Abstract
In 1970, a kinase activity that phosphorylated a minor species of seryl-tRNA to form phosphoseryl-tRNA was found in rooster liver [Maenpaa, P. H. & Bernfield, M. R. (1970) Proc. Natl. Acad. Sci. USA 67, 688–695], and a minor seryl-tRNA that decoded the nonsense UGA was detected in bovine liver. The phosphoseryl-tRNA and the minor UGA-decoding seryl-tRNA were subsequently identified as selenocysteine (Sec) tRNA[Ser]Sec, but the kinase activity remained elusive. Herein, by using a comparative genomics approach that searched completely sequenced archaeal genomes for a kinase-like protein with a pattern of occurrence similar to that of components of Sec insertion machinery, we detected a candidate gene for mammalian phosphoseryl-tRNA[Ser]Sec kinase (pstk). Mouse pstk was cloned, and the gene product (PSTK) was expressed and characterized. PSTK specifically phosphorylated the seryl moiety on seryl-tRNA[Ser]Sec and, in addition, had a requirement for ATP and Mg2+. Proteins with homology to mammalian PSTK occur in Drosophila, Caenorhabditis elegans, Methanopyrus kandleri, and Methanococcus jannaschii, suggesting a conservation of its function across archaea and eukaryotes that synthesize selenoproteins and the absence of this function in bacteria, plants, and yeast. The fact that PSTK has been highly conserved in evolution suggests that it plays an important role in selenoprotein biosynthesis and/or regulation.
Selenocysteine (Sec) has its own code word, UGA, and its own tRNA, and therefore is viewed as the 21st amino acid in the genetic code (reviewed in refs. 1–4). Although UGA usually codes for the termination of protein synthesis, it also specifies Sec if specific requirements are met. The presence of a stem–loop structure downstream of UGA, called a Sec insertion sequence (SECIS) element, is the critical component in seleno-protein mRNAs that dictates UGA to code for Sec (reviewed in ref. 5). In mammals, the SECIS element occurs in the 3′ untranslated region of selenoprotein mRNAs. A specific elongation factor, EFsec, specifically recognizes selenocysteyl-tRNA[Ser]Sec (6, 7), and a SECIS element binding protein, SBP2, binds specifically to the SECIS element (8), directing the insertion of Sec into protein in response to UGA.
It has been known for several years that the biosynthesis of Sec occurs on its tRNA in both bacteria (9) and mammals (10) after the tRNA is initially aminoacylated with serine by seryl-tRNA synthetase. In Escherichia coli, the pathway for the biosynthesis of Sec has been completely established (reviewed in ref. 1). After the aminoacylation of bacterial tRNA[Ser]Sec with serine, the hydroxyl group is removed from the seryl moiety to yield an aminoacrylyl intermediate, and this step is catalyzed by a pyridoxal phosphate-dependent Sec synthase. The aminoacrylyl intermediate serves as the acceptor for the activated selenium donor, monoselenophosphate, which is synthesized from selenite and ATP in the presence of selenophosphate synthetase (reviewed in ref. 1). Once selenium is donated to the intermediate, the biosynthesis of Sec on tRNA[Ser]Sec is complete.
In eukaryotes, however, the biosynthesis of Sec has not been established, but several components have been identified over the years that play a role in this process. For example, in 1970, a minor seryl-tRNA was reported to form phosphoseryl-tRNA by a kinase activity from rooster liver (11), and a minor seryl-tRNA from bovine, rabbit, and chicken livers was reported to specifically decode the nonsense codon UGA (12). Although it was subsequently shown that both the minor seryl-tRNA that formed phosphoseryl-tRNA and the one that decoded UGA were the same tRNA (13), it was not known at the time these components were discovered that they played instrumental roles in the biosynthesis of Sec and its incorporation into protein as the 21st amino acid in the genetic code. The minor serine tRNA was subsequently found to be selenocysteyl-tRNA[Ser]Sec (10), but the role of the putative kinase that phosphorylated seryl-tRNA[Ser]Sec has not been resolved. Higher vertebrates contain two Sec tRNA[Ser]Sec isoforms that differ from each other by a single methyl group that occurs at the 2′-hydroxyl of ribose in the wobble position of the anticodon. One isoform, containing 5-methylcarboxymethyluridine (mcm5U), is the precursor of the other, which contains 5-methylcarboxymethyluridine-2′-O-methylribose (mcm5Um) (reviewed in ref. 2).
In the present study, a computational search of several archaeal and eukaryotic genomes for the presence of a kinase-like gene that occurred in those organisms containing the Sec insertion machinery but not in those organisms lacking this machinery revealed a candidate phosphoseryl-tRNA[Ser]Sec kinase gene (designated pstk). The pstk gene product (PSTK) was expressed and characterized. It indeed specifically phosphorylates seryl-tRNA[Ser]Sec. These and other properties of PSTK are reported herein.
Methods
Materials. Selenium-75 (specific activity 1,000 Ci/mmol; 1 Ci = 37 GBq) was obtained from the Research Reactor Facility, University of Missouri, Columbia. [γ-32P]ATP (specific activity ≈6,000 Ci/mmol), [α-32P]ATP (specific activity ≈3,000 Ci/mmol), and [3H]serine (specific activity 29 Ci/mmol) were obtained from Amersham Biosciences. The following were purchased: Ni-NTA (nitrilotriacetate) agarose from Qiagen (Valencia, CA); Pfu DNA polymerase and pBluescript II from Stratagene; the pET-16b expression vector, the pET-32b vector (encoding 109-aa thioredoxin with a His-tag), and BL21(DE3) competent cells from Novagen; polynucleotide kinase and Nu-PAGE Bis-Tris gels from Invitrogen; alkaline phosphatase from New England Biolabs; T7 RiboMAX Express Large Scale RNA Production Systems and Wizard Genomic DNA Purification Kit from Promega; and puReTaq Ready-To-Go PCR Beads and Hybond-N+ nylon membranes from Amersham Biosciences. Mung bean nuclease was purchased from New England Biolabs, and P81 phosphocellulose filter paper (catalog no. 3698325) was purchased from Whatman. I.M.A.G.E. Consortium Clone ID 654975 encoding pstk was obtained from the American Type Culture Collection. All other reagents were commercial products of the highest grade available.
Computational Search for a Candidate PSTK. The strategy to search for the Sec kinase was to identify all predicted proteins in the Methanopyrus kandleri genome for which close homologs were present in Methanococcus jannaschii but were absent in 12 other completely sequenced archaeal genomes that did not code for Sec insertion machinery (Aeropyrum pernix, Archaeoglobus fulgidus, Halobacterium, Methanobacterium thermoautotrophicum, Pyrobaculum aerophilum, Pyrococcus abyssi, Pyrococcus furiosus, Pyrococcus horikoshii, Sulfolobus solfataricus, Sulfolobus toko-daii, Thermoplasma acidophilum, and Thermoplasma volcanium). To achieve this goal, each of the 1,687 annotated Methanopyrus kandleri proteins was searched against complete, annotated archaeal proteomes with the blastp program. A candidate protein was considered to have a close homolog in the genome if two criteria were satisfied for this protein: (i) a hit with an E-value below 10–12 was obtained and (ii) when this protein hit was searched against a full set of predicted Methanopyrus kandleri proteins, the closest homolog was either the original Methanopyrus kandleri protein or the protein that, when compared with the original protein by using the National Center for Biotechnology Information bl2seq program, had an E-value below 10–20. By using this algorithm, 27 candidate ORFs were identified, which were further screened for homology to known kinase domains and proteins annotated in the National Center for Biotechnology Information nonredundant database as kinases. This analysis resulted in two proteins (gi: 20093704 and 20095115), which were further analyzed for the presence of homologs in completely sequenced eukaryotic genomes. Only the protein annotated as “predicted nucleotide kinase” (gi: 20095115) had homologs in eukaryotes. Furthermore, these homologs were present in organisms with a functional Sec insertion system and were absent in eukaryotes lacking this system.
Cloning, Expression, and Purification of PSTK. pstk cDNA was amplified by using Pfu DNA polymerase and a forward primer, 5′-GGGAATTCCATATGGACTACAAGGACGACGATGACAAAATGAAGACCGCGGCGGCTCGTG-3′, to generate seven bases missing at the amino terminus (underlined bases), an immediate upstream Flag-tag (bolded bases), and an NdeI restriction site (italicized bases). A reverse primer, 5′-CGCGGATCCTTAATGTTGCTTTGAAAAATACTTC, was used to generate a BamHI restriction site (italicized bases) immediately downstream of the termination signal (italicized and underlined bases) at the carboxyl terminus. The resulting product was cloned into the pET-16b vector at the NdeI–BamHI cloning site in which the vector contained a His-tag immediately upstream of, and in-frame with, the Flag-tag and the pstk ORF. The sequence of the cDNA construct was confirmed by sequencing the product generated from the T7 promoter and T7 termination primers encoded in the pET-16b vector. BL21(DE3) cells were transformed with the construct. For expressing PSTK, a colony was inoculated in 200 ml of LB containing 100 μg/ml ampicillin. When the OD600 of the culture reached 0.6, 0.5 mM isopropyl β-d-thiogalactoside was added and expression was induced for 4 h. The cells were harvested and Ni-NTA agarose was used to purify PSTK in its native condition, according to the vendor's instructions. PSTK was eluted, dialyzed against 1× PBS, and stored at –20°C in 50% glycerol.
Isolation, Aminoacylation, and Fractionation of Natural and Synthetic tRNA. Total tRNA was isolated from bovine liver and then aminoacylated with 20 unlabeled amino acids or 19 unlabeled amino acids and [3H]serine in the presence of a fresh preparation of rabbit reticulocyte aminoacyl-tRNA synthetases containing all 20 active synthetases (14). The resulting aminoacyl-tRNAs were fractionated on an RPC-5 column (15) and tRNA was precipitated from individual fractions and collected as described in ref. 14. Both Sec tRNA[Ser]Sec isoforms and serine 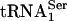 were purified to homogeneity, as described in ref. 16. The 32P-labeled probe used in detecting seryl-tRNA[Ser]Sec or tRNA[Ser]Sec is described in ref. 17. Synthetic tRNA[Ser]Sec was prepared from the pBluescript II expression vector that encoded the mouse Sec tRNA[Ser]Sec gene. The gene had been amplified by using a PCR and then cloned into the vector. The 3′ end of the Sec tRNA[Ser]Sec gene was cut with HindIII and blunt-ended with mung bean nuclease, Sec tRNA[Ser]Sec was transcribed by using the T7 RiboMAX Express Large Scale RNA Production System, and the resulting product was purified according to the manufacturer's instructions. The 5′ triphosphate that was present on synthetic tRNA[Ser]Sec was removed by incubation of the freshly synthesized RNA with alkaline phosphatase and isolation of tRNA[Ser]Sec after extraction with phenol and precipitation in ethanol (16).
were purified to homogeneity, as described in ref. 16. The 32P-labeled probe used in detecting seryl-tRNA[Ser]Sec or tRNA[Ser]Sec is described in ref. 17. Synthetic tRNA[Ser]Sec was prepared from the pBluescript II expression vector that encoded the mouse Sec tRNA[Ser]Sec gene. The gene had been amplified by using a PCR and then cloned into the vector. The 3′ end of the Sec tRNA[Ser]Sec gene was cut with HindIII and blunt-ended with mung bean nuclease, Sec tRNA[Ser]Sec was transcribed by using the T7 RiboMAX Express Large Scale RNA Production System, and the resulting product was purified according to the manufacturer's instructions. The 5′ triphosphate that was present on synthetic tRNA[Ser]Sec was removed by incubation of the freshly synthesized RNA with alkaline phosphatase and isolation of tRNA[Ser]Sec after extraction with phenol and precipitation in ethanol (16).
PSTK Assay. Total aminoacyl-tRNA, fractionated aminoacyl-tRNAs, purified 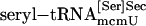 , purified
, purified 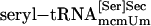 , synthetic seryl-tRNA[Ser]Sec, or purified
, synthetic seryl-tRNA[Ser]Sec, or purified 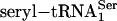 was used as a substrate for PSTK in 50 μl of reaction mixture [20 mM Tris·HCl (pH 7.4)/0.01 mM EGTA/1 mM DTT/10 mM MgCl2/50 μM ATP/1 μl of [γ-32P]ATP (final specific activity ≈600 Ci/mmol)/0.5 μg of pure PSTK]. Coupled aminoacylation–phosphorylation reactions involved the same components plus rabbit reticulocyte aminoacyl-tRNA synthetases (14). Reactions were incubated for 20 min or at time intervals designated in figures at 30°C and were spotted on P81 phosphocellulose filter paper. The filters were washed four times with 0.75% phosphoric acid (≈50 ml per wash) and dried, and radio-activity was measured in a liquid scintillation counter. [32P]Phospho[3H]seryl-tRNA was deacylated, and the products were identified as described in ref. 13. The phosphorylated adenosine product of the kinase reaction was identified by using [α-32P]ATP and analyzing the 32P-labeled product by TLC. The reversibility of PSTK was assessed by assaying aliquots as described above in reactions with and without ADP. The level of γ-32P-labeled ATP generated from ADP and O-[32P]phosphoseryl-tRNA[Ser]Sec at the end of the incubation period and the amount of remaining O-[32P]phosphoserine (after deacylation of O-[32P]phosphoseryl-tRNA[Ser]Sec) were determined by analyzing the 32P counts in the products ([γ-32P]ATP and O-[32P]phosphoserine) by cutting out filter strips after TLC (18).
was used as a substrate for PSTK in 50 μl of reaction mixture [20 mM Tris·HCl (pH 7.4)/0.01 mM EGTA/1 mM DTT/10 mM MgCl2/50 μM ATP/1 μl of [γ-32P]ATP (final specific activity ≈600 Ci/mmol)/0.5 μg of pure PSTK]. Coupled aminoacylation–phosphorylation reactions involved the same components plus rabbit reticulocyte aminoacyl-tRNA synthetases (14). Reactions were incubated for 20 min or at time intervals designated in figures at 30°C and were spotted on P81 phosphocellulose filter paper. The filters were washed four times with 0.75% phosphoric acid (≈50 ml per wash) and dried, and radio-activity was measured in a liquid scintillation counter. [32P]Phospho[3H]seryl-tRNA was deacylated, and the products were identified as described in ref. 13. The phosphorylated adenosine product of the kinase reaction was identified by using [α-32P]ATP and analyzing the 32P-labeled product by TLC. The reversibility of PSTK was assessed by assaying aliquots as described above in reactions with and without ADP. The level of γ-32P-labeled ATP generated from ADP and O-[32P]phosphoseryl-tRNA[Ser]Sec at the end of the incubation period and the amount of remaining O-[32P]phosphoserine (after deacylation of O-[32P]phosphoseryl-tRNA[Ser]Sec) were determined by analyzing the 32P counts in the products ([γ-32P]ATP and O-[32P]phosphoserine) by cutting out filter strips after TLC (18).
Binding of tRNA to PSTK. PSTK or thioredoxin containing a His-tag, which was used as a control protein, was added in a total volume of 100 μl containing reaction mixture (see above) with or without 50 μM unlabeled ATP and 500 ng of a highly purified sample of a mixture of both tRNA[Ser]Sec isoforms or highly purified  ,
, 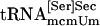 ,
, 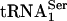 , or synthetic tRNA[Ser]Sec for 30 min at room temperature. Ni-NTA agarose (20 μl) was added to pull down PSTK or the control protein. After washing three times with 1 ml of 1× TBS/0.1% Tween, the beads were suspended in 40 μl of TBE-urea loading buffer (90 mM Tris/64.6 mM boric acid/2.5 mM EDTA/3.5 M urea, pH 8.3), and 5 μl of each sample was loaded onto a 15% TBE-urea gel. After electrophoresis and transfer of the RNA to a nylon membrane, RNA was detected by Northern blotting with the appropriate probe (17).
, or synthetic tRNA[Ser]Sec for 30 min at room temperature. Ni-NTA agarose (20 μl) was added to pull down PSTK or the control protein. After washing three times with 1 ml of 1× TBS/0.1% Tween, the beads were suspended in 40 μl of TBE-urea loading buffer (90 mM Tris/64.6 mM boric acid/2.5 mM EDTA/3.5 M urea, pH 8.3), and 5 μl of each sample was loaded onto a 15% TBE-urea gel. After electrophoresis and transfer of the RNA to a nylon membrane, RNA was detected by Northern blotting with the appropriate probe (17).
PSTK Antibodies. Polyclonal antibodies against PSTK were generated in rabbits by Spring Valley Laboratories (Woodbine, MD) by using a peptide encoding a conjugated predicted amino-terminal epitope region (GATRRDGQPKLGLC-OH) as antigen.
Results
Computational Search for a Sec Kinase. The requirements for Sec biosynthesis appear to be similar in eukaryotes and archaea in regard to the location of SECIS elements in the 3′ untranslated region and the separation of functions for SECIS binding and Sec insertion in separate proteins in these organisms (2–4). In contrast, in bacteria, the SECIS elements are located immediately downstream of the UGA Sec codon, and SECIS binding and Sec insertion functions are carried out by a single protein, designated SelB (1). We therefore hypothesized that the formation of phosphoserine on Sec tRNA might also be conserved between eukaryotes and archaea.
To test this hypothesis, we attempted to identify a putative kinase-like protein that is encoded in Methanopyrus kandleri and Methanococcus jannaschii genomes, the two archaea that encode the Sec insertion machinery (19, 20), but is absent in 12 other completely sequenced archaeal genomes that do not code for the Sec insertion machinery. The search, performed as described in Methods, yielded only 27 predicted ORFs common to Methanopyrus kandleri and Methanococcus jannaschii genomes and absent in other archaeal genomes. Of these, two proteins were homologous to kinases and were annotated as “predicted sugar kinase” (gi: 20093704) and “predicted nucleotide kinase” (gi: 20095115). The first protein had no homologs in eukaryotes and thus could not be a Sec kinase common to archaea and eukaryotes. The second protein had homologs in several eukaryotic organisms but no homologs in bacteria. Interestingly, the occurrence of homologs in eukaryotes closely matched the presence of known components of eukaryotic Sec insertion machinery (selenophosphate synthetase, SBP2, and EFSec). For example, the kinase homologs were present in C. elegans, Drosophila, and mammals, which are known to have selenoproteins, but were absent in yeast and higher plants, which do not synthesize selenoproteins and lack the components of the Sec insertion machinery.
The predicted protein sequences of the candidate mouse and human PSTKs are shown in Fig. 1 and are compared to orthologs in Drosophila, C. elegans, and the archaea Methanopyrus kandleri and Methanococcus jannaschii. The mouse and human PSTKs have 80% identity. The predicted ATP-binding domain appears to be located at residues 25–32, and it has a high degree of homology in each organism, as shown in Fig. 1.
Fig. 1.
Alignment of PSTK sequences. Accession numbers for PSTK sequences are as follows: Mouse, XP_133874; human, NP_699167; C. elegans, CAB11556; Drosophila, NP_788937; Methanococcus jannaschii, NP_248546; and Methanopyrus kandleri, NP_614962. Consensus PSTK sequence is shown below the alignment.
Sequence of pstk. The I.M.A.G.E. Consortium Clone ID 6654975 was sequenced and found to contain the complete coding sequence of pstk, with the exception of seven bases at the amino terminus. As described in Methods, the missing bases were inserted at the 5′ end (Fig. 2A). The pstk coding sequence is 1,080 nucleotides in length and encodes a 359-aa protein.
Fig. 2.
Sequence and expression of pstk. (A) Nucleotide sequence. The cloned pstk sequence is shown. The ATG initiation and TAA termination codons are indicated in bold. (B) Expression of PSTK in bacteria. Lane 1 shows Coomassie-blue-stained gel, and lane 2 shows Western blotting. PSTK was expressed in E. coli cells as a His-tagged protein; the recombinant protein was isolated and probed with polyclonal antibodies against the amino-terminal peptide of PSTK.
Expression and Isolation of PSTK. PSTK, after its expression and purification (see Methods), was run on a denaturing gel, and, after electrophoresis, the gel was either stained with Coomassie blue (Fig. 2B, lane 1) or exposed to antibodies generated against an amino-terminal peptide of mouse PSTK (Fig. 2B, lane 2). This highly purified sample of PSTK was used for subsequent characterization.
PSTK Activity. The rates of aminoacylation of tRNA[Ser]Sec and phosphorylation of seryl-tRNA[Ser]Sec as independent reactions and as a coupled reaction are shown in Fig. 3 A and B, respectively. The aminoacylation of tRNA[Ser]Sec proceeded slowly and reached a maximal level at ≈4 min, and then the seryl-tRNA[Ser]Sec product began to deacylate. The rapid decline in product was most certainly due to the amount of ATP used in reaction mixtures, which was much lower than that previously used in generating seryl-tRNA[Ser]Sec with mammalian seryl-tRNA synthetases (14). Phosphorylation of seryl-tRNA[Ser]Sec occurred rapidly and reached a plateau in ≈3–4 min (Fig. 3A). As expected, in the coupled reaction, the rate of phosphorylation depended on the rate of aminoacylation. The coupled reaction initially proceeded rapidly, and then phosphorylation occurred more slowly over the course of the reaction period (Fig. 3B). The data demonstrated that the candidate PSTK phosphorylated seryl-tRNA[Ser]Sec.
Fig. 3.
PSTK activity. (A) Aminoacylation and phosphorylation as independent reactions. Aminoacylation of synthetic tRNA[Ser]Sec with [3H]serine and phosphorylation of [3H]seryl-tRNA[Ser]Sec with [γ-32P]ATP were carried out. (B) Aminoacylation and phosphorylation as coupled reactions. Coupled reactions in which aminoacylation and phosphorylation occurred in the same reaction with [3H]serine and [γ-32P]ATP were carried out. Use of thioredoxin as a control protein in place of PSTK in the kinase reaction showed no phosphorylation activity of tRNA[Ser]Sec.(C) Divalent metal cation requirement. Reactions for determining the cofactor requirement were carried out with increasing concentrations of divalent cation. (D) Reversibility of the reaction. Aliquots were removed from reactions with or without ADP, and the amount of radioactivity remaining in O-[32P]phosphoseryl-tRNA[Ser]Sec was determined by the phosphocellulose filter assay at the time intervals shown. At the end of the reaction, the remaining sample was deacylated, and the radioactivity present in ATP and in O-phosphoserine was determined by measuring strips that were cut out from a developed TLC chromatogram. The reaction was reversed 26.6%, as determined by the radioactivity present in ATP (8.25 × 104 cpm) divided by the total counts present in ATP and O-phosphoserine (3.105 × 105 cpm). Details of the reactions involving aminoacylation, phosphorylation, cofactor requirement, and reversibility are given in Methods.
Several divalent cations were examined to assess their requirement for kinase activity (Fig. 3C). Although there was no activity at 0.01 mM with any of the three divalent cations examined, a small amount of activity occurred at 0.1 mM. At higher levels, Mg2+ was found to be essential for PSTK activity, and the maximal activity level occurred at 1.0 mM. Mn2+ could replace Mg2+ as a divalent metal cation, but it was not as efficient as Mg2+.
To examine the reversibility of the kinase reaction, O-[32P]phosphoseryl-tRNA[Ser]Sec was incubated in a mixture with Mg2+ and PSTK and with or without 50 μM ADP, and the loss of 32P was monitored over a 20-min period (Fig. 3D). In the absence of ADP, ≈18% of the total O-phosphoseryl-tRNA[Ser]Sec was lost, whereas ≈45% was lost in the presence of ADP. These data suggest that ≈30% of O-phosphoseryl-tRNA[Ser]Sec was converted to seryl-tRNA[Ser]Sec and ATP during the course of the reaction. This result was confirmed by determining the 32P radioactivity present in these two components at the end of the incubation period (Fig. 3D). Comparison of the reversibility of the aminoacylation and phosphorylation reactions suggests that attachment of the phosphate moiety to serine enhances the stability of this aminoacyl group on tRNA[Ser]Sec.
PSTK Reaction Products. As shown above, the starting components in the PSTK reaction are seryl-tRNA[Ser]Ser, ATP, and Mg2+. However, it was important to also determine the products in the reaction. Clearly, the γ-phosphate group on ATP was transferred to ser yl-tRNA[Ser]Sec to yield O-phosphoser yl-tRNA[Ser]Sec. To verify that the adenosine phosphate product is ADP, we used [α-32P]ATP as a substrate in place of [γ-32P]ATP and identified the adenosine phosphate product as ADP by TLC (see Methods; data not shown).
Furthermore, deacylation of [32P]phospho[3H]seryl-tRNA[Ser]Sec and chromatography of the resulting amino acid before and after treatment with alkaline phosphatase demonstrated that the deacylated product migrated with O-phosphoserine, whereas the alkaline phosphatase-treated products migrated with inorganic phosphate (32P) and serine (3H) (data not shown).
Because tRNA[Ser]Sec is the only known tRNA to begin transcription within the coding sequence (2) and therefore has a5′ triphosphate, we examined whether the presence or absence of the 5′ triphosphate had any effect on PSTK activity and whether PSTK could possibly replace a phosphate group at the 5′ end. The presence or absence of the 5′ triphosphate had no effect on PSTK activity, and PSTK did not replace the 5′ phosphate group (data not shown).
PSTK Specificity. To determine the specificity of PSTK, total aminoacyl-tRNA was fractionated on an RPC-5 column as shown in Fig. 4. The two major Sec tRNA[Ser]Sec isoforms are known to be hydrophobic and elute late from the column (see ref. 17 and references therein). Their elution in the latter portion of the column run was confirmed by assaying individual fractions with a 32P-labeled probe that specifically recognized Sec tRNA[Ser]Sec. Both isoforms eluted within column fractions 50 and 72. Assay of the column fractions with PSTK and [γ-32P]ATP demonstrated that only the two Sec tRNA[Ser]Sec isoforms became labeled, providing strong evidence that the kinase is specific for these tRNAs. In addition, seryl-tRNASer1 did not serve as a substrate for PSTK.
Fig. 4.
Specificity of PSTK. Total calf liver tRNA that had been aminoacylated with all 20 unlabeled amino acids in the presence of rabbit reticulocyte synthetases was fractionated on an RPC-5 column (total of 150 A260 units). The aminoacyl-tRNAs in individual fractions were precipitated, the precipitants were collected and redissolved, and an aliquot was spotted on a nitrocellulose filter and hybridized with a specific Sec tRNA[Ser]Sec probe or added to a reaction containing PSTK and the components for measuring kinase activity. Arbitrary units represent either the 32P determined in the kinase reaction by measuring radioactivity of samples in a scintillation counter (e.g., 10,000 cpm = 10,000 arbitrary units) or the PhosphorImager units measured with 32P-labeled probe in a PhosphorImager (Molecular Dynamics) (e.g., 10,000 PhosphorImager units = 400 arbitrary units).
Binding of Acylated and Unacylated tRNA[Ser]Sec to PSTK. The ability of tRNA[Ser]Sec, seryl-tRNA[Ser]Sec, and phosphoseryl-tRNA[Ser]Sec to bind to PSTK in the presence or absence of ATP was examined (Fig. 5A) by Northern-blot analysis of the tRNA with the appropriate labeled tRNA probes. Thioredoxin was used as a control protein in the binding assay. Synthetic seryl-tRNA[Ser]Sec bound to PSTK (lane 2), but addition of ATP to the reaction, which resulted in the formation of O-phosphoseryl-tRNA[Ser]Sec, reduced the binding (lane 3). The double bands observed in lanes 2 and 3 most likely represent acylated and unacylated tRNA[Ser]Sec. Synthetic tRNA[Ser]Sec also bound to PSTK, and the binding appeared to be enhanced by the addition of ATP (compare lanes 5 and 6). A mixture of  and
and  bound to PSTK, but when ATP was added to the reaction the amount of tRNA binding was reduced (compare lanes 8 and 9), as was observed with synthetic seryl-tRNA[Ser]Sec (see lanes 2 and 3). Purified
bound to PSTK, but when ATP was added to the reaction the amount of tRNA binding was reduced (compare lanes 8 and 9), as was observed with synthetic seryl-tRNA[Ser]Sec (see lanes 2 and 3). Purified 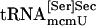 and
and 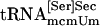 also bound (lanes 11 and 12, respectively), but the former isoform appeared to bind more efficiently. tRNASer1 did not bind to PSTK (data not shown). Neither the naturally occurring isoforms nor the synthetic form bound to the control protein (lanes 1, 4, 7, and 10).
also bound (lanes 11 and 12, respectively), but the former isoform appeared to bind more efficiently. tRNASer1 did not bind to PSTK (data not shown). Neither the naturally occurring isoforms nor the synthetic form bound to the control protein (lanes 1, 4, 7, and 10).
Fig. 5.
Interaction of tRNA[Ser]Sec and PSTK. (A) Binding of tRNA[Ser]Sec to PSTK. Synthetic seryl-tRNA[Ser]Sec was used in lanes 1–3, synthetic tRNA[Ser]Sec in lanes 3–6, a mixture of both naturally occurring seryl-tRNA[Ser]Sec isoforms in lanes 7–9, a mixture of both naturally occurring tRNA[Ser]Sec isoforms in lane 10, 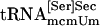 in lane 11, and
in lane 11, and 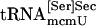 in lane 12. The control protein was thioredoxin. Reaction mixtures were incubated, the proteins were pulled down with Ni-NTA agarose, and Northern blots were prepared as described in Methods. Each reaction was carried out at least three times, yielding similar results, and representative samples are shown. (B) Effect of tRNA[Ser]Sec on phosphorylation. Synthetic [3H]seryl-tRNA[Ser]Sec (2 μg) was added to PSTK reactions with increasing concentrations of synthetic tRNA[Ser]Sec as indicated. The percent aminoacylation used in reactions was 62%, and this amount of unacylated tRNA[Ser]Sec (0.76 μg) was taken into account, as indicated in the initial reaction.
in lane 12. The control protein was thioredoxin. Reaction mixtures were incubated, the proteins were pulled down with Ni-NTA agarose, and Northern blots were prepared as described in Methods. Each reaction was carried out at least three times, yielding similar results, and representative samples are shown. (B) Effect of tRNA[Ser]Sec on phosphorylation. Synthetic [3H]seryl-tRNA[Ser]Sec (2 μg) was added to PSTK reactions with increasing concentrations of synthetic tRNA[Ser]Sec as indicated. The percent aminoacylation used in reactions was 62%, and this amount of unacylated tRNA[Ser]Sec (0.76 μg) was taken into account, as indicated in the initial reaction.
Because tRNA[Ser]Sec bound to PSTK, we examined how efficiently unacylated tRNA[Ser]Sec inhibited the kinase reaction (Fig. 5B). Phosphorylation of the seryl moiety was dramatically inhibited by increasing the amount of unacylated tRNA[Ser]Sec in the reaction, suggesting that tRNA[Ser]Sec was indeed a potent inhibitor of the reaction. The fact that tRNA[Ser]Sec is a potent inhibitor of the kinase reaction precluded the determination of the Km or affinity constant for the two isoforms.
Discussion
PSTK was originally discovered >30 years ago when a kinase activity was detected in rooster liver that catalyzed the formation of phosphoseryl-tRNA on a minor isoform of the total seryl-tRNA population (11). Since this original report, the identity, isolation, and cellular role of PSTK, as well as phosphoseryl-tRNA[Ser]Sec, have remained largely obscure. The minor phosphoseryl-tRNA and another minor seryl-tRNA identified in bovine, rabbit, and chicken livers that decoded the stop codon UGA (12) were shown to be Sec tRNA[Ser]Sec (13). It was subsequently reported that phosphoseryl-tRNA[Ser]Sec was the intermediate in the biosynthesis of Sec in mammals (21, 22) and in bacteria (23). However, after the identification of aminoacryl-tRNA[Ser]Sec as the correct intermediate in bacteria (reviewed in ref. 1), it was reported that phosphoseryl-tRNA[Ser]Sec was not an intermediate in Sec biosynthesis in mammals and that Sec biosynthesis proceeded by the same pathway as that in bacteria (24, 25). The intermediate in the latter studies, however, was not identified, nor was phosphoseryl-tRNA[Ser]Sec ruled out as a possible intermediate. Thus, the biosynthesis of Sec in mammalian cells and the role of phosphoseryl-tRNA[Ser]Sec have not been resolved. It should be noted that the identity elements in Sec tRNA[Ser]Sec for PSTK have been identified and found to reside primarily in the secondary structure and length of the D-stem (26).
In the present study, PSTK was identified by using an algorithm that searched for a kinase-like protein gene in the genomes of archaea that are known to encode the Sec insertion machinery vs. those that do not and then comparing the purported kinase gene sequences to genes manifesting homology in eukaryotes that do and do not contain the Sec insertion machinery. The single candidate gene that was found in the genomes of two archaea and in eukaryotes containing the ability to insert Sec into proteins, but not in those that do not, was further characterized. After its expression in bacteria, it was isolated as a pure protein, and in vitro assays confirmed that this purported kinase was indeed PSTK. PSTK was found to be specific for seryl-tRNA[Ser]Sec, whereas no activity with other tRNAs was detected. PSTK catalyzed the formation of phosphoseryl-tRNA[Ser]Sec from seryl-tRNA[Ser]Sec and ATP in the presence of Mg2+. Although Mn2+ can also serve as a divalent metal cation for the reaction, it was not as effective as Mg2+. The products were identified as O-phosphoseryl-tRNA[Ser]Sec and ADP, and the reaction was found to be reversible. Therefore, the reaction can be written as
 |
In a binding assay, either tRNA[Ser]Sec or seryl-tRNA[Ser]Sec bound to PSTK, but inclusion of ATP with the two tRNA substrates had different effects. ATP appeared to enhance the binding of tRNA[Ser]Sec but decrease the binding of seryl-tRNA[Ser]Sec. The decreased binding of seryl-tRNA[Ser]Sec was likely due to its conversion to phosphoseryl-tRNA[Ser]Sec, which was found to have low affinity for PSTK. Interestingly, tRNA[Ser]Sec was a potent inhibitor of the phosphorylation reaction, and although the significance of this inhibition is unclear, PSTK will likely be inhibited significantly when selenoprotein biosynthesis is down-regulated and the unacylated Sec isoforms are enriched.
PSTK has been highly conserved in evolution in genomes that encode the machinery for Sec insertion into protein, because it was found in archaea and numerous lower and higher animals containing this machinery. What, then, is the possible role of PSTK and phosphoseryl-tRNA[Ser]Sec in cellular metabolism? It has been speculated that phosphoseryl-tRNA[Ser]Sec may serve as “an active storage form” which can, after its dephosphorylation, be regenerated as seryl-tRNA[Ser]Sec for the biosynthesis of Sec (27). Several lines of evidence argue against this proposal. For example, the poor rate of dephosphorylation of phosphoseryl-tRNA[Ser]Sec by PSTK observed in the present study suggests that the phosphate group cannot readily be removed, as has similarly been found for other kinases, unless the unlikely possibility of a specific phosphatase might exist. Furthermore, seryl-tRNA[Ser]Sec is an authentic suppressor tRNA that decodes UGA stop codons in vivo (28), and phosphoseryl-tRNA[Ser]Sec is capable of decoding UGA in vitro (29), suggesting that these forms of tRNA[Ser]Sec are likely maintained intracellularly only transiently as substrates for other reactions. Seryl-tRNA[Ser]Sec is a substrate for PSTK, and if the product of this reaction is not an intermediate in the biosynthesis of Sec, then seryl-tRNA[Ser]Sec would also be used in Sec biosynthesis by another route. In any case, it is easy to visualize that seryl-tRNA[Ser]Sec is, and must be, ephemeral.
The fact that PSTK has been conserved in evolution as one of the components of the Sec insertion machinery also strongly argues that it must play an important role in Sec incorporation into protein and that it has not been conserved solely for converting seryl-tRNA[Ser]Sec to a storage form. If phosphoseryl-tRNA[Ser]Sec is used ephemerally, as proposed above for seryl-tRNA[Ser]Sec, then it would most likely be used as an intermediate in the biosynthesis of Sec (reviewed in ref. 2). Indeed, the phosphate moiety of phosphoseryl-tRNA[Ser]Sec would make an excellent leaving group for replacement with selenium. If it is an intermediate in the biosynthesis of Sec, then it, too, would likely exist only ephemerally intracellularly. In the event phosphoseryl-tRNA[Ser]Sec is not an intermediate in the biosynthesis of Sec, and if it does not exist as a storage form for Sec-tRNA[Ser]Sec, additional hypotheses must be advanced to address its function. Like the serylated form of tRNA[Ser]Sec (6), phosphoseryl-tRNA[Ser]Sec does not bind to EFsec (Z. Stoytcheva and M.J.B., unpublished work), and, therefore, it cannot be used directly in incorporating phosphoserine into protein through EFsec and UGA. Although several important steps in the identity and characterization of pstk and PSTK have been carried out in the present study, the function of this kinase and its biosynthetic product, phosphoseryl-tRNA[Ser]Sec, must await future studies.
Acknowledgments
This paper is dedicated to the memory of Merton R. Bernfield, April 9, 1939–March 18, 2002, who originally detected phosphoseryl-tRNA[Ser]Sec kinase activity in 1970. This work was funded in part by National Institutes of Health Grant GM061603 (to V.N.G.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Sec, selenocysteine; mcm5U, 5-methylcarboxymethyluridine; mcm5Um, 5-methylcarboxymethyluridine-2′-O-methylribose; PSTK, phosphoseryl-tRNA[Ser]Sec kinase; SECIS, Sec insertion sequence.
References
- 1.Bock, A. (2001) in Selenium: Its Molecular Biology and Role in Human Health, ed. Hatfield, D. L. (Kluwer, Boston), pp. 7–22.
- 2.Hatfield, D. L. & Gladyshev, V. N. (2002) Mol. Cell. Biol. 22, 3565–3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birringer, M., Pilawa, S. & Flohe, L. (2002) Nat. Prod. Rep. 19, 693–718. [DOI] [PubMed] [Google Scholar]
- 4.Driscoll, D. M. & Copeland, P. R. (2003) Annu. Rev. Nutr. 23, 17–40. [DOI] [PubMed] [Google Scholar]
- 5.Low, S. C. & Berry M. J. (1996) Trends Biochem. Sci. 21, 203–208. [PubMed] [Google Scholar]
- 6.Tujebajeva, R. M., Copeland, P. R., Xu, X.-M., Carlson, B. A., Harney, J. W., Driscoll, D. M., Hatfield, D. L. & Berry, M. J. (2000) EMBO Rep. 1, 158–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fagegaltier, D., Hubert, N., Yamada, K., Mizutani, T., Carbon, P. & Krol, A. (2000) EMBO J. 19, 4796–4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Copeland, P. R., Fletcher, J. E., Carlson, B. A., Hatfield, D. L. & Driscoll, D. M. (2000) EMBO J. 19, 306–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leinfelder, W., Stadtman, T. C. & Bock, A. J. (1989) J. Biol. Chem. 264, 9720–9723. [PubMed] [Google Scholar]
- 10.Lee, B. J., Worland, P. J., Davis, J. N., Stadtman, T. C. & Hatfield, D. L. (1989) J. Biol. Chem. 264, 9724–9727. [PubMed] [Google Scholar]
- 11.Maenpaa, P. H. & Bernfield, M. R. (1970) Proc. Natl. Acad. Sci. USA 67, 688–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hatfield, D. & Portugal, F. H. (1970) Proc. Natl. Acad. Sci. USA 67, 1200–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hatfield, D., Diamond, A. & Dudock, B. (1982) Proc. Natl. Acad. Sci. USA 79, 6215–6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hatfield, D. L., Matthews, C. R. & Rice, M. (1979) Biochim. Biophys. Acta 564, 414–423. [DOI] [PubMed] [Google Scholar]
- 15.Kelmers, A. D. & Heatherly, D. E. (1971) Anal. Biochem. 44, 486–495. [DOI] [PubMed] [Google Scholar]
- 16.Diamond, A. M., Choi, I. S., Crain, P. F., Hashizume, T., Pomerantz, S. C., Cruz, R., Steer, C. J., Hill, K. E., Burk, R. F., McCloskey, J. A. & Hatfield, D. L. (1993) J. Biol. Chem. 268, 14215–14223. [PubMed] [Google Scholar]
- 17.Moustafa, M. E., Carlson, B. A., El-Saadani, M. A., Kryukov, G. V., Sun, Q. A., Harney, J. W., Hill, K. E., Combs, G. F., Feigenbaum, L., Mansur, D. B., et al. (2001) Mol. Cell. Biol. 21, 3840–3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hatfield, D. L., Thorgeirsson, S. S., Copeland, T. D., Oroszlan, S. & Bustin, M. (1988) Biochemistry 27, 1179–1183. [DOI] [PubMed] [Google Scholar]
- 19.Slesarev, A. I., Mezhevaya, K. V., Makarova, K. S., Polushin, N. N., Shcherbinina, O. V., Shakhova, V. V., Belova, G. I., Aravind, L., Natale, D. A., Rogozin, I. B., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 4644–4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bult, C. J., White, O., Olsen, G. J., Zhou, L., Fleischmann, R. D., Sutton, G. G., Blake, J. A., Fitzgerald, L. M., Clayton, R. A., Gocayne, J. D., et al. (1996) Science 273, 1058–1073. [DOI] [PubMed] [Google Scholar]
- 21.Mizutani, T. & Hitaka, T. (1988) FEBS Lett. 232, 243–248. [DOI] [PubMed] [Google Scholar]
- 22.Mizutani, T. (1989) FEBS Lett. 250, 142–146. [DOI] [PubMed] [Google Scholar]
- 23.Mizutani, T., Maruyama, N., Hitaka, T. & Sukenaga, Y. (1989) FEBS Lett. 247, 345–348. [DOI] [PubMed] [Google Scholar]
- 24.Mizutani, T., Kurata, H. & Yamada, K. (1991) FEBS Lett. 289, 59–63. [DOI] [PubMed] [Google Scholar]
- 25.Mizutani, T., Kurata, H., Yamada, K. & Totsuka, T. (1992) Biochem. J. 284, 827–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu, X. Q. & Gross, H. J. (1993) Nucleic Acids Res. 21, 5589–5594, and erratum (1994) 22, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu, X. Q. & Gross, H. J. (1994) EMBO J. 13, 241–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chittum, H. S., Lane, W. S., Carlson, B. A., Roller, P. P., Lung, F. D., Lee, B. J. & Hatfield, D. L. (1998) Biochemistry 37, 10866–10870. [DOI] [PubMed] [Google Scholar]
- 29.Mizutani, T. & Tachibana, Y. (1986) FEBS Lett. 207, 162–166. [DOI] [PubMed] [Google Scholar]







