Abstract
The Gram-positive cell envelope serves as a molecular platform for surface display of capsular polysaccharides, wall teichoic acids (WTAs), lipoteichoic acids (LTAs), lipoproteins, surface proteins and pili. WTAs, LTAs, and sortase-assembled pili are a few features that make the Gram-positive cell envelope distinct from the Gram-negative counterpart. Interestingly, a set of LytR-CpsA-Psr family proteins, found in all Gram-positives but limited to a minority of Gram-negative organisms, plays divergent functions, while decorating the cell envelope with glycans. Furthermore, a phylum of Gram-positive bacteria, the actinobacteria, appear to employ oxidative protein folding as the major folding mechanism, typically occurring in an oxidizing environment of the Gram-negative periplasm. These distinctive features will be highlighted, along with recent findings in the cell envelope biogenesis.
Introduction
The biochemical, biological, and protective nature of bacterial cell envelopes defines what they are. On the basis of membranous enclosure, Gram-positive bacteria are monoderm, i.e. single membrane, lacking the outer membrane seen in the Gram-negative counterparts. They are further classified into Firmicutes and Actinobacteria, based on the low and high GC content of their genomes, respectively. Intriguingly, several actinobacteria, such as Corynebacterium glutamicum and Mycobacterium tuberculosis, produce mycolic acids that constitute the mycobacterial outer membrane, often referred as the mycomembrane [1]; whether or not this mycomembrane would provide actinobacteria an oxidizing environment for oxidative protein folding (discussed below) is up for debate. By definition, the cytoplasmic membrane, thick layers of peptidoglycan, and accessory factors are part of the cell envelope of Gram-positive bacteria. Recent discoveries of the Escherichia coli flippase, a transporter protein that translocates lipid II across the cytoplasmic membrane, provide insights into the mechanism of peptidoglycan biosynthesis not only in Gram-negative but also Gram-positive bacteria. Considerably equivalent to the Gram-negative bacterial LPS, wall teichoic acids (WTAs) and lipoteichoic acids (LTAs) are cell wall- and membrane-anchored anionic polymers, respectively, that play various cellular functions. Likewise, capsular polysaccharides form another class of anionic polymers that give rise to many serotypes of streptococci. Readers are referred to several recent reviews describing these topics in detail [2–4]. Perhaps there is only a single class of surface proteins that are covalently anchored to the cell wall of Gram-positive bacteria, those with the cell wall sorting signal (CWSS). The biosynthesis pathway of these proteins will be discussed at length below, along with flippase that helps build the cell wall for surface proteins to attach to, folding of these proteins, and some new aspects of surface glycoproteins that are also covalently linked to the cell wall.
Building the Cell Wall
Early investigations into the mechanism of action of penicillin might have marked the beginning of molecular studies of bacterial cell wall biogenesis [5,6]. Work in Staphylococcus aureus has essentially provided the basic principle of cell wall synthesis in Gram-positive bacteria, which can be divided into three stages: the cytoplasmic synthesis of Park’s nucleotide (UDP-MurNAc-L-Ala-D-iGlu-L-Lys-D-Ala-D-Ala), generation of lipid II in the membrane, and assembly of peptidoglycan. The details of each stage have been well described in several excellent reviews [7–9], which are briefly summarized here. Initially, Park’s nucleotide is synthesized in the cytoplasm. At the cytoplasmic face of the membrane, Park’s nucleotide is then attached to an undecaprenyl-pyrophosphate carrier molecule (C55-PP) to form lipid I. The attachment of N-acetyl-glucosamine to lipid I yields lipid II. In S. aureus, a pentaglycyl cross-bridge is added to lipid II by a set of peptidyl transferases, called FemA, FemB and FemX [10,11], before the resulting product is flipped to the outer leaflet of the cytoplasmic membrane. Finally, peptidoglycan is built from lipid II by the action of transglycosylases and transpeptidases (also called penicillin binding proteins).
While the above pathway is widely acknowledged, the exact identity of a factor that flips Lipid II across the membrane has been controversial [12]. In the Gram-negative bacterium E. coli, in vitro assays have implicated FtsW to act as a flippase [13], contrary to the previous view that MurJ is likely the lipid II flippase [14]. More recently, with an in vivo assay for lipid II flippase activity that takes advantage of ColM, which cleaves lipid II once it is flipped to the periplasmic face of the inner membrane, Sham and colleagues elegantly demonstrate that MurJ is the lipid II flippase in E. coli [15]. On the other hand, a quest for flippase(s) in Gram-positive bacteria has been less bumpy. In C. glutamicum, RodA, a SEDS (shape, elongation, division and sporulation) family protein highly similar to FtsW, appears to be the flippase of this organism [16]. In Bacillus subtilis, amj, identified to form a synthetic lethal pair with murJ, encodes a novel lipid II flippase named Amj that is a functional alternative to MurJ [17]. The observation that Amj expression is under the control of σM and the activity of σM increases in the absence of MurJ raises the possibility that Amj provides the bacterium a countermeasure in case that MurJ is antagonized by inhibitors by other soil-dwelling organisms [17]. It is interesting to know if non-soil-dwelling Gram-positive bacteria would possess this capability.
Attaching Surface Proteins and Pilus Polymers to the Bacterial Cell Wall
In Gram-positive bacteria, less than 10% exported proteins are covalently attached to the bacterial cell wall. The precursor form of these proteins contains a signal peptide and a C-terminal cell wall sorting signal (CWSS), which is comprised of a LPXTG motif, followed by a hydrophobic domain and a short tail of positively charged residues [18]. The CWSS is necessary and sufficient for cell wall attachment mediated by a conserved transpeptidase enzyme named sortase SrtA first discovered in S. aureus [19,20]. Results from structural studies and in vivo studies of SrtA [21,22], as well as in vitro reconstitution of cell wall anchoring [22,23], put forth a well-known model of sortase-catalyzed cell wall anchoring of surface proteins [18,24,25], whereby the membrane-bound sortase SrtA enzyme cleaves the LPXTG motif between threonine and glycine and links the cleaved substrate to the amino-group of the pentaglycyl cross-bridge within lipid II; the generated product is ultimately incorporated into the cell wall. The enzymatic activity of sortase requires two conserved residues Cys and His [26], which constitute a catalytic pocket within the β-barrel structure of SrtA [21].
SrtA is not the only sortase that has cell wall anchoring activities; among six classes of sortase enzymes reported to date,, i.e. A–F with S. aureus SrtA as the prototype class A sortase [27], S. aureus SrtB (class B sortase) and two class E sortases, Corynebacterium diphtheriae SrtF and Actinomyces oris SrtA, have been shown to mediate cell wall anchoring of surface proteins. In S. aureus, SrtB specifically catalyzes cell wall anchoring of IsdC, a heme transport protein, which harbors an NPQTN motif [28]. In C. diphtheriae, SrtF recognizes a slightly different sorting signal motif, LAXTG, present in pilus base proteins [29,30]. On the other hand, the housekeeping SrtA enzyme of A. oris appears to accommodate both LPXTG- and LAXTG-containing surface proteins and pilins [31,32]. It is interesting to know what determines this broad range of substrates; the availability of A. oris SrtA structures would be helpful in this regard.
Compared to class A sortases, and possibly other sortases of the remaining classes, class C sortases are structurally and functionally distinct. While class C sortases have a typical sortase fold, i.e. β-barrel, they possess a lid that covers their catalytic pocket [33]. They are the only sortase enzymes that have polymerizing activities or pilus polymerization, i.e. linking monomeric proteins into covalently bonded polymers, hence called pilus-specific sortase [18,27]. Sortase-mediated pilus polymerization was first described in C. diphtheriae with the SpaA pilus, which is comprised of the pilus shaft SpaA, pilus tip SpaC and pilus base SpaB, with all pilins containing the CWSS [34]. In addition, SpaA harbors a pilin motif with the conserved lysine residue participating in pilus crosslinking reactions [34,35]. In principle, pilus polymerization is the same as the transpeptidation reaction resulting in cell wall anchoring of surface proteins (see Fig. 1 for detail). Currently, it is not known how the switch between pilus polymerization and termination is determined, but it may be well related to pilin stoichiometry. Consistent with this view, overexpression of SpaA leads to formation of exceedingly long pili [34], whereas overexpression of SpaB results in short pili [30]; the finding also implies that SpaB expression in cells is subject to regulation. Another possibility is that incorporation of SpaB into the pilus base may force the pilus system to switch from pilus polymerization to cell wall anchoring because the housekeeping sortase SrtF has high affinity for SpaB [30].
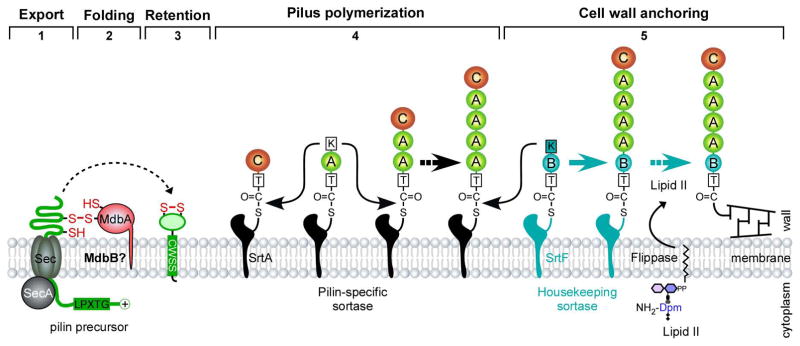
Figure 1.
Pilus assembly in Gram-positive bacteria – modeled after the SpaA pilus of C. diphtheriae. The SpaA pilus, encoded by the pilus gene cluster spaA-srtA-spaB-spaC, is built from the SpaA shaft pilins, with SpaC located at the tip and SpaB at the pilus base. Pilus assembly begins with the synthesis of pilin precursors in the cytoplasm, followed by the Sec-mediated transport of the precursors in an unfolded state. A membrane-bound thiol-disulfide oxidoreductase named MdbA catalyzes post-translocational protein folding, permitting membrane insertion of the precursors, which are polymerized by a pilus-specific sortase via lysine-mediated transpeptidation. Pilus polymerization is terminated when SpaB enters to the pilus base in a similar transpeptidation reaction mediated by a lysine residue [30]. Subsequently, the housekeeping sortase catalyzes the linkage of SpaB to lipid II; the resulting polymer is incorporated into the cell wall [29]. Of note, flipping of lipid II across the membrane is presumably mediated by unknown flippase.
To understand the molecular nature of pilin linkages, Baker and colleagues have solved a three-dimensional structure of the C. diphtheriae shaft pilin SpaA by X-ray crystallization [36], revealing conserved features found in other Gram-positive pilins [37,38]. SpaA is composed of three tandem IgG-like domains, with the middle and the C-terminal domains containing an intramolecular Lys–Asn isopeptide bond [36]. These isopeptide bonds contribute to the protein stability that sustains mechanical forces up to 525 pN, making SpaA one of the most mechanically stable proteins known [39]. Intriguingly, the C-domain also contains a disulfide bond that is essential for post-translocational protein folding [40] (see below). Disulfide bonds appear to be a common feature in only Actinobacterial pilins [41], as they are completely absent in Firmicutes pilins reported to date [42]. Currently, there is no three-dimensional structure of SpaB or SpaC, but based on the protein sequence of each pilin, one could predict that SpaC may possess disulfide bonds, but SpaB does not.
Not all pili of Gram-positive bacteria are heterotrimeric as is the case of the C. diphtheriae SpaABC pili. Pili of Actinomyces spp., Bacillus cereus, and Streptococcus suis are heterodimeric, i.e. only shaft and tip pilins [43–45]; hence the last shaft pilin subunit serves as the pilus base [43,46,47]. This poses some intriguing possibilities. Since there is no SpaB that acts as a switch, termination of pilus polymerization may be dependent on the cell wall anchoring sortase. Secondly, pilus-specific sortases in these organisms may be able to efficiently anchor pilus polymers to the cell wall. Recent studies of the housekeeping sortase SrtA in A. oris lend some truth to these possibilities; it was observed that extremely long pili remain attached to the cell surface in the absence of srtA [32]. It remains to be determined if A. oris SrtA is indeed involved in regulation of pilus length and possesses additional features that gives rise its dual enzymatic activities.
Sugar Coating the Cell Envelope
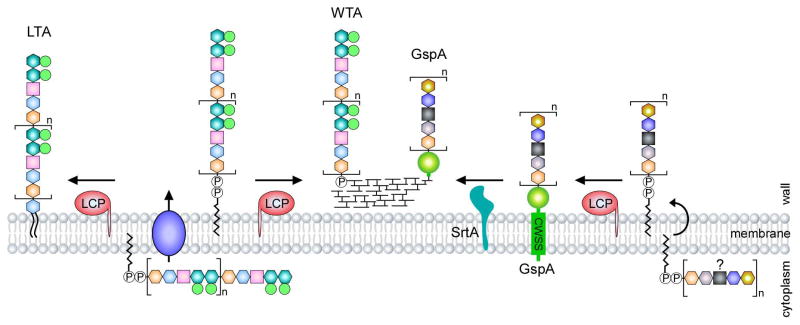
Gram-positive bacteria decorate their cell envelope with various forms of sugars and sugar derivatives, such as capsular polysaccharides and WTAs and LTA glycopolymers; the latter are found variably within Gram-positive species. WTAs are composed of phosphodiester-linked polyol repeat units (polyribitol phosphate – polyRboP or polyglycerol phosphate – polyGroP) and a disaccharide linkage unit, which is covalently attached to peptidoglycan via a phosphodiester bond to the C6 hydroxyl of N-acetyl muramic acid (MurNAc) [3]. LTAs are basically polyGroP polymers tethered to the membrane via a diacylglycerol lipid, and many types of LTAs based on the chemical structures of polyGroP polymers have been classified [2]. Of particular interest is the type IV LTA of Streptococcus pneumoniae that has the backbone of pseudopentasaccharide repeating units comprised of 2-acetamido-4-amino- 2,4,6-trideoxy-D-galactose, glucose, ribitol phosphate, and two N-acetylgalatosamine molecules [2,48]. The backbone is built on undecaprenyl phosphate in the cytoplasm and flipped across the membrane by TacF. The resulting backbone is then either linked to a glycolipid lipid anchor or peptidoglycan by LytR-CpsA-Psr (LCP) family enzymes to generate LTAs or WTAs, respectively [2] (Fig. 2). First functionally demonstrated in B. subtilis [49], LCP proteins are widespread in Gram-positive bacteria and often present in multiple copies [50]. In B. anthracis, for example, six LCP enzymes are presumably involved in transferring of cell wall polysaccharides to discrete sites within the cell envelope [51]. Interestingly, the role of the LCP enzymes seems to extend beyond WTA and LTA biosynthesis. In A. oris, an LCP protein is thought to catalyze the transfer of an unknown glycopolymer to an LPXTG-containing protein called GspA, before it is attached to the cell wall by the housekeeping sortase SrtA [32] (Fig. 2). The nature of this GspA glycosylation by LCP is currently unknown, so is the conservation of this pathway in other Gram-positive bacteria.
Figure 2.
LCP-mediated glycosylation in Gram-positive bacteria. The LytR-CpsA-Psr (LCP) family proteins are widespread in Gram-positive bacteria. In S. pneumoniae, LCP enzymes are involved in WTA and LTA biosynthesis, whereby they catalyze the joining of glycopolymers to peptidoglycan or a glycolipid anchor, hence generating WTAs and LTAs, respectively (modified after Percy and Grundling [2]). In A. oris, it is proposed that an LCP enzyme mediates the transfer of an unknown glycan chain to a LPXTG-containing protein named GspA, which is then anchored to the cell wall by the housekeeping sortase SrtA. It is unclear about the constituents of this glycan chain.
Life after Membrane Translocation
Unlike proteins exported by the twin-arginine translocation (TAT) system, Gram-positive bacterial proteins with a signal peptide are translocated by the Sec machine in an unfolded state. How these translocated proteins acquire their native conformation has been an intriguing question. More than 15 years ago, Beckwith and Henriques-Normark groups reported that the secretome of Actinobacteria, such as Corynebacterium glutamicum and Mycobacterium tuberculosis, comprises a high number of proteins with two or more cysteine residues; in contrast, the secretome of Firmicutes, such as S. aureus and B. subtilis, is largely devoid of incorporated cysteine residues [52,53]. In line with this, recent studies in the two actinobacteria A. oris and C. diphtheriae support the notion that Actinobacteria employ disulfide bond formation as the major mechanism of post-translocational protein folding [54]. In both actinobacteria, a membrane-bound thiol-disulfide oxidoreductase called MdbA catalyzes disulfide bond formation in a manner that is similar to E. coli DsbA [40,41]. In fact, crystal studies reveal that both MdbA enzymes harbor a DsbA-like fold, i.e. a thioredoxin-like domain and an extended α-helical domain, despite little sequence similarity with DsbA and known DsbA-like proteins [40,41]. Given that more than 60% secreted proteins in A. oris and C. diphtheriae contain two or more cysteine residues, it has been proposed that MdbA is the primary thiol-disulfide oxidoreductase that catalyzes oxidative protein folding in these two organisms [40,41] (see Fig. 1). Consistent with this notion, mutant strains devoid of mdbA fail to form adhesive pili [40,41] and are attenuated in virulence [40]. In addition, deletion of mdbA is lethal in A. oris [41] and in C. diphtheriae at 37°C [40]. The basis of this essentiality may be that components vital to cell envelope biogenesis, e.g. penicillin-binding proteins, require MdbA-mediated postranslocational protein folding since mdbA-depleted strains exhibit aberrant cell morphology and growth arrest [54]. If this is the case, inhibitors of MdbA may augment the effectiveness of cell envelope-targeting antibiotics and lessen antibiotic resistance.
The essentiality of MdbA-mediated oxidative protein folding and cysteine inclusion of exported proteins in the actinobacteria raises an interesting question of how post-translocational folding of exported proteins lacking cysteines in the actinobacteria and those in the firmicutes takes place. One possibility is that chaperones, foldases and peptidylprolyl isomerases, such as PsrA and PpiA [55], may provide an alternative pathway.
Concluding Remarks
Cell envelope biogenesis in Gram-positive bacteria has been subject to extensive investigations for more than six decades. It started with molecular studies of how antibiotics affect the cell envelope, and great progress has been made to unlock various steps of cell envelope biogenesis. Future experiments may be directed at examining the exoplasmic compartment surrounding the Gram-positive cytoplasmic membrane and what makes it suitable for oxidative protein folding in actinobacteria (Fig. 3), as well as how folded proteins are translocated across the thick layer of Gram-positive peptidoglycan. In this regard, recent technical advances in cryo-electron microscopy may allow detailed examinations of the cell envelope at high resolution in the native state. Finally, the protective and immunological nature of the cell envelope may present a challenge as to how its constituents of individual bacteria and microbiome as a whole impact the host organism, yet it offers tremendous opportunities as targets for antimicrobial development.
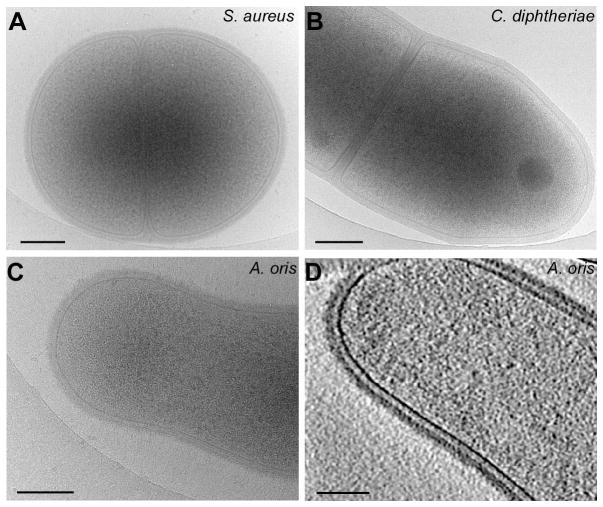
Figure 3.
Electron microscopic analysis of Gram-positive bacteria. Fresh bacterial cultures of the firmicutes S. aureus (strain Newman) (A) and the actinobacteria C. diphtheriae (strain NCTC 13129) (B) and A. oris (strain MG1) (C) were deposited onto holey carbon grids, which were rapidly frozen in liquid ethane. Cryo-EM images of the frozen-hydrated specimens were taken on a direct electron detector (Gatan K2) on a cryo-electron microscope (FEI Polara). (D) Shown is an A. oris cell obtained by three-dimensional reconstructions by cryo-electron tomography [56]. Note a less dense layer between the membrane and peptidoglycan; a scale bar of 0.2 μm.
Highlights.
Many Gram-positive bacteria assemble on the cell envelope covalently linked pili that are polymerized by a conserved transpeptidase called pilus-specific sortase.
LytR-CpsA-Psr family proteins present in all Gram-positive bacteria are involved not only in transferring teichoic acids to peptidoglycan or glycolipid anchors but also linking glycans to a cell wall-anchored protein.
The high GC content bacteria or actinobacteria utilize a membrane-bound thiol-disulfide oxidoreductase as the major protein folding machine for exported proteins.
Acknowledgments
We thank our lab members for the critical review and discussion of this work. H.T.-T. acknowledges funding support from the National Institute of Dental and Craniofacial Research of the National Institutes of Health (DE017382 and DE025015). J.L. is supported in part by the Welch Foundation Grant (AU-1714).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and Recommended Reading
- 1.Marrakchi H, Laneelle MA, Daffe M. Mycolic acids: structures, biosynthesis, and beyond. Chem Biol. 2014;21:67–85. doi: 10.1016/j.chembiol.2013.11.011. [DOI] [PubMed] [Google Scholar]
- 2•.Percy MG, Grundling A. Lipoteichoic acid synthesis and function in gram-positive bacteria. Annu Rev Microbiol. 2014;68:81–100. doi: 10.1146/annurev-micro-091213-112949. An excellent review on the synthesis and function of Gram-positive lipoteichoic acids. [DOI] [PubMed] [Google Scholar]
- 3.Brown S, Santa Maria JP, Jr, Walker S. Wall teichoic acids of gram-positive bacteria. Annu Rev Microbiol. 2013;67:313–336. doi: 10.1146/annurev-micro-092412-155620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Geno KA, Gilbert GL, Song JY, Skovsted IC, Klugman KP, Jones C, Konradsen HB, Nahm MH. Pneumococcal Capsules and Their Types: Past, Present, and Future. Clin Microbiol Rev. 2015;28:871–899. doi: 10.1128/CMR.00024-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park JT, Johnson MJ. Accumulation of labile phosphate in Staphylococcus aureus grown in the presence of penicillin. J Biol Chem. 1949;179:585–592. [PubMed] [Google Scholar]
- 6.Strominger JL, Tipper DJ. Bacterial cell wall synthesis and structure in relation to the mechanism of action of penicillins and other antibacterial agents. Am J Med. 1965;39:708–721. doi: 10.1016/0002-9343(65)90093-8. [DOI] [PubMed] [Google Scholar]
- 7.Navarre WW, Schneewind O. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol Mol Biol Rev. 1999;63:174–229. doi: 10.1128/mmbr.63.1.174-229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhavsar AP, Brown ED. Cell wall assembly in Bacillus subtilis: how spirals and spaces challenge paradigms. Mol Microbiol. 2006;60:1077–1090. doi: 10.1111/j.1365-2958.2006.05169.x. [DOI] [PubMed] [Google Scholar]
- 9.Silhavy TJ, Kahne D, Walker S. The bacterial cell envelope. Cold Spring Harb Perspect Biol. 2010;2:a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ton-That H, Labischinski H, Berger-Bachi B, Schneewind O. Anchor structure of staphylococcal surface proteins. III. Role of the FemA, FemB, and FemX factors in anchoring surface proteins to the bacterial cell wall. J Biol Chem. 1998;273:29143–29149. doi: 10.1074/jbc.273.44.29143. [DOI] [PubMed] [Google Scholar]
- 11.Rohrer S, Berger-Bachi B. FemABX peptidyl transferases: a link between branched-chain cell wall peptide formation and beta-lactam resistance in gram-positive cocci. Antimicrob Agents Chemother. 2003;47:837–846. doi: 10.1128/AAC.47.3.837-846.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruiz N. Lipid Flippases for Bacterial Peptidoglycan Biosynthesis. Lipid Insights. 2015;8:21–31. doi: 10.4137/LPI.S31783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mohammadi T, van Dam V, Sijbrandi R, Vernet T, Zapun A, Bouhss A, Diepeveen-de Bruin M, Nguyen-Disteche M, de Kruijff B, Breukink E. Identification of FtsW as a transporter of lipid-linked cell wall precursors across the membrane. EMBO J. 2011;30:1425–1432. doi: 10.1038/emboj.2011.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruiz N. Bioinformatics identification of MurJ (MviN) as the peptidoglycan lipid II flippase in Escherichia coli. Proc Natl Acad Sci U S A. 2008;105:15553–15557. doi: 10.1073/pnas.0808352105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15••.Sham LT, Butler EK, Lebar MD, Kahne D, Bernhardt TG, Ruiz N. Bacterial cell wall. MurJ is the flippase of lipid-linked precursors for peptidoglycan biogenesis. Science. 2014;345:220–222. doi: 10.1126/science.1254522. An elegant study using an in vivo flippase activity assay to identify MurJ as the flippase of E. coli. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sieger B, Schubert K, Donovan C, Bramkamp M. The lipid II flippase RodA determines morphology and growth in Corynebacterium glutamicum. Mol Microbiol. 2013;90:966–982. doi: 10.1111/mmi.12411. [DOI] [PubMed] [Google Scholar]
- 17•.Meeske AJ, Sham LT, Kimsey H, Koo BM, Gross CA, Bernhardt TG, Rudner DZ. MurJ and a novel lipid II flippase are required for cell wall biogenesis in Bacillus subtilis. Proc Natl Acad Sci U S A. 2015;112:6437–6442. doi: 10.1073/pnas.1504967112. Using a synthetic lethal screen this study identifies a second flippase that is specific to Gram-positive bacteria. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siegel SD, Reardon ME, Ton-That H. Anchoring of LPXTG-Like Proteins to the Gram-Positive Cell Wall Envelope. Curr Top Microbiol Immunol. 2016 doi: 10.1007/82_2016_8. [DOI] [PubMed] [Google Scholar]
- 19.Mazmanian SK, Liu G, Ton-That H, Schneewind O. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science. 1999;285:760–763. doi: 10.1126/science.285.5428.760. [DOI] [PubMed] [Google Scholar]
- 20.Schneewind O, Model P, Fischetti VA. Sorting of protein A to the staphylococcal cell wall. Cell. 1992;70:267–281. doi: 10.1016/0092-8674(92)90101-h. [DOI] [PubMed] [Google Scholar]
- 21.Ilangovan U, Ton-That H, Iwahara J, Schneewind O, Clubb RT. Structure of sortase, the transpeptidase that anchors proteins to the cell wall of Staphylococcus aureus. Proc Natl Acad Sci U S A. 2001;98:6056–6061. doi: 10.1073/pnas.101064198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ton-That H, Liu G, Mazmanian SK, Faull KF, Schneewind O. Purification and characterization of sortase, the transpeptidase that cleaves surface proteins of Staphylococcus aureus at the LPXTG motif. Proc Natl Acad Sci U S A. 1999;96:12424–12429. doi: 10.1073/pnas.96.22.12424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ton-That H, Mazmanian SK, Faull KF, Schneewind O. Anchoring of surface proteins to the cell wall of Staphylococcus aureus. Sortase catalyzed in vitro transpeptidation reaction using LPXTG peptide and NH(2)-Gly(3) substrates. J Biol Chem. 2000;275:9876–9881. doi: 10.1074/jbc.275.13.9876. [DOI] [PubMed] [Google Scholar]
- 24.Ton-That H, Marraffini LA, Schneewind O. Protein sorting to the cell wall envelope of Gram-positive bacteria. Biochim Biophys Acta. 2004;1694:269–278. doi: 10.1016/j.bbamcr.2004.04.014. [DOI] [PubMed] [Google Scholar]
- 25.Mazmanian SK, Ton-That H, Schneewind O. Sortase-catalysed anchoring of surface proteins to the cell wall of Staphylococcus aureus. Mol Microbiol. 2001;40:1049–1057. doi: 10.1046/j.1365-2958.2001.02411.x. [DOI] [PubMed] [Google Scholar]
- 26.Ton-That H, Mazmanian SK, Alksne L, Schneewind O. Anchoring of surface proteins to the cell wall of Staphylococcus aureus. Cysteine 184 and histidine 120 of sortase form a thiolate-imidazolium ion pair for catalysis. J Biol Chem. 2002;277:7447–7452. doi: 10.1074/jbc.M109945200. [DOI] [PubMed] [Google Scholar]
- 27.Spirig T, Weiner EM, Clubb RT. Sortase enzymes in Gram-positive bacteria. Mol Microbiol. 2011;82:1044–1059. doi: 10.1111/j.1365-2958.2011.07887.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mazmanian SK, Ton-That H, Su K, Schneewind O. An iron-regulated sortase anchors a class of surface protein during Staphylococcus aureus pathogenesis. Proc Natl Acad Sci U S A. 2002;99:2293–2298. doi: 10.1073/pnas.032523999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swaminathan A, Mandlik A, Swierczynski A, Gaspar A, Das A, Ton-That H. Housekeeping sortase facilitates the cell wall anchoring of pilus polymers in Corynebacterium diphtheriae. Mol Microbiol. 2007;66:961–974. doi: 10.1111/j.1365-2958.2007.05968.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mandlik A, Das A, Ton-That H. The molecular switch that activates the cell wall anchoring step of pilus assembly in gram-positive bacteria. Proc Natl Acad Sci U S A. 2008;105:14147–14152. doi: 10.1073/pnas.0806350105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31••.Reardon-Robinson ME, Wu C, Mishra A, Chang C, Bier N, Das A, Ton-That H. Pilus hijacking by a bacterial coaggregation factor critical for oral biofilm development. Proc Natl Acad Sci U S A. 2014;111:3835–3840. doi: 10.1073/pnas.1321417111. First description of an LPXTG-containing adhesin that hijacks the pilus assembly machine to be optimally displayed on the Gram-positive bacterial surface. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32•.Wu C, Huang IH, Chang C, Reardon-Robinson ME, Das A, Ton-That H. Lethality of sortase depletion in Actinomyces oris caused by excessive membrane accumulation of a surface glycoprotein. Mol Microbiol. 2014;94:1227–1241. doi: 10.1111/mmi.12780. Presents the first evidence that the houskeeping sortase gene is essential in A. oris and that a LCP protein is involved glycosylation of a cell wall-anchored protein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manzano C, Izore T, Job V, Di Guilmi AM, Dessen A. Sortase activity is controlled by a flexible lid in the pilus biogenesis mechanism of gram-positive pathogens. Biochemistry. 2009;48:10549–10557. doi: 10.1021/bi901261y. [DOI] [PubMed] [Google Scholar]
- 34.Ton-That H, Schneewind O. Assembly of pili on the surface of Corynebacterium diphtheriae. Mol Microbiol. 2003;50:1429–1438. doi: 10.1046/j.1365-2958.2003.03782.x. [DOI] [PubMed] [Google Scholar]
- 35.Ton-That H, Marraffini LA, Schneewind O. Sortases and pilin elements involved in pilus assembly of Corynebacterium diphtheriae. Mol Microbiol. 2004;53:251–261. doi: 10.1111/j.1365-2958.2004.04117.x. [DOI] [PubMed] [Google Scholar]
- 36.Kang HJ, Paterson NG, Gaspar AH, Ton-That H, Baker EN. The Corynebacterium diphtheriae shaft pilin SpaA is built of tandem Ig-like modules with stabilizing isopeptide and disulfide bonds. Proc Natl Acad Sci U S A. 2009;106:16967–16971. doi: 10.1073/pnas.0906826106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kang HJ, Coulibaly F, Clow F, Proft T, Baker EN. Stabilizing isopeptide bonds revealed in gram-positive bacterial pilus structure. Science. 2007;318:1625–1628. doi: 10.1126/science.1145806. [DOI] [PubMed] [Google Scholar]
- 38.Krishnan V, Gaspar AH, Ye N, Mandlik A, Ton-That H, Narayana SV. An IgG-like domain in the minor pilin GBS52 of Streptococcus agalactiae mediates lung epithelial cell adhesion. Structure. 2007;15:893–903. doi: 10.1016/j.str.2007.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Echelman DJ, Alegre-Cebollada J, Badilla CL, Chang C, Ton-That H, Fernandez JM. CnaA domains in bacterial pili are efficient dissipaters of large mechanical shocks. Proc Natl Acad Sci U S A. 2016;113:2490–2495. doi: 10.1073/pnas.1522946113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reardon-Robinson ME, Osipiuk J, Jooya N, Chang C, Joachimiak A, Das A, Ton-That H. A thiol-disulfide oxidoreductase of the Gram-positive pathogen Corynebacterium diphtheriae is essential for viability, pilus assembly, toxin production and virulence. Mol Microbiol. 2015;98:1037–50. doi: 10.1111/mmi.13172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41••.Reardon-Robinson ME, Osipiuk J, Chang C, Wu C, Jooya N, Joachimiak A, Das A, Ton-That H. A Disulfide Bond-forming Machine Is Linked to the Sortase-mediated Pilus Assembly Pathway in the Gram-positive Bacterium Actinomyces oris. J Biol Chem. 2015;290:21393–21405. doi: 10.1074/jbc.M115.672253. Describes the conserved oxidative protein folding pathway in Actinobacteria that is invovled in post-translocational folding of exported proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krishnan V. Pilins in gram-positive bacteria: A structural perspective. IUBMB Life. 2015;67:533–543. doi: 10.1002/iub.1400. [DOI] [PubMed] [Google Scholar]
- 43.Mishra A, Das A, Cisar JO, Ton-That H. Sortase-Catalyzed Assembly of Distinct Heteromeric Fimbriae in Actinomyces naeslundii. J Bacteriol. 2007;189:3156–3165. doi: 10.1128/JB.01952-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Budzik JM, Marraffini LA, Schneewind O. Assembly of pili on the surface of Bacillus cereus vegetative cells. Mol Microbiol. 2007;66:495–510. doi: 10.1111/j.1365-2958.2007.05939.x. [DOI] [PubMed] [Google Scholar]
- 45.Okura M, Osaki M, Fittipaldi N, Gottschalk M, Sekizaki T, Takamatsu D. The Minor Pilin Subunit Sgp2 Is Necessary for Assembly of the Pilus Encoded by the srtG Cluster of Streptococcus suis. J Bacteriol. 2011;193:822–831. doi: 10.1128/JB.01555-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Budzik JM, Oh SY, Schneewind O. Cell wall anchor structure of BcpA pili in Bacillus anthracis. J Biol Chem. 2008;283:36676–36686. doi: 10.1074/jbc.M806796200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mishra A, Wu C, Yang J, Cisar JO, Das A, Ton-That H. The Actinomyces oris type 2 fimbrial shaft FimA mediates co-aggregation with oral streptococci, adherence to red blood cells and biofilm development. Mol Microbiol. 2010;77:841–854. doi: 10.1111/j.1365-2958.2010.07252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Denapaite D, Bruckner R, Hakenbeck R, Vollmer W. Biosynthesis of teichoic acids in Streptococcus pneumoniae and closely related species: lessons from genomes. Microb Drug Resist. 2012;18:344–358. doi: 10.1089/mdr.2012.0026. [DOI] [PubMed] [Google Scholar]
- 49.Kawai Y, Marles-Wright J, Cleverley RM, Emmins R, Ishikawa S, Kuwano M, Heinz N, Bui NK, Hoyland CN, Ogasawara N, et al. A widespread family of bacterial cell wall assembly proteins. EMBO J. 2011;30:4931–4941. doi: 10.1038/emboj.2011.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hubscher J, Luthy L, Berger-Bachi B, Stutzmann Meier P. Phylogenetic distribution and membrane topology of the LytR-CpsA-Psr protein family. BMC Genomics. 2008;9:617. doi: 10.1186/1471-2164-9-617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liszewski Zilla M, Lunderberg JM, Schneewind O, Missiakas D. Bacillus anthracis lcp Genes Support Vegetative Growth, Envelope Assembly, and Spore Formation. J Bacteriol. 2015;197:3731–3741. doi: 10.1128/JB.00656-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Daniels R, Mellroth P, Bernsel A, Neiers F, Normark S, von Heijne G, Henriques-Normark B. Disulfide bond formation and cysteine exclusion in gram-positive bacteria. J Biol Chem. 2010;285:3300–3309. doi: 10.1074/jbc.M109.081398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53•.Dutton RJ, Boyd D, Berkmen M, Beckwith J. Bacterial species exhibit diversity in their mechanisms and capacity for protein disulfide bond formation. Proc Natl Acad Sci U S A. 2008;105:11933–11938. doi: 10.1073/pnas.0804621105. Provides a comprehensive analysis of bacterial disulfide bond-forming proteins and their potential pathway and associated substrates. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reardon-Robinson ME, Ton-That H. Disulfide-Bond-Forming Pathways in Gram-Positive Bacteria. J Bacteriol. 2016;198:746–754. doi: 10.1128/JB.00769-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sarvas M, Harwood CR, Bron S, van Dijl JM. Post-translocational folding of secretory proteins in Gram-positive bacteria. Biochim Biophys Acta. 2004;1694:311–327. doi: 10.1016/j.bbamcr.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 56•.Hu B, Margolin W, Molineux IJ, Liu J. The bacteriophage t7 virion undergoes extensive structural remodeling during infection. Science. 2013;339:576–579. doi: 10.1126/science.1231887. An elegant study by cryo-electron microscopy to capture successive stages of phage T7 infection of E. coli. [DOI] [PMC free article] [PubMed] [Google Scholar]