Abstract
A highly active, monomeric chorismate mutase, obtained by topological redesign of a dimeric helical bundle enzyme from Methanococcus jannaschii, was investigated by NMR and various other biochemical techniques, including H/D exchange. Although structural disorder is generally considered to be incompatible with efficient catalysis, the monomer, unlike its natural counterpart, unexpectedly possesses all of the characteristics of a molten globule. Global conformational ordering, observed upon binding of a transition state analog, indicates that folding can be coupled to catalysis with minimal energetic penalty. These results support the suggestion that many modern enzymes might have evolved from molten globule precursors. Insofar as their structural plasticity confers relaxed substrate specificity and/or catalytic promiscuity, molten globules may also be attractive starting points for the evolution of new catalysts in the laboratory.
Conformational diversity is essential to the function of many proteins. For example, numerous regulatory proteins in the cell are intrinsically unfolded, adopting a well defined structure only in the presence of a suitable target molecule (1). In the immune system, conformational plasticity appears to provide antibodies with enhanced functional diversity, enabling their binding to multiple antigens through different preexisting conformers (2). Moreover, enzyme activity is often fine-tuned through local substrate-induced conformational changes (“induced-fit” mechanisms) and allosteric transitions (3). Nevertheless, excessive conformational diversity can lead to protein dysfunction. The etiology of certain diseases, most notably those involving amyloid formation such as prion disorders, is intimately linked to pronounced structural changes in protein molecules (4), and the unfolded states of proteins, especially those of enzymes, are completely inactive (5–7).
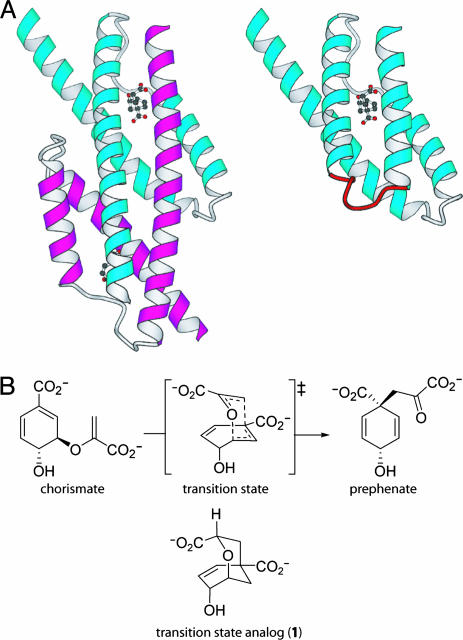
The sensitivity of enzyme activity to even seemingly minor perturbations in active-site structure (6) makes the (re)design of protein catalysts a major challenge. This challenge is increasingly being met through the application of evolutionary strategies that do not require detailed prior knowledge of structure–function relationships (8). For instance, we have successfully exploited genetic selection in combination with structure-based design to convert an entwined dimeric enzyme [the AroQ chorismate mutase from Methanococcus jannaschii (MjCM)] into a monomeric four-helix-bundle protein with near-native enzymatic activity (Fig. 1A) (9). The monomer mMjCM was isolated from a library of 206 variants containing random hinge-loop sequences inserted into the dimer-spanning H1 helix of the parent enzyme. It contains the interhelical turn Ala-Arg-Trp-Pro-Trp-Ala-Glu-Lys and efficiently catalyzes the rearrangement of chorismate into prephenate with a kcat value (3.2 s–1) that is indistinguishable from that of the parent dimeric enzyme and a Km value (170 μM) that is elevated by only 3-fold (9).
Fig. 1.
Topological redesign of the enzyme MjCM. (A) The thermostable MjCM homodimer (Left) was converted into a monomer (mMjCM, Right) by inserting a flexible hinge loop (red) into the long H1 helix (9). The models are based on the x-ray structure (20) of a related E. coli chorismate mutase domain complexed with a transition-state analog, 1 (14), which is shown in the models in ball-and-stick representation. (B) Both enzymes efficiently catalyze the rearrangement of chorismate to prephenate.
Here, we present evidence that the resting state of mMjCM, unlike that of its dimeric precursor, has the properties of a molten globule, a nonnative fluxional state that is normally associated with protein-folding intermediates and functional inactivity (5, 6, 10, 11). The surprising finding that this highly active catalyst is structurally disordered has potentially general implications for understanding how catalysis occurs in natural biological systems and for future progress toward the design of novel enzymes.
Materials and Methods
Chorismate Mutase Production and Purification. Truncated versions of mMjCM and MjCM, in which the six C-terminal amino acids were replaced by LEHHHHHH, were used for all experiments (9, 12). We produced 15N-labeled proteins in Escherichia coli strain KA13 (12) by using 20% Bioexpress growth medium (Cambridge Isotope Laboratories, Cambridge, MA) supplemented with M9 salts (containing 15NH4Cl) and glucose. The enzymes were purified by using standard methods (9, 12).
NMR Spectroscopy. All spectra were obtained at 20°C with a 600-MHz Bruker NMR spectrometer (Bruker, Billerica, MA); the proteins were in PBS buffer (20 mM sodium phosphate/50 mM NaCl, pH 6.5). Backbone resonances for the complex between mMjCM and a transition-state analog (1, Fig. 1B) were assigned by conventional 3D NMR techniques, including transverse relaxation-optimized spectroscopy (TROSY), using 13C,15N-labeled mMjCM.
CD Spectroscopy. Near-UV spectra of protein samples (0.15 mM plus 0.28 mM compound 1) were collected at 20°C by using an Aviv 202 spectropolarimeter (Aviv Associates, Lakewood, NJ). Thermal denaturation of proteins (16 μM) in the absence and presence of 1 (80 μM) was monitored at 222 nm.
Binding of 1-Anilinonaphthalene 8-Sulfonate (ANS). Proteins (2 μM) in the absence and presence of 1 (60 μM) were analyzed in PBS containing 2 μM ANS at 20°C with an Aminco-Bowman series-2 luminescence spectrometer (excitation wavelength, 370 nm).
H/D Exchange and MS. Isotopic exchange was initiated at room temperature by 20-fold dilution of an aqueous protein solution (25 μM protein in 2.5 mM sodium phosphate/40 mM NaCl, pH 6.7, in the absence and presence of 0.8 mM 1) into D2O (pD 7.4). At appropriate times after dilution, 150 μl-aliquots were removed, quenched by decreasing the pD to 2.6 (with the addition of 150 μl of acetonitrile, containing 0.6% formic acid) and by lowering the temperature to 0°C, and analyzed on a TSQ 7000 electrospray-ionization MS spectrometer (Finnigan-MAT, San Jose, CA) (13). The monomer contains 109 amino acid residues and a total number of 254 labile protons, whereas each dimer subunit, which lacks the 8-aa engineered loop, has 101 residues and 231 labile H atoms. The calculation of the percentage of protected protons [percentage of protected H atoms = (MD – M)/(MD – MH) × 100%, where MH, M, and MD are the average molecular masses of the nondeuterated, partially deuterated, and fully deuterated protein, respectively] allows direct comparison of the H/D exchange behavior of the monomer and the wild-type dimer.
Results
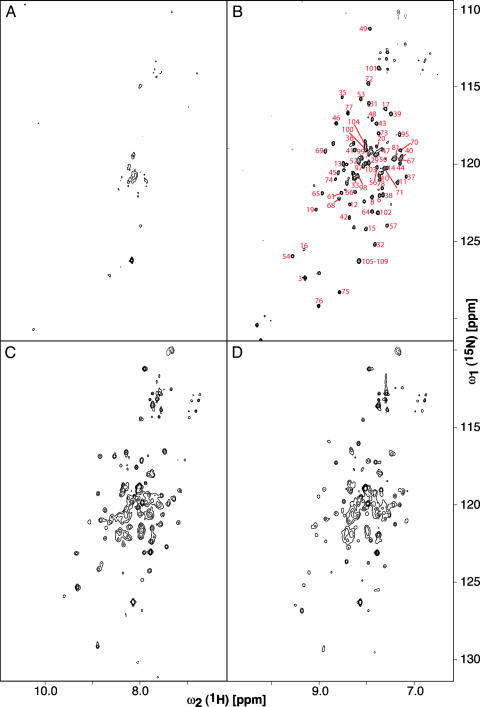
Structural studies were initiated to gain insight into the properties of mMjCM. The topologically redesigned enzyme is highly helical, as judged by CD spectroscopy, and its monomeric topology has been confirmed by gel filtration and analytical ultracentrifugation (9). Because attempts to crystallize the monomer have been unsuccessful to date, we produced it in labeled form for NMR spectroscopic analysis. Surprisingly, the [15N, 1H]-TROSY spectrum of the monomer exhibits far fewer signals than expected for a native-like protein, and the dispersion of the observed peaks is poor (Fig. 2A). These features are characteristic of a poorly packed protein with multiple energetically accessible conformations. However, addition of the bicyclic dicarboxylic acid 1 (Fig. 1B) (14), a transition-state analog inhibitor for the enzyme (Ki = 2.5 μM), induces a dramatic transition from a disordered to an ordered state. At saturating concentrations of ligand, a well resolved spectrum is obtained with signal dispersion, which is typical for a conventionally folded protein (Fig. 2B). This behavior is unique to the designed monomer because the NMR data show that the parent MjCM dimer has native-like structure both in the absence and presence of ligand (Fig. 2 C and D).
Fig. 2.
NMR spectra of 15N-mMjCM and 15N-MjCM in the absence and presence of 1.(A) The [15N, 1H]-TROSY spectrum of 40 μM mMjCM. Peak dispersion did not change appreciably when the protein concentration was varied in the range of 10 μM to 0.6 mM, ruling out oligomerization-induced line-broadening effects. (B) The [15N, 1H]-TROSY spectrum of the ligand-bound mMjCM (sample from A, supplemented with 1.2 mM 1). The red numbers next to the peaks indicate the corresponding residues in the protein. The secondary structure of the monomer, assessed based on the chemical shifts of N, Cα, and Cβ spins (P. Anikeeva and K.P., unpublished results), is in agreement with the proposed structural model (Fig. 1A). Residues 105–109 comprise part of the C-terminal His tag, which is unstructured and gives rise to degenerate resonances. (C) The [15N, 1H]-TROSY spectrum of 0.6 mM MjCM. (D) The [15N, 1H]-TROSY spectrum of the ligand-bound MjCM (sample from C, supplemented with 2 mM 1).
Spectra of the monomer obtained at intermediate concentrations of ligand feature two sets of resonances corresponding to a superposition of the spectra measured with and without saturating amounts of ligand (see Fig. 5, which is published as supporting information on the PNAS web site). The amplitudes of the resonances attributed to the ordered state are directly proportional to the concentration of protein present as a complex with the transition-state analog. This observation supports the existence of a dynamic equilibrium between two distinct states of the protein, disordered and ordered, which is a function of ligand concentration and is slow on the NMR time scale. The poor dispersion of resonances in the disordered state can be attributed to rapid interconversion of an ensemble of (many) energetically accessible conformations, possibly including a fraction of native-like species. Assuming that native structures would exhibit a similar chemical-shift dispersion as the complex with the transition-state analog, we attempted to stabilize them in the absence of ligand by changing the temperature (see Fig. 5), adding osmolytes such as sodium sulfate and tetramethylammonium chloride, and reducing protein concentration (to minimize dimerization of monomers). Because only minor changes were evident in the resulting NMR spectra, we conclude that the fraction of native-like conformers in the population of unliganded enzyme is small.
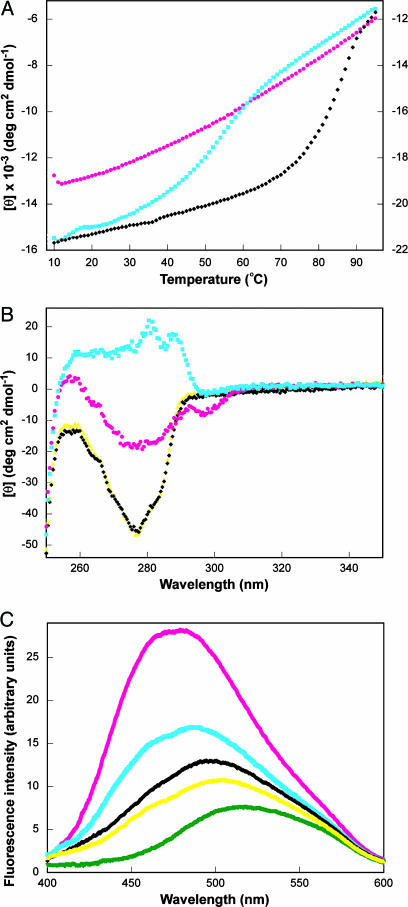
Various additional biophysical methods confirm that unliganded mMjCM is disordered but becomes more structured upon ligand binding. First, thermal denaturation of the protein is noncooperative in the absence of 1 but shows modest cooperativity in its presence (Tm = 55°C) (Fig. 3A). For comparison, the unliganded MjCM dimer unfolds with a high degree of cooperativity (Tm = 88°C) (12). Second, large ligand-induced changes in the near-UV CD spectrum of mMjCM are observed (15); analogous spectral changes are not evident in MjCM by this technique (Fig. 3B). Third, as is typical for molten globules, the engineered monomer binds the environmentally sensitive hydrophobic dye ANS, giving rise to significantly enhanced fluorescence with a characteristic blue shift of the emission maximum (Fig. 3C) (15, 16). ANS, which does not inhibit the enzyme, also binds to mMjCM in the presence of 1 to some extent, suggesting that the mMjCM complex itself retains considerable dynamic character. By comparison, the fluorescence increase seen for ANS in the presence of the wild-type dimer is more typical for a conventionally folded protein (17).
Fig. 3.
Characterization of mMjCM in the absence (magenta circles) and in the presence (cyan squares) of 1, and of MjCM in the absence (black diamonds) and in the presence (yellow triangles) of ligand. (A) Thermal denaturation curves for mMjCM with and without 1. The published (12) denaturation curve for MjCM is provided for comparison (scale on the right). (B) Near-UV CD spectra of the free and ligand-bound forms of mMjCM and MjCM. The well resolved peaks (280.5 and 287.5 nm) for the mMjCM·1 complex indicate that the two tryptophan residues located in the engineered loop are in a more asymmetric environment compared with the free protein (31). In the near-UV CD spectrum of MjCM (which contains no tryptophans) these peaks are not observed. (C) Fluorescence emission spectra of the free and ligand-bound proteins in the presence of ANS and of ANS alone (green triangles).
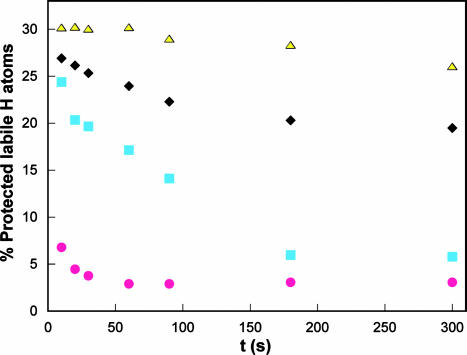
Further compelling evidence for the high conformational flexibility of the monomer comes from H/D exchange experiments (18). The rate of exchange was monitored by electrosprayionization MS in the presence and absence of the transition-state analog (Fig. 4 and Table 1, which is published as supporting information on the PNAS web site). Shortly after initiation of exchange (10 s), all but 17% of the labile protons in mMjCM become deuterated. Addition of 1 initially protects a significant fraction of the protons in the monomer from exchange (≈35%), but within 5 min the degree of deuteration in the complex is comparable with that for the unliganded enzyme. Thus, these results provide additional evidence for a highly mobile structure of mMjCM, even in the presence of ligand. In contrast, the wild-type dimer is considerably more resistant to H/D exchange. Of the exchangeable protons in the MjCM dimer, ≈35% are shielded from exchange at the initial time point; ≈25% of the labile protons do not exchange for several hours. As in the case of the monomer, ligand binding to the dimer protects additional NH groups from exchange, but the magnitude of the effect (≈3–5%) is much smaller.
Fig. 4.
H/D exchange of mMjCM (magenta circles), mMjCM·1 (cyan squares), MjCM (black diamonds), and MjCM·1 (yellow triangles). The substrate chorismate protects mMjCM from exchange to a similar extent as 1 (data not shown).
Discussion
Poor amide-signal dispersion, noncooperative thermal denaturation, ANS binding, and rapid H/D exchange are the hallmarks of molten globules, relatively compact states of proteins having near-native secondary structure but fluctuating tertiary structure (5, 10, 11, 17, 19). The monomeric mutase apparently exists as an ensemble of conformers in dynamic equilibrium and adopts an ordered structure only in the presence of ligand. Although native-like species may be present to some extent in the absence of ligand, they are not detected by NMR or near-UV CD, suggesting that they constitute only a small fraction of the total population.
The molten globular character of unliganded mMjCM may be attributable to the absence of a well packed hydrophobic core. The interior of mMjCM must accommodate the highly polar active site (20). The large number of charged residues at the core of the protein likely destabilizes a native-like folded state in the absence of ligand. However, the transition-state analog or substrate can fill the pocket, providing a template around which the protein can “crystallize.” The extensive H-bonding and electrostatic interactions that are formed between 1 and the protein can then propagate globally to improve packing throughout the structure. Because the parent MjCM dimer contains a conventional hydrophobic core between its two active sites (20), it is considerably more stable than mMjCM, even in the absence of 1, and substantially less structural ordering is required for catalysis.
The enhanced flexibility of mMjCM compared with MjCM has an amazingly small energetic cost. The monomer and dimer differ in catalytic efficiency (kcat/Km) only by a factor of three (≈0.6 kcal/mol) entirely because of differences in Km (9). Preliminary calorimetric measurements show that the unfavorable entropy associated with preorganizing the enzyme is offset by enthalpic gains from the noncovalent interactions between 1 and mMjCM (K.V. and I. Jelesarov, unpublished results), in accord with the proposal of Williams et al. (21) that catalytic efficiency can originate from improved packing interactions in the transition state. In a sense, the structural ordering seen here can be considered an extension of the classic induced-fit model of enzyme catalysis (22) in which the substrate causes structural rearrangement at the active site that lead to a high affinity complex. In the case of mMjCM, however, the substrate appears to induce global ordering, rather than local restructuring, taking the classical model to new extremes. Although precise positioning of catalytic residues is generally considered to be a prerequisite for efficient catalysis, our findings suggest that there is little penalty associated with locking the catalytically active conformation of mMjCM in place as the reaction proceeds.
The absence of a significant population of natively structured protein in the absence of ligand argues against an alternative “preequilibrium” mechanism (23) in which addition of ligand shifts the equilibrium between the molten globule and a scarcely populated native-like species competent for ligand binding and catalysis in favor of the native state. The high activity of the catalyst would require that ≈30% of the free protein be ordered, which is not observed. However, presteady-state kinetic measurements will be necessary to exclude this mechanistic possibility definitively.
Many de novo designed proteins have turned out to be molten globules (17). In fact, protein engineers have expended considerable effort over the years to convert these molecules into native-like proteins (17, 24–27). The tacit assumption has been that a preorganized native-like structure is required for biological function. However, our results with mMjCM show that the molten globular state does not necessarily preclude efficient catalysis. Although the molten globules formed during the (un)folding of many naturally occurring enzymes are completely inactive (5, 6), there have been hints in the literature that other molten globules might retain function under some conditions. For example, two circularly permuted dihydrofolate reductase variants, although less active than the parent enzyme, were shown to possess some properties of the molten globule state and recover native-like tertiary structure in the presence of NADPH and methotrexate (28). A staphylococcal nuclease mutant also displays certain features of the molten globule state and adopts a functional structure upon binding Ca2+ and thymidine 3′, 5′-bisphosphate (29). Additional characterization of these systems is clearly needed, but in light of our findings, the available data suggest that the coupling of folding and catalysis may be a more general phenomenon than has been previously appreciated.
Conceivably, many if not most modern-day enzymes could have evolved from molten globules (30). Insofar as structural plasticity confers relaxed substrate specificity and/or catalytic promiscuity (23), a molten globule could have had a significant evolutionary advantage in the primordial world, performing several functions in the primitive cell. The similar activities of mMjCM and MjCM, and the ability of the monomer to assume the metabolic role of the dimer in the cell (9), suggest that catalytic efficiency may not have been the (sole) driving force for the evolution of native protein structures. Instead, the need to control individual functions separately, minimize intrinsic instability, or prevent aggregation, proteolytic degradation, or improper cellular trafficking would have favored the well defined structures of modern enzymes. Given their possible evolutionary heritage, it is tempting to speculate that molten globules might be superior to highly evolved enzymes as starting points for the directed evolution of tailored catalysts in the laboratory. By applying random mutagenesis and powerful selection or screening technologies to molecules like mMjCM, it may be possible to channel their inherent conformational diversity (23) down multiple evolutionary trajectories and generate a variety of interesting new structures and functions.
Supplementary Material
Acknowledgments
We thank R. Pulido for synthesizing the transition-state analog, K. Woycechowsky for careful reading of the manuscript, and the reviewers for valuable comments. This work was supported by the Schweizerischer Nationalfonds and the ETH Zurich.
Abbreviations: ANS, 1-anilinonaphthalene 8-sulfonate; TROSY, transverse relaxation-optimized spectroscopy.
References
- 1.Dyson, H. J. & Wright, P. E. (2002) Curr. Opin. Struct. Biol. 12, 54–60. [DOI] [PubMed] [Google Scholar]
- 2.James, L. C., Roversi, P. & Tawfik, D. S. (2003) Science 299, 1362–1367. [DOI] [PubMed] [Google Scholar]
- 3.Hammes, G. G. (2002) Biochemistry 41, 8221–8228. [DOI] [PubMed] [Google Scholar]
- 4.Dobson, C. M. (1999) Trends Biochem. Sci. 24, 329–332. [DOI] [PubMed] [Google Scholar]
- 5.Ptitsyn, O. B., Pain, R. H., Semisotnov, G. V., Zerovnik, E. & Razgulyaev, O. I. (1990) FEBS Lett. 262, 20–24. [DOI] [PubMed] [Google Scholar]
- 6.Tsou, C. (1995) Biochim. Biophys. Acta 1253, 151–162. [DOI] [PubMed] [Google Scholar]
- 7.Dunker, A. K., Brown, C. J., Lawson, D. J., Iakoucheva, L. M. & Obradovic, Z. (2002) Biochemistry 41, 6573–6582. [DOI] [PubMed] [Google Scholar]
- 8.Taylor, S. V., Kast, P. & Hilvert, D. (2001) Angew. Chem. Int. Ed. Engl. 40, 3310–3335. [DOI] [PubMed] [Google Scholar]
- 9.MacBeath, G., Kast, P. & Hilvert, D. (1998) Science 279, 1958–1961. [DOI] [PubMed] [Google Scholar]
- 10.Dolgikh, D. A., Gilmanshin, R. I., Brazhnikov, E. V., Bychkova, V. E., Semisotnov, G. V., Venyaminov, S. Y. & Ptitsyn, O. B. (1981) FEBS Lett. 136, 311–315. [DOI] [PubMed] [Google Scholar]
- 11.Kuwajima, K. (1989) Proteins Struct. Funct. Genet. 6, 87–103.2695928 [Google Scholar]
- 12.MacBeath, G., Kast, P. & Hilvert, D. (1998) Biochemistry 37, 10062–10073. [DOI] [PubMed] [Google Scholar]
- 13.Smith, D. L., Deng, Y. & Zhang, Z. (1997) J. Mass Spectrom. 32, 135–146. [DOI] [PubMed] [Google Scholar]
- 14.Bartlett, P. A. & Johnson, C. R. (1985) J. Am. Chem. Soc. 107, 7792–7793. [Google Scholar]
- 15.Demarest, S. J., Boice, J. A., Fairman, R. & Raleigh, D. P. (1999) J. Mol. Biol. 294, 213–221. [DOI] [PubMed] [Google Scholar]
- 16.Semisotnov, G. V., Rodionova, N. A., Razgulyaev, O. I., Uversky, V. N., Gripas, A. F. & Gilmanshin, R. I. (1991) Biopolymers 31, 119–128. [DOI] [PubMed] [Google Scholar]
- 17.Betz, S. F., Raleigh, D. P. & DeGrado, W. F. (1993) Curr. Opin. Struct. Biol. 3, 601–610. [Google Scholar]
- 18.Englander, S. W., Mayne, L., Bai, Y. & Sosnick, T. R. (1997) Protein Sci. 6, 1101–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ohgushi, M. & Wada, A. (1983) FEBS Lett. 164, 21–24. [DOI] [PubMed] [Google Scholar]
- 20.Lee, A. Y., Karplus, P. A., Ganem, B. & Clardy, J. (1995) J. Am. Chem. Soc. 117, 3627–3628. [Google Scholar]
- 21.Williams, D. H., Stephens, E. & Zhou, M. (2003) J. Mol. Biol. 329, 389–399. [DOI] [PubMed] [Google Scholar]
- 22.Koshland, D. E. (1994) Angew. Chem. Int. Ed. Engl. 33, 2375–2378. [Google Scholar]
- 23.James, L. C. & Tawfik, D. S. (2003) Trends Biochem. Sci. 28, 361–368. [DOI] [PubMed] [Google Scholar]
- 24.Bryson, J. W., Betz, S. F., Lu, H. S., Suich, D. J., Zhou, H. X., O'Neil, K. T. & DeGrado, W. F. (1995) Science 270, 935–941. [DOI] [PubMed] [Google Scholar]
- 25.Cordes, M. H. J., Davidson, A. R. & Sauer, R. T. (1996) Curr. Opin. Struct. Biol. 6, 3–10. [DOI] [PubMed] [Google Scholar]
- 26.Gibney, B. R., Rabanal, F., Skalicky, J. J., Wand, A. J. & Dutton, P. L. (1999) J. Am. Chem. Soc. 121, 4952–4960. [Google Scholar]
- 27.Wei, Y., Kim, S., Fela, D., Baum, J. & Hecht, M. H. (2003) Proc. Natl. Acad. Sci. USA 100, 13270–13273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uversky, V. N., Kutyshenko, V. P., Protasova, N. Y., Rogov, V. V., Vassilenko, K. S. & Gudkov, A. T. (1996) Protein Sci. 5, 1844–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, Y. & Jing, G. (2000) J. Biochem. 128, 739–744. [DOI] [PubMed] [Google Scholar]
- 30.DeGrado, W. F. (1993) Nature 365, 488–489. [DOI] [PubMed] [Google Scholar]
- 31.Woody, W. R. & Dunker, A. K. (1996) Circular Dichroism and the Conformational Analysis of Biomolecules (Plenum, New York).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.