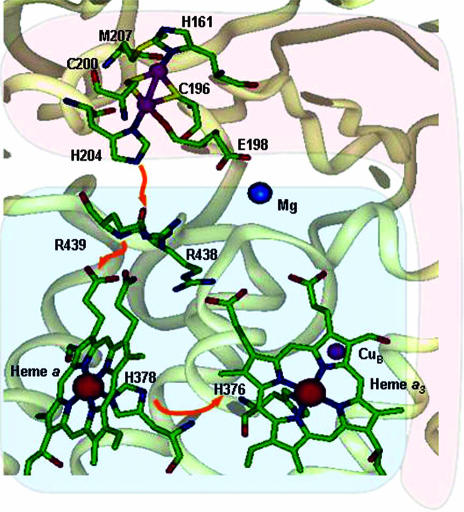
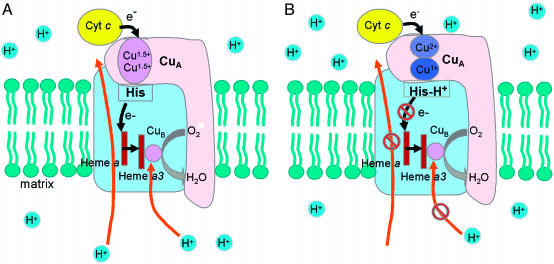
Structural studies of CcO have revealed an extended hydrogen bonding network, including a number of water molecules at the interface between subunit I and II (54–56). One of the copper-bound histidines (His-204, bovine heart CcO numbering) that corresponds to the C-terminal His-120 in CuA azurin plays an important role in this hydrogen bonding network (Fig. 5). This structural feature supports the hypothesis that this histidine is a critical residue along the major ET pathway between CuA and heme a (57, 58). Therefore, we propose that the protonation of the histidine may be a mechanism to regulate proton-coupled electron transfer from the CuA center to heme a (Fig. 6). When the C-terminal histidine is deprotonated, the CuA center is in its class III delocalized mixed valence state, whose reduction potential is low enough to transfer electron from CuA to heme a (Fig. 6A). This ET results in O2 reduction and proton pumping. The pumped proton may result in protonation of the C-terminal histidine, which then causes valence localization of the CuA center, higher reduction potential to shut down the ET, and thus to stop proton pumping (Fig. 6B). Once the pumped proton is released into the outer membrane space where the CuA resides, the histidine is then deprotonated and ready to start the ET process again. Previously, different redox centers, such as binuclear heme a3-CuB (10, 59, 60), heme a (61–64), and/or the CuA center (65–67), have been proposed to be involved in proton-coupled electron transfer in CcO. Our findings and proposal here may lend support for CuA being involved in this process.
Fig. 5.
Structure of the three metal sites in CcO. The figure was generated by using insight ii (Accelrys, San Diego) with coordinates from bovine heart CcO (1OCC). Subunit I and subunit II are colored in light blue and pink, respectively. Orange arrows indicate the suggested electron transfer pathway.
Fig. 6.
Schematic model of valence localized (A) and valence delocalized (B) states of CuA center in CcO. Subunit I is in light blue, and subunit II is in pink. Black arrows represent the flow of electrons, and orange arrows represent the flow of protons.
To our knowledge, no similar pH-dependent behavior has been reported in native CuA in either the CcO or a soluble fragment because most studies have focused on transition to a type 2 copper at high pH. It would be very interesting to find out whether similar pH-dependent behavior to what we found here also occurs in native CuA at low pH. Interestingly, electron transfer studies in the H260N mutant of CuA center in CcO from Rhodobacter sphaeroides corroborate our proposal that protonation of the C-terminal histidine resulted in a change in the valence state and reduction potential and thus reduced ET rate. His-260 is the C-terminal histidine for the CuA center in CcO from R. sphaeroides. The rate constant for ET from the CuA center to heme a for this mutant decreased by over four orders of magnitude compared with the WT (68). From the rapid kinetic assays, the reduction potential of the CuA center in H260N CcO was estimated to be increased by 90 mV with respect to the WT. From this study, it was concluded that His-260 plays a significant role in electron transfer between CuA and heme a. Our results presented here further define possible roles of this histidine; its protonation can result in dramatic changes in mixed valence states and reduction potentials of the CuA center.
In summary we have demonstrated an example of reversible transitions between localized and delocalized mixed valence binuclear copper by simply changing the pH. We further identified protonation of the C-terminal histidine ligand as the trigger of the transition through site-directed mutagenesis. This result strongly suggests that this histidine plays an electronic role by maintaining the equivalency of two copper atoms in CuA centers. We have also demonstrated that the protonation resulted in dramatic changes in the driving force such that it would be unfavorable to transfer an electron from the CuA center to heme a (Fig. 6). Because the corresponding histidine is in the major ET pathway from CuA to heme a, our findings may suggest that CuA and the histidine may play an important role in regulating proton-coupled electron transfer.
Supplementary Material
Acknowledgments
This material is based on work supported by National Science Foundation Grant CHE-0139203.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CcO, cytochrome c oxidase; Uv-vis, UV-visible; ET, electron transfer.
References
- 1.Babcock, G. T. & Wikstrom, M. (1992) Nature 356, 301–309. [DOI] [PubMed] [Google Scholar]
- 2.García-Horsman, J. A., Barquera, B., Rumbley, J., Ma, J. & Gennis, R. B. (1994) J. Bacteriol. 176, 5587–5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferguson-Miller, S. & Babcock, G. T. (1996) Chem. Rev. 96, 2889–2907. [DOI] [PubMed] [Google Scholar]
- 4.Robin, M. B. & Day, P. (1967) Adv. Inorg. Chem. Radiochem. 10, 247–422. [Google Scholar]
- 5.Beinert, H. (1997) Eur. J. Biochem. 245, 521–532. [DOI] [PubMed] [Google Scholar]
- 6.Solomon, E. I., Randall, D. W. & Glaser, T. (2000) Coord. Chem. Rev. 200–202, 595–632. [Google Scholar]
- 7.Gray, H. B., Malmström, B. G. & Williams, R. J. P. (2000) J. Biol. Inorg. Chem. 5, 551–559. [DOI] [PubMed] [Google Scholar]
- 8.Vila, A. J. & Fernández, C. O. (2001) in Handbook on Metalloproteins, eds. Bertini, I., Sigel, A. & Sigel, H. (Dekker, New York), pp. 813–856.
- 9.Lu, Y. (2004) in Biocoordination Chemistry, eds. Que, L., Jr., & Tolman, W. B., Comprehensive Coordination Chemistry II: From Biology to Nanotechnology, eds. McCleverty, J. A. & Meyer, T. J. (Elsevier, Oxford), Vol. 8, pp 91–122. [Google Scholar]
- 10.Iwata, S., Ostermeier, C., Ludwig, B. & Michel, H. (1995) Nature 376, 660–669. [DOI] [PubMed] [Google Scholar]
- 11.Wilmanns, M., Lappalainen, P., Kelly, M., Sauer-Eriksson, E. & Saraste, M. (1995) Proc. Natl. Acad. Sci. USA. 92, 11955–11959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robinson, H., Ang, M. C., Gao, Y.-G., Hay, M. T., Lu, Y. & Wang, A. H. J. (1999) Biochemistry 38, 5677–5683. [DOI] [PubMed] [Google Scholar]
- 13.Williams, P. A., Blackburn, N. J., Sanders, D., Bellamy, H., Stura, E. A., Fee, J. A. & McRee, D. E. (1999) Nat. Struct. Biol. 6, 509–516. [DOI] [PubMed] [Google Scholar]
- 14.Brown, K., Tegoni, M., Prudencio, M., Pereira, A. S., Besson, S., Moura, J. J., Moura, I. & Cambillau, C. (2000) Nat. Struct. Biol. 7, 191–195. [DOI] [PubMed] [Google Scholar]
- 15.Hathaway, B. J. (1987) in Comprehensive Coordination Chemistry: The Synthesis, Reactions, Properties & Application of Coordination Compounds, eds. Wilkinson, G., Gillard, R. D. & McCleverty, J. A. (Pergamon, Oxford), Vol. 5, pp. 533–774. [Google Scholar]
- 16.Dunaj-Jurco, M., Ondrejovic, G., Melnik, M. & Garaj, J. (1988) Coord. Chem. Rev. 83, 1–28. [Google Scholar]
- 17.Harding, C., McKee, V. & Nelson, J. (1991) J. Am. Chem. Soc. 113, 9684–9685. [Google Scholar]
- 18.Barr, M. E., Smith, P. H., Antholine, W. E. & Spencer, B. (1993) J. Chem. Soc. Chem. Commun., 1649–1652.
- 19.Harding, C., Nelson, J., Symons, M. C. R. & Wyatt, J. (1994) J. Chem. Soc. Chem. Commun., 2499–2500.
- 20.Houser, R. P., Young, V. G., Jr., & Tolman, W. B. (1996) J. Am. Chem. Soc. 118, 2101–2102. [Google Scholar]
- 21.LeCloux, D. D., Davydov, R. & Lippard, S. J. (1998) Inorg. Chem. 37, 6814–6826. [DOI] [PubMed] [Google Scholar]
- 22.He, C. & Lippard, S. J. (2000) Inorg. Chem. 39, 5225–5231. [DOI] [PubMed] [Google Scholar]
- 23.Gupta, R., Zhang, Z. H., Powell, D., Hendrich, M. P. & Borovik, A. S. (2002) Inorg. Chem. 41, 5100–5106. [DOI] [PubMed] [Google Scholar]
- 24.Zhang, X.-M., Tong, M.-L. & Chen, X.-M. (2002) Angew. Chem. Int. Ed. 41, 1029–1031. [DOI] [PubMed] [Google Scholar]
- 25.Gagne, R. R., Koval, C. A. & Smith, T. J. (1977) J. Am. Chem. Soc. 99, 8367–8368. [DOI] [PubMed] [Google Scholar]
- 26.Long, R. C. & Hendrickson, D. N. (1983) J. Am. Chem. Soc. 105, 1513–1521. [Google Scholar]
- 27.Hay, M., Richards, J. H. & Lu, Y. (1996) Proc. Natl. Acad. Sci. USA 93, 461–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hay, M. T., Ang, M. C., Gamelin, D. R., Solomon, E. I., Antholine, W. E., Ralle, M., Blackburn, N. J., Massey, P. D., Wang, X., Kwon, A. H. & Lu, Y. (1998) Inorg. Chem. 37, 191–198. [Google Scholar]
- 29.Hwang, H. J., Ang, M. C. & Lu, Y. (2004) J. Biol. Inorg. Chem. 9, 489–494. [DOI] [PubMed] [Google Scholar]
- 30.Jeuken, L. J. C. & Armstrong, F. A. (2001) J. Phys. Chem. B 105, 5271–5282. [Google Scholar]
- 31.Bard, A. J. & Faulkner, L. R. (1980) Electrochemical Methods: Fundamentals and Applications (Wiley, New York)
- 32.Lappalainen, P., Aasa, R., Malmström, B. G. & Saraste, M. (1993) J. Biol. Chem. 268, 26416–26421. [PubMed] [Google Scholar]
- 33.Immoos, C., Hill, M. G., Sanders, D., Fee, J. A., Slutter, C. E., Richards, J. H. & Gray, H. B. (1996) J. Biol. Inorg. Chem. 1, 529–531. [Google Scholar]
- 34.Himmelwright, R. S., Eickman, N. C., LuBien, C. D. & Solomon, E. I. (1980) J. Am. Chem. Soc. 102, 5378–5388. [Google Scholar]
- 35.Gupta, S., Warne, A., Saraste, M. & Mazumdar, S. (2001) Biochemistry 40, 6180–6189. [DOI] [PubMed] [Google Scholar]
- 36.Salgado, J., Warmerdam, G., Bubacco, L. & Canters, G. W. (1998) Biochemistry 37, 7378–7389. [DOI] [PubMed] [Google Scholar]
- 37.Scott, B. & Willett, R. (1991) Inorg. Chem. 30, 110–113. [Google Scholar]
- 38.Scott, B., Willett, R., Porter, L. & Williams, J. (1992) Inorg. Chem. 31, 2483–2492. [Google Scholar]
- 39.Farrar, J. A., Neese, F., Lappalainen, P., Kroneck, P. M. H., Saraste, M., Zumft, W. G. & Thomson, A. J. (1996) J. Am. Chem. Soc. 118, 11501–11514. [Google Scholar]
- 40.Gamelin, D. R., Randall, D. W., Hay, M. T., Houser, R. P., Mulder, T. C., Canters, G. W., de Vries, S., Tolman, W. B., Lu, Y. & Solomon, E. I. (1998) J. Am. Chem. Soc. 120, 5246–5263. [Google Scholar]
- 41.Tsukihara, T., Aoyama, H., Yamashita, E., Tomizaki, T., Yamaguchi, H., Shinzawa-Itoh, K., Nakashima, R., Yaono, R. & Yoshikawa, S. (1995) Science 269, 1069–1074. [DOI] [PubMed] [Google Scholar]
- 42.Dutton, P. L., Wilson, D. F. & Lee, C. P. (1970) Biochemistry 9, 5077–5082. [DOI] [PubMed] [Google Scholar]
- 43.Blair, D. F., Ellis, W. R., Jr., Wang, H., Gray, H. B. & Chan, S. I. (1986) J. Biol. Chem. 261, 11524–11537. [PubMed] [Google Scholar]
- 44.Ellis, W. R., Jr., Wang, H., Blair, D. F., Gray, H. B. & Chan, S. I. (1986) Biochemistry 25, 161–167. [DOI] [PubMed] [Google Scholar]
- 45.Van de Kamp, M., Canters, G. W., Andrew, C. R., Sanders-Loehr, J., Bender, C. J. & Peisach, J. (1993) Eur. J. Biochem. 218, 229–238. [DOI] [PubMed] [Google Scholar]
- 46.St. Clair, C. S., Ellis, W. R., Jr., & Gray, H. B. (1992) Inorg. Chim. Acta 191, 149–155. [Google Scholar]
- 47.Nar, H., Messerschmidt, A., Huber, R., van der Kamp, M. & Canters, G. W. (1991) J. Mol. Biol. 221, 765–772. [DOI] [PubMed] [Google Scholar]
- 48.Sykes, A. G. (1985) Chem. Soc. Rev. 14, 283–315. [Google Scholar]
- 49.Guss, J. M., Harrowell, P. R., Murata, M., Norris, V. A. & Freeman, H. C. (1986) J. Mol. Biol. 192, 361–387. [DOI] [PubMed] [Google Scholar]
- 50.Lommen, A., Canters, G. W. & Van Beeumen, J. (1988) Eur. J. Biochem. 176, 213–223. [DOI] [PubMed] [Google Scholar]
- 51.Lommen, A. & Canters, G. W. (1990) J. Biol. Chem. 265, 2768–2774. [PubMed] [Google Scholar]
- 52.Dennison, C., Kohzuma, T., McFarlane, W., Suzuki, S. & Sykes, A. G. (1994) Inorg. Chem. 33, 3299–3305. [Google Scholar]
- 53.Shepard, W. E. B., Anderson, B. F., Lewandoski, D. A., Norris, G. E. & Baker, E. N. (1990) J. Am. Chem. Soc. 112, 7817–7819. [Google Scholar]
- 54.Ostermeier, C., Harrenga, A., Ermler, U. & Michel, H. (1997) Proc. Natl. Acad. Sci. USA 94, 10547–10553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Svensson-Ek, M., Abramson, J., Larsson, G., Tornroth, S., Brzezinski, P. & Iwata, S. (2002) J. Mol. Biol. 321, 329–339. [DOI] [PubMed] [Google Scholar]
- 56.Tsukihara, T., Shimokata, K., Katayama, Y., Shimada, H., Muramoto, K., Aoyama, H., Mochizuki, M., Shinzawa-itoh, K., Yamashita, E., Yao, M., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 15304–15309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Regan, J. J., Ramirez, B. E., Winkler, J. R., Gray, H. B. & Malmström, B. G. (1998) J. Bioenerg. Biomembr. 30, 35–39. [DOI] [PubMed] [Google Scholar]
- 58.Ramirez, B. E., Malmström, B. G., Winkler, J. R. & Gray, H. B. (1995) Proc. Natl. Acad. Sci. USA 92, 11949–11951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wikstrom, M. (2000) Biochim. Biophys. Acta 1458, 188–198. [DOI] [PubMed] [Google Scholar]
- 60.Rich, P. R., Breton, J., Junemann, S. & Heathcote, P. (2000) Biochim. Biophys. Acta 1459, 475–480. [DOI] [PubMed] [Google Scholar]
- 61.Artzatbanov, V. Y., Konstantinov, A. A. & Skulachev, V. P. (1978) FEBS Lett. 87, 180–185. [DOI] [PubMed] [Google Scholar]
- 62.Rousseau, D. L., Sassaroli, M., Ching, Y. C. & Dasgupta, S. (1988) Ann. N.Y. Acad. Sci. 550, 223–237. [DOI] [PubMed] [Google Scholar]
- 63.de Paula, J. C., Peiffer, W. E., Ingle, R. T., Centeno, J. A., Ferguson-Miller, S. & Babcock, G. T. (1990) Biochemistry 29, 8702–8706. [DOI] [PubMed] [Google Scholar]
- 64.Papa, S., Capitanio, N. & Villani, G. (1998) FEBS Lett. 439, 1–8. [DOI] [PubMed] [Google Scholar]
- 65.Chan, S. I. & Li, P. M. (1990) Biochemistry 29, 1–12. [DOI] [PubMed] [Google Scholar]
- 66.Capitanio, N., Capitanio, G., Minuto, M., De Nitto, E., Palese, L. L., Nicholls, P. & Papa, S. (2000) Biochemistry 39, 6373–6379. [DOI] [PubMed] [Google Scholar]
- 67.Capitanio, N., Capitanio, G., Boffoli, D. & Papa, S. (2000) Biochemistry 39, 15454–15461. [DOI] [PubMed] [Google Scholar]
- 68.Wang, K., Geren, L., Zhen, Y., Ma, L., Ferguson-Miller, S., Durham, B. & Millett, F. (2002) Biochemistry 41, 2298–2304. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.