Abstract
Cell growth requires the removal of proteins that are unwanted or toxic. In bacteria, AAA+ proteases like the Clp family and Lon selectively destroy proteins defined by intrinsic specificity or adaptors. Caulobacter crescentus is a gram-negative bacterium that undergoes an obligate developmental transition every cell division cycle. Here we highlight recent work that reveals how a hierarchy of adaptors targets the degradation of key proteins at specific times during this cell cycle, integrating protein destruction with other cues. We describe recent insight into how Caulobacter manages DNA replication and repair through Lon and Clp proteases. Because proteases must manage a broad substrate repertoire there must be methods to compensate for protease saturation and we discuss these scenarios.
Introduction
The regulated destruction of proteins is crucial for bacterial growth and development during normal and stress conditions. In the bacterium Caulobacter crescentus, regulated degradation of key proteins drives the cell cycle and depletion of replication factors during stress responses allows cells time to recover from these damages. In this review, we will give an overview of protein degradation in Caulobacter focusing on recent studies showing how regulated proteolysis by the Clp and Lon family of proteases impacts both normal and stress related growth. The common and unique substrate profiles of these proteases allow them to robustly provide for normal growth and respond to stress. Comparison of several different bacterial systems allows us to determine common themes reflecting broad responses to stress.
Energy dependent proteases in Caulobacter
Like most bacteria, regulated proteolysis in Caulobacter is accomplished by several energy dependent proteases. Although they differ in specific protein subunits, these ring-shaped proteases generally function by recognizing targets, then unfolding them using energy captured from ATP hydrolysis, ultimately threading these polypeptides into a chamber harboring active sites for peptide bond hydrolysis. Because of their design, these chambered peptidases cannot normally degrade folded or full-length polypeptides on their own and are solely dependent upon the active delivery of the target. The responsibility of target recognition falls on ATP-dependent chaperones that are encoded on separate proteins (such as the case with ClpXP) or domains (such as the case for the Lon protease) from the peptidase (Figure 1a).
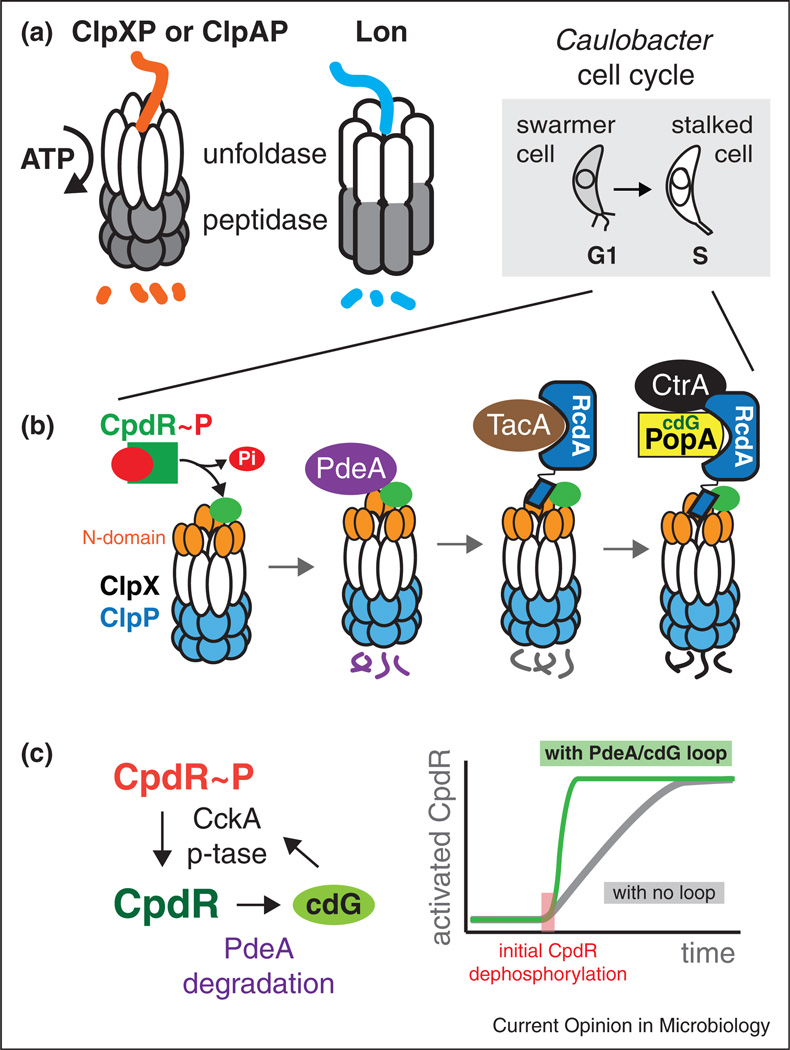
Figure 1.
Protein degradation by energy dependent proteases can be shaped by hierarchical adaptors. (a) The Clp family of proteases are composed of unfoldases (ClpX or ClpA) paired with the ClpP peptidase. The Lon protease is a single polypeptide with these activities contained in different domains. (b) The G1-S transition in Caulobacter is accompanied by morphological changes from a motile swarmer cell to a sessile stalked cell. At this transition, the dephosphorylation of CpdR initiates the assembly of an adaptor hierarchy that results in staged degradation of substrates. (c) CpdR phosphorylation (and its activity) is ultimately controlled by CckA. CckA is a histidine kinase, but high levels of cdG cause it to switch to a phosphatase, resulting in increased dephosphorylation of CpdR. CpdR is directly responsible for delivering the cdG phosphodiesterase PdeA to the ClpXP protease. This sets up a putative positive feedback loop in which initial CpdR dephosphorylation can catalyze full conversion of the CpdR pool to an activated state.
With important exceptions, these proteases completely and processively degrade their targets once engaged without obvious sequence preference [1,2]. Therefore the initial specificity of these proteases is the major determinant of how they will impact the cell. Protease target recognition arises from a combination of the intrinsic specificity of the protease and the use of adaptor proteins to further tune proteolytic range [2]. Because protein degradation is irreversible, understanding this initial step of recognition is crucial.
Regulated proteolysis during the cell cycle
In Caulobacter, regulated protein degradation during the cell cycle drives replication and developmental transitions. The essential regulator CtrA controls transcription of many cell cycle genes and is also a replication inhibitor. Removal of CtrA activity through degradation or post-translational changes is therefore necessary so that cells can initiate replication during the G1-S transition. Genetic and cell biology studies during the last ten years revealed that degradation of CtrA requires the ClpXP protease, the auxiliary factors CpdR, RcdA and PopA, and the second messenger cyclic di-GMP [3–7]. Interestingly, these factors were not solely dedicated to CtrA degradation. For example, proteolysis of the chemotaxis protein McpA and the cyclic di-GMP phosphodiesterase PdeA during the G1-S transition required CpdR and ClpXP but not RcdA or PopA [5,8]. How these inputs collectively resulted in degradation of specific substrates at specific times was an outstanding question.
The pacemaker of proteolytic control during cell cycle is the cyclic phosphorylation of the CpdR adaptor. CpdR is phosphorylated by the same kinase cascade responsible for CtrA phosphorylation [9], but the outcome of this posttranslational modification is opposite for the two proteins. Like canonical response regulators, phosphorylation of CtrA activates it as a transcription factor, while phosphorylation of CpdR inhibits its ability to stimulate the ClpXP protease [5]. Thus, activation of CtrA also results in its stabilization. Similarly, inactivation of CtrA through dephosphorylation also catalyzes CtrA destruction because the same pathway dephosphorylates and activates CpdR [5,9].
Adaptor hierarchies drive class specific substrate degradation
Recent biochemical work shows that CpdR, RcdA and PopA act as adaptors that hierarchically assemble to deliver substrates dependent on the degree of assembly [10••,11•,12•] (Figure 1b). Reconstitution experiments using highly purified proteins showed that CpdR binds the ClpXP protease, priming it for recognition of substrates such as PdeA and McpA. Phosphorylation of CpdR causes it to release from ClpXP providing a simple mechanism for its control [11•]. In addition to improved substrate recognition, CpdR-primed ClpXP could now bind RcdA, which was shown to bind several cargos [10••]. RcdA could then deliver its bound target substrates, for example the developmental regulator TacA, to the CpdR-primed ClpXP [10••]. Finally, RcdA also binds PopA and in the presence of cyclic di-GMP these proteins form a complex with CtrA [4,12•]. Importantly, formation of this final complex promotes the robust degradation of CtrA by a CpdR-primed ClpXP, especially apparent in conditions where CtrA degradation by ClpXP alone is poor [10••,12•,13]. This model rationalized the importance of each of these proteins in the final degradation of CtrA as well as supported the need for cyclic di-GMP (Figure 1a).
Interestingly, it was recently shown that high levels of cyclic di-GMP causes dephosphorylation of CpdR/CtrA by switching the CckA kinase into a phosphatase [14•]. As PdeA is a phosphodiesterase that limits cyclic di-GMP accumulation, it is tempting to speculate that degradation of PdeA upon dephosphorylation of CpdR can further stimulate CpdR activation by increasing levels of cyclic di-GMP. The advantage of this positive feedback is that activation of a subpopulation of CpdR would catalytically induce the conversion of the entire pool in short order, resulting in a sharper switch for proteolytic activation during the G1-S transition. These and other types of feedback regulation are likely needed for the robust transition between cell cycle stages crucial for normal development and growth (Figure 1c).
Although the specific example given above has been shown in Caulobacter, adaptor hierarchies are likely to be found in other bacterial systems. For example, during sporulation in Bacillus subtilis, degradation of the SpoIVA regulator by the ClpXP protease eliminates defective cells. This process requires the small protein CmpA that binds directly to ClpXP, but genetic evidence suggests the need for additional factors [15•]. In this light, CmpA could be acting as part of an adaptor hierarchy that ensures only high quality spores endure. It also stands to reason that additional adaptor-dependent protease pathways will emerge that control sporulation, given the irreversible and critical nature of this developmental decision.
Finally, it is worth remarking that finding additional adaptor hierarchies is particularly challenging as protease adaptors are defined by their ability to stimulate substrate degradation by the protease. Yet biochemical validation of a protease substrate requires the prior knowledge of the adaptor in order to fully reconstitute this activity. Thus, addressing the circular challenge of novel adaptor/substrate identification is an outstanding question.
ClpXP balances critical aspects of DNA metabolism
The ClpXP protease is essential in Caulobacter. The wide range of potential ClpP protease substrates leads one to assume that pleotropic penalty paid by the loss of ClpXP would result in cell death [16]. However, a suppressor screen showed that a single toxin protein, SocB, was responsible for the truly essential nature of ClpXP [17]. The SocB toxin binds replication clamps and blocks replication elongation presumably by competing with DNA polymerase III for clamp [17] (Figure 2b). SocB activity is limited by the SocA antitoxin, which acts as an adaptor to deliver SocB to ClpXP. Like other adaptors, this activity requires the N-terminal domain of ClpX and the removal of the SocB toxin appears to be a major function of the ClpXP protease during normal growth conditions. SocB is upregulated in the presence of DNA damaging agents [17,18], suggesting that clamp inhibition may be an important aspect of the normal DNA damage response program. It is worth noting that although deletion of SocB toxin allows for strains to survive without ClpX, these cells have highly aberrant morphologies with dramatic reductions in fitness and growth, consistent with a larger role for ClpX beyond the need for degrading this single toxin.
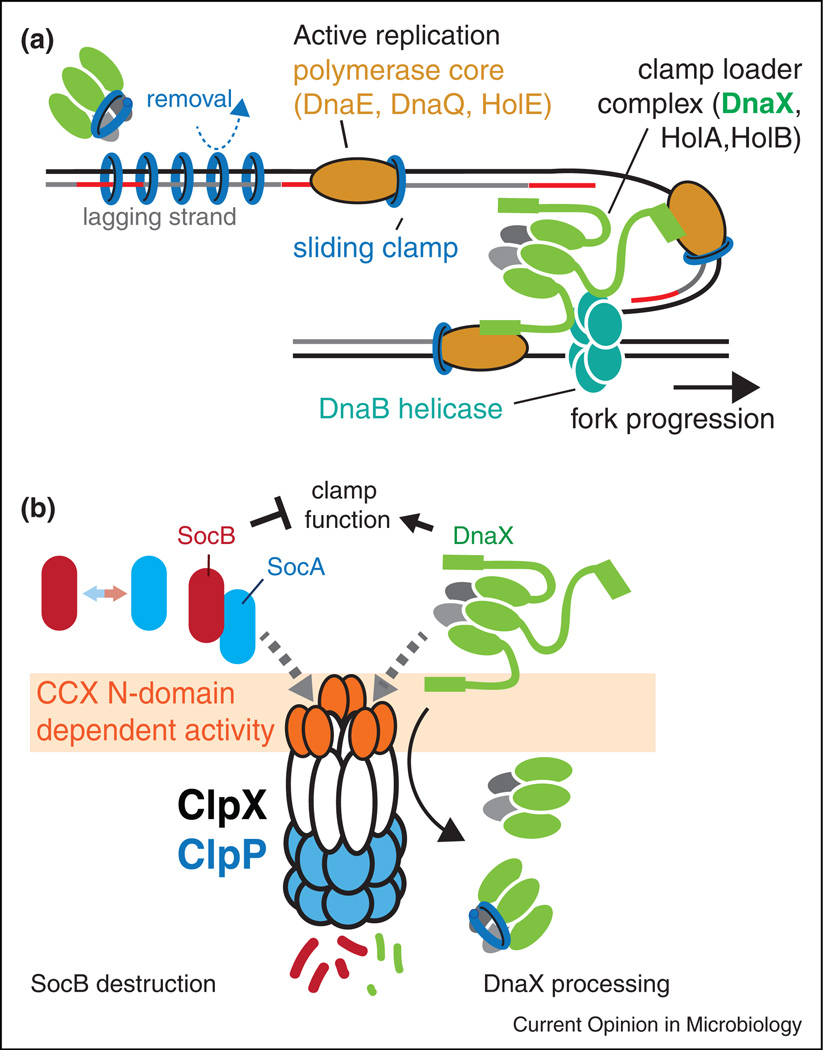
Figure 2.
Replication clamp activity is controlled in part by the ClpXP protease. (a) DnaX is the protein in clamp loader that delivers the mechanical force needed for clamp opening. The full-length clamp loader is tethered to the replication fork by interactions between the C-terminus of DnaX, DNA helicase (DnaB) and the DnaE component of the polymerase. Removal of the C-terminus would release a shortened version of the clamp loader that could act as an unloader away from the replication fork. (b) Under normal circumstances, both DnaX and SocB (with SocA acting as an adaptor) are degraded by ClpXP in an N-domain dependent manner. If the N-domain of ClpX is perturbed during damaging conditions (either by competition or direct damage) both SocB and full-length DnaX levels would rise providing compensating effects. Alternatively, if SocAB levels rise dramatically, this could itself compete for ClpXP, slowing the processing of DnaX and ensuring retention of clamp loading activity at the replication fork or damaged sites.
Another direct link between ClpXP and DNA metabolism in Caulobacter was identified in a proteomic approach that revealed the widespread nature of ClpP substrates [16]. DnaX is the ATP hydrolyzing core subunit of the clamp loader needed for the loading and unloading of the replication clamps (Figure 2a). DnaX was first identified in Escherichia coli, where it was found to exist in two forms generated through programmed ribosomal frameshifting [19–21]. DnaX also exists as multiple forms in Caulobacter but these shorter forms are generated upon partial proteolysis by ClpXP [22]. Both forms are essential and strains engineered to express two DnaX variants locked in either long or short forms are viable. However, these strains are deficient in DNA damage tolerance, suggesting that dynamics of DnaX processing are important for this stress response [22]. Like the SocB example, processing of DnaX also requires the N-domain of ClpX (Figure 2b).
Recently, it was shown that the short form of DnaX is also important for DNA damage tolerance in E. coli, although it is dispensable for viability [23,24•]. What is the short form doing? Prior work found that the short form is sufficient to load/unload clamps but lacks the regions needed to tether the full-length clamp loader to the replication fork [25] (Figure 2a). A tempting hypothesis is that the shorter DnaX clamp loader is dedicated to unloading [26], a feature that could be particularly useful during damaging conditions. Along these lines, a dedicated replication clamp unloader (Elg1) in yeast was recently described in yeast and shown to be important in DNA damage tolerance [27]. Another possibility is that the longer form of DnaX limits exchange of mutagenic polymerases due to the increased interactions with other pol III components [24•]. In this manner, processing of DnaX by ClpXP may assist in polymerase exchange during damaging conditions.
Integration of signals through the ClpX N-domain
From the above results, an intriguing speculation is that ClpXP may help balance clamp dynamics in Caulobacter by degrading an inhibitor of clamp and processing the clamp loader to generate an essential isoform (Figure 2b). Both these pathways rely on the unique N-domain of ClpX [17,22], a domain critical for adaptor binding [10••,11•,28]. If ClpXP activity is compromised due to direct damage or competition from other partners, protein levels of SocB would rapidly increase and DnaX would be less processed, resulting in more clamp inhibition and restricting clamp loader to the replication fork (Figure 2b). Such an occurrence could be beneficial during damaging conditions in order to prepare for quick restart of replication after repair and clamps were freed. Interestingly, direct damage to the ClpX N-domain has been suggested to underlie the transient stabilization of ClpXP substrates in B. subtilis during disulfide stress [29]. The use of a common protease for both loss of clamp function and activation of clamp activity would allow both these activities to change in concert if ClpXP is saturated by a surge in protease substrates. Thus, the N-domain of ClpXP would serve to integrate substrate load as an input with clamp dynamics as an output.
Lon degrades both folded and misfolded substrates
The Lon protease has long been known to be crucial for degrading misfolded or damaged proteins during stress conditions. In Caulobacter, Lon is responsible for both normal and stress related degradation of a several important regulators (Figure 3a). For example, the CcrM methylase is responsible for epigenetic regulation of a number of cell cycle genes [30] and its levels are partially managed by Lon-dependent proteolysis [31]. The SciP protein is a cofactor for CtrA that prevents activation of CtrA controlled genes during the G1 phase of the cell cycle [32,33]. Lon degradation of SciP during the cell cycle is important to remove SciP so that CtrA regulated genes can be activated [13]. Lon was recently shown to degrade the replication initiator DnaA, a function particularly important during proteotoxic stress and starvation conditions [34,35].
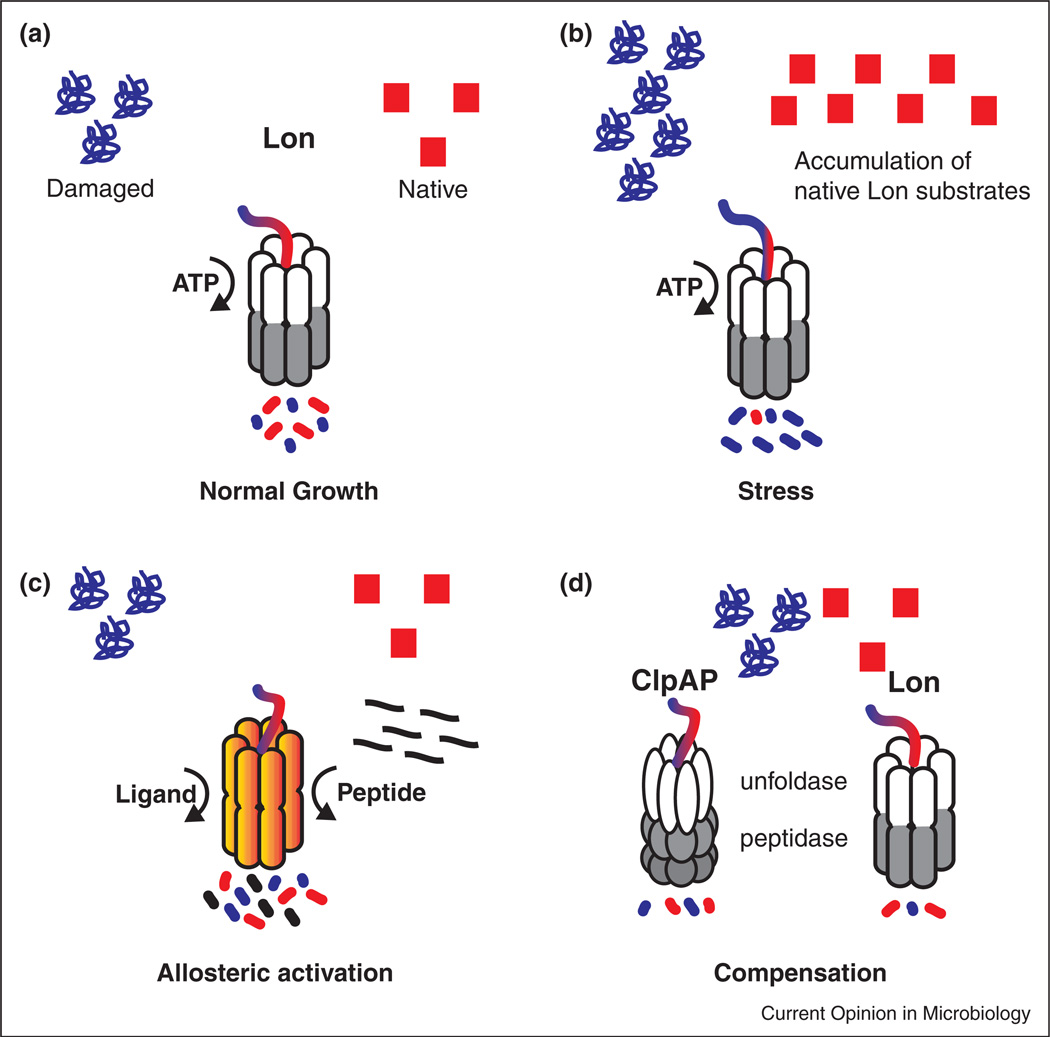
Figure 3.
Lon balances normal degradation with stress related duties. (a) Lon maintains homeostasis by balancing two main functions: protein quality control and destruction of natively folded substrates. (b) Stress results in the accumulation of native or damaged proteins that may saturate Lon and result in a subsequent build up of other substrates. (c). Allosteric activation of Lon by ligands or misfolded proteins can compensate for the extra protease demand in stress conditions. (d) Saturation of Lon can also be compensated for through upregulation of other proteases, such as ClpAP, which has some overlapping specificity with Lon.
The promiscuous nature of Lon has costs and benefits. Because any protein can misfold, a protease that eliminates misfolded proteins must have rather broad specificity. In fact, Lon is thought to recognize features of misfolded proteins such as exposed hydrophobic elements for most of its quality control targets, rather than specific sequences [36]. That said, Lon also clearly recognizes specific substrates even when they are folded, such as DnaA [34]. In addition, Lon specificity can be augmented by adaptors, as shown recently in B. subtilis where adaptor-dependent Lon degradation controls cell motility upon surface contact [37•]. Because Lon must recognize so many targets, it stands to reason that saturation of this protease might readily occur during damaging or stress conditions. Cases of protease saturation have been described and in some cases leveraged for synthetic biology [38–40]. Nonetheless, the unconstrained increase in substrates (either damaged or native) could lead to harm if left unchecked (Figure 3b).
Cellular responses to protease saturation
How could protease systems respond to this toxic consequence? One way is to increase protease capacity through increased production or increased activity. In fact, allosteric stimulation of Lon has been demonstrated in vitro and in vivo where substrate recognition by Lon stimulates the degradation of a second substrate (Figure 3c) [34,41]. In Caulobacter, stimulation of Lon by misfolded protein substrates is thought to underlie the loss of DnaA that arrests the cell cycle during proteotoxic stress [34]. Another method to combat protein overflow is by upregulating other protease activities (Figure 3d). For example, like Lon, the ClpAP protease also degrades poorly folded substrates such as casein [42] and in Caulobacter, the ClpP family of proteases has been implicated in degradation of DnaA [43]. Consistent with this compensation model, overexpression of ClpAP protects against the toxic accumulation of DnaA in cells lacking Lon (unpublished; JL, PC). Given the widespread nature of ClpAP and Lon in gram-negative bacteria, perhaps similar compensation will be found elsewhere.
Finally, we note that Lon has long been known to be involved in DNA damage tolerance. Lon’s ability to regulate DNA damage dates back to Evelyn Witkin’s initial genetic studies of a UV sensitive, naturally Lon-deficient, B strain of E. coli [44]. In addition to radiation sensitivity, it was shown that Lon mutants also showed stabilization of β-galactosidase fragments leading to the discovery of suppressors of Lon (Sul) [45]. These suppressors were mapped to SulA, an FtsZ inhibitor upregulated in response to UV stress [46,47]. When Lon is absent SulA accumulates and irreversibly blocks cell division, ultimately resulting in cell death [48]. Whether Lon regulates DNA damage responses across bacteria is unclear, particularly in bacteria without obvious SulA homologs. In C. crescentus no SulA homolog exists. However, the small proteins SidA and DidA block cell division during DNA damage by inhibiting the divisome proteins FtsW and FtsN, respectively [18,49•]. It remains to be seen if Lon plays a role in the turnover of these cell division inhibitors.
Conclusions
Regulated protein degradation is critical to all life. For AAA+ proteases such as ClpXP and Lon, ensuring specificity is crucial because degradation is irreversible. Substrate choice can be controlled by the intrinsic activity of the protease and/or tuned by adaptor proteins. In some cases, such as during the Caulobacter cell cycle, adaptor proteins assemble into hierarchies that deliver different substrate classes depending on their degree of assembly [10••,11•,12•].
The ClpXP protease in Caulobacter seems to play a fundamental role in replication as it degrades both an inhibitor of replication clamps and generates an essential isoform of the clamp loader complex through partial proteolysis. The Lon protease degrades both misfolded proteins of varying sequences and specific folded proteins, but must balance this breadth of substrate recognition with the cost of being readily saturated. Saturation of one protease type can be balanced by the increased protease activity of the same type (either through increased synthesis or stimulation) or by compensation through upregulation of another protease with overlapping specificity. In some cases, this saturation might result in additional regulation, such as when two opposing factors are recognized through the same protease. Under-standing these increasingly diverse mechanisms of protease regulation and their impact on cell growth or stress response are a rich topic for future exploration.
Acknowledgments
Funding for work in the Chien lab was provided in part by a grant from the National Institutes of Health (R01GM111706) to PC and from a Chemistry Biology Interface Program Training Grant (NIH T32GM08515) to RHV and RZ.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Kraut DA, Israeli E, Schrader EK, Patil A, Nakai K, Nanavati D, Inobe T, Matouschek A. Sequence- and species-dependence of proteasomal processivity. ACS Chem Biol. 2012;7:1444–1453. doi: 10.1021/cb3001155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sauer RT, Baker TA. AAA+ proteases: ATP-fueled machines of protein destruction. Annu Rev Biochem. 2011;80:587–612. doi: 10.1146/annurev-biochem-060408-172623. [DOI] [PubMed] [Google Scholar]
- 3.Jenal U, Fuchs T. An essential protease involved in bacterial cell-cycle control. EMBO J. 1998;17:5658–5669. doi: 10.1093/emboj/17.19.5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duerig A, Abel S, Folcher M, Nicollier M, Schwede T, Amiot N, Giese B, Jenal U. Second messenger-mediated spatiotemporal control of protein degradation regulates bacterial cell cycle progression. Genes Dev. 2009;23:93–104. doi: 10.1101/gad.502409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iniesta AA, McGrath PT, Reisenauer A, McAdams HH, Shapiro L. A phospho-signaling pathway controls the localization and activity of a protease complex critical for bacterial cell cycle progression. Proc Natl Acad Sci U S A. 2006;103:10935–10940. doi: 10.1073/pnas.0604554103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McGrath PT, Iniesta AA, Ryan KR, Shapiro L, McAdams HH. A dynamically localized protease complex and a polar specificity factor control a cell cycle master regulator. Cell. 2006;124:535–547. doi: 10.1016/j.cell.2005.12.033. [DOI] [PubMed] [Google Scholar]
- 7.Taylor JA, Wilbur JD, Smith SC, Ryan KR. Mutations that alter RcdA surface residues decouple protein localization and CtrA proteolysis in Caulobacter crescentus. J Mol Biol. 2009;394:46–60. doi: 10.1016/j.jmb.2009.08.076. [DOI] [PubMed] [Google Scholar]
- 8.Abel S, Chien P, Wassmann P, Schirmer T, Kaever V, Laub MT, Baker TA, Jenal U. Regulatory cohesion of cell cycle and cell differentiation through interlinked phosphorylation and second messenger networks. Mol Cell. 2011;43:550–560. doi: 10.1016/j.molcel.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biondi EG, Reisinger SJ, Skerker JM, Arif M, Perchuk BS, Ryan KR, Laub MT. Regulation of the bacterial cell cycle by an integrated genetic circuit. Nature. 2006;444:899–904. doi: 10.1038/nature05321. [DOI] [PubMed] [Google Scholar]
- 10. Joshi KK, Berge M, Radhakrishnan SK, Viollier PH, Chien P. An adaptor hierarchy regulates proteolysis during a bacterial cell cycle. Cell. 2015;163:419–431. doi: 10.1016/j.cell.2015.09.030. Reveals how an adaptor hierarchy assembles to stage degradation of cell cycle substrates.
- 11. Lau J, Hernandez-Alicea L, Vass RH, Chien P. A phosphosignaling adaptor primes the AAA+ protease ClpXP to drive cell cycle-regulated proteolysis. Mol Cell. 2015;59:104–116. doi: 10.1016/j.molcel.2015.05.014. Shows that binding of a cell cycle adaptor binding to ClpXP prepares it for substrate recognition.
- 12. Smith SC, Joshi KK, Zik JJ, Trinh K, Kamajaya A, Chien P, Ryan KR. Cell cycle-dependent adaptor complex for ClpXP-mediated proteolysis directly integrates phosphorylation and second messenger signals. Proc Natl Acad Sci U S A. 2014;111:14229–14234. doi: 10.1073/pnas.1407862111. Shows that formation of a multi-protein complex is responsible for CtrA degradation.
- 13.Gora KG, Cantin A, Wohlever M, Joshi KK, Perchuk BS, Chien P, Laub MT. Regulated proteolysis of a transcription factor complex is critical to cell cycle progression in Caulobacter crescentus. Mol Microbiol. 2013;87:1277–1289. doi: 10.1111/mmi.12166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lori C, Ozaki S, Steiner S, Bohm R, Abel S, Dubey BN, Schirmer T, Hiller S, Jenal U. Cyclic di-GMP acts as a cell cycle oscillator to drive chromosome replication. Nature. 2015;523:236–239. doi: 10.1038/nature14473. Demonstrates that cyclic di-GMP switches a cell cycle kinase to a phophatase.
- 15. Tan IS, Weiss CA, Popham DL, Ramamurthi KS. A quality-control mechanism removes unfit cells from a population of sporulating bacteria. Dev Cell. 2015;34:682–693. doi: 10.1016/j.devcel.2015.08.009. Shows how adaptor-mediated degradation controls spore quality in B. subtilis.
- 16.Bhat NH, Vass RH, Stoddard PR, Shin DK, Chien P. Identification of ClpP substrates in Caulobacter crescentus reveals a role for regulated proteolysis in bacterial development. Mol Microbiol. 2013;88:1083–1092. doi: 10.1111/mmi.12241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aakre CD, Phung TN, Huang D, Laub MT. A bacterial toxin inhibits DNA replication elongation through a direct interaction with the beta sliding clamp. Mol Cell. 2013;52:617–628. doi: 10.1016/j.molcel.2013.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Modell JW, Hopkins AC, Laub MT. A DNA damage checkpoint in Caulobacter crescentus inhibits cell division through a direct interaction with FtsW. Genes Dev. 2011;25:1328–1343. doi: 10.1101/gad.2038911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blinkowa AL, Walker JR. Programmed ribosomal frameshifting generates the Escherichia coli DNA polymerase III gamma subunit from within the tau subunit reading frame. Nucleic Acids Res. 1990;18:1725–1729. doi: 10.1093/nar/18.7.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flower AM, McHenry CS. The gamma subunit of DNA polymerase III holoenzyme of Escherichia coli is produced by ribosomal frameshifting. Proc Natl Acad Sci U S A. 1990;87:3713–3717. doi: 10.1073/pnas.87.10.3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsuchihashi Z, Kornberg A. Translational frameshifting generates the gamma subunit of DNA polymerase III holoenzyme. Proc Natl Acad Sci U S A. 1990;87:2516–2520. doi: 10.1073/pnas.87.7.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vass RH, Chien P. Critical clamp loader processing by an essential AAA+ protease in Caulobacter crescentus. Proc Natl Acad Sci U S A. 2013;110:18138–18143. doi: 10.1073/pnas.1311302110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blinkova A, Hervas C, Stukenberg PT, Onrust R, O’Donnell ME, Walker JR. The Escherichia coli DNA polymerase III holoenzyme contains both products of the dnaX gene, tau and gamma, but only tau is essential. J Bacteriol. 1993;175:6018–6027. doi: 10.1128/jb.175.18.6018-6027.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dohrmann PR, Correa R, Frisch RL, Rosenberg SM, McHenry CS. The DNA polymerase III holoenzyme contains gamma and is not a trimeric polymerase. Nucleic Acids Res. 2016;44:1285–1297. doi: 10.1093/nar/gkv1510. Shows a biological role for the short DnaX form in DNA damage tolerance in E. coli.
- 25.Kim S, Dallmann HG, McHenry CS, Marians KJ. tau couples the leading- and lagging-strand polymerases at the Escherichia coli DNA replication fork. J Biol Chem. 1996;271:21406–21412. doi: 10.1074/jbc.271.35.21406. [DOI] [PubMed] [Google Scholar]
- 26.Leu FP, Hingorani MM, Turner J, O’Donnell M. The delta subunit of DNA polymerase III holoenzyme serves as a sliding clamp unloader in Escherichia coli. J Biol Chem. 2000;275:34609–34618. doi: 10.1074/jbc.M005495200. [DOI] [PubMed] [Google Scholar]
- 27.Johnson C, Gali VK, Takahashi TS, Kubota T. PCNA retention on DNA into G2/M phase causes genome instability in cells lacking Elg1. Cell Rep. 2016 doi: 10.1016/j.celrep.2016.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chowdhury T, Chien P, Ebrahim S, Sauer RT, Baker TA. Versatile modes of peptide recognition by the ClpX N domain mediate alternative adaptor-binding specificities in different bacterial species. Protein Sci. 2010;19:242–254. doi: 10.1002/pro.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garg SK, Kommineni S, Henslee L, Zhang Y, Zuber P. The YjbH protein of Bacillus subtilis enhances ClpXP-catalyzed proteolysis of Spx. J Bacteriol. 2009;191:1268–1277. doi: 10.1128/JB.01289-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fioravanti A, Fumeaux C, Mohapatra SS, Bompard C, Brilli M, Frandi A, Castric V, Villeret V, Viollier PH, Biondi EG. DNA binding of the cell cycle transcriptional regulator GcrA depends on N6-adenosine methylation in Caulobacter crescentus and other Alphaproteobacteria. PLoS Genet. 2013;9:e1003541. doi: 10.1371/journal.pgen.1003541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wright R, Stephens C, Zweiger G, Shapiro L, Alley MR. Caulobacter Lon protease has a critical role in cell-cycle control of DNA methylation. Genes Dev. 1996;10:1532–1542. doi: 10.1101/gad.10.12.1532. [DOI] [PubMed] [Google Scholar]
- 32.Gora KG, Tsokos CG, Chen YE, Srinivasan BS, Perchuk BS, Laub MT. A cell-type-specific protein-protein interaction modulates transcriptional activity of a master regulator in Caulobacter crescentus. Mol Cell. 2010;39:455–467. doi: 10.1016/j.molcel.2010.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tan MH, Kozdon JB, Shen X, Shapiro L, McAdams HH. An essential transcription factor, SciP, enhances robustness of Caulobacter cell cycle regulation. Proc Natl Acad Sci U S A. 2010;107:18985–18990. doi: 10.1073/pnas.1014395107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jonas K, Liu J, Chien P, Laub MT. Proteotoxic stress induces a cell-cycle arrest by stimulating Lon to degrade the replication initiator DnaA. Cell. 2013;154:623–636. doi: 10.1016/j.cell.2013.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leslie DJ, Heinen C, Schramm FD, Thuring M, Aakre CD, Murray SM, Laub MT, Jonas K. Nutritional control of DNA replication initiation through the proteolysis and regulated translation of DnaA. PLoS Genet. 2015;11:e1005342. doi: 10.1371/journal.pgen.1005342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gur E, Sauer RT. Recognition of misfolded proteins by Lon, a AAA(+) protease. Genes Dev. 2008;22:2267–2277. doi: 10.1101/gad.1670908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mukherjee S, Bree AC, Liu J, Patrick JE, Chien P, Kearns DB. Adaptor-mediated Lon proteolysis restricts Bacillus subtilis hyperflagellation. Proc Natl Acad Sci U S A. 2015;112:250–255. doi: 10.1073/pnas.1417419112. Identification of the first Lon protease adaptor.
- 38.Fredriksson A, Ballesteros M, Peterson CN, Persson O, Silhavy TJ, Nystrom T. Decline in ribosomal fidelity contributes to the 80 Growth and development: prokaryotes accumulation and stabilization of the master stress response regulator sigmaS upon carbon starvation. Genes Dev. 2007;21:862–874. doi: 10.1101/gad.409407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cookson NA, Mather WH, Danino T, Mondragon-Palomino O, Williams RJ, Tsimring LS, Hasty J. Queueing up for enzymatic processing: correlated signaling through coupled degradation. Mol Syst Biol. 2011;7:561. doi: 10.1038/msb.2011.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prindle A, Selimkhanov J, Li H, Razinkov I, Tsimring LS, Hasty J. Rapid and tunable post-translational coupling of genetic circuits. Nature. 2014;508:387–391. doi: 10.1038/nature13238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gur E, Sauer RT. Degrons in protein substrates program the speed and operating efficiency of the AAA+ Lon proteolytic machine. Proc Natl Acad Sci U S A. 2009;106:18503–18508. doi: 10.1073/pnas.0910392106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Katayama-Fujimura Y, Gottesman S, Maurizi MR. A multiple-component, ATP-dependent protease from Escherichia coli. J Biol Chem. 1987;262:4477–4485. [PubMed] [Google Scholar]
- 43.Gorbatyuk B, Marczynski GT. Regulated degradation of chromosome replication proteins DnaA and CtrA in Caulobacter crescentus. Mol Microbiol. 2005;55:1233–1245. doi: 10.1111/j.1365-2958.2004.04459.x. [DOI] [PubMed] [Google Scholar]
- 44.Witkin EM, Sicurella NA, Bennett GM. Photoreversibility of induced mutations in a nonphotoreactivable strain of Escherichia Coli. Proc Natl Acad Sci U S A. 1963;50:1055–1059. doi: 10.1073/pnas.50.6.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gottesman S, Zipser D. Deg phenotype of Escherichia coli lon mutants. J Bacteriol. 1978;133:844–851. doi: 10.1128/jb.133.2.844-851.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lutkenhaus J, Sanjanwala B, Lowe M. Overproduction of FtsZ suppresses sensitivity of lon mutants to division inhibition. J Bacteriol. 1986;166:756–762. doi: 10.1128/jb.166.3.756-762.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schoemaker JM, Gayda RC, Markovitz A. Regulation of cell division in Escherichia coli: SOS induction and cellular location of the sulA protein, a key to lon-associated filamentation and death. J Bacteriol. 1984;158:551–561. doi: 10.1128/jb.158.2.551-561.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mukherjee A, Cao C, Lutkenhaus J. Inhibition of FtsZ polymerization by SulA, an inhibitor of septation in Escherichia coli. Proc Natl Acad Sci U S A. 1998;95:2885–2890. doi: 10.1073/pnas.95.6.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Modell JW, Kambara TK, Perchuk BS, Laub MT. A DNA damage-induced, SOS-independent checkpoint regulates cell division in Caulobacter crescentus. PLoS Biol. 2014;12:e1001977. doi: 10.1371/journal.pbio.1001977. Identification of a novel cell division inhibitor upregulated during DNA damage.