Abstract
Cytoplasmic dynein is a minus-end-directed microtubule motor involved in numerous essential processes within eukaryotic cells, such as nuclear segregation and trafficking of intracellular particles. The motor domain of the dynein heavy chain comprises six tandemly linked AAA (ATPase associated with diverse cellular activities) modules (AAA1–AAA6). The first four modules include nucleotide-binding sites (Walker A or P-loop motifs), and each of the four sites appears to bind ATP. However, the role and the function of each binding site are unknown. Especially, the question of which P-loops are ATP-hydrolyzing sites has not been answered, because it is difficult to measure the ATPase activity of each P-loop. Here, we purified several truncated Saccharomyces cerevisiae cytoplasmic dynein fragments and their mutants expressed in Escherichia coli and then measured their ATPase activities. Our results suggest that there are multiple ATP-binding sites that have abilities to hydrolyze ATP in cytoplasmic dynein. Furthermore, a single AAA module is insufficient for ATP hydrolysis, and the adjacent module facing the ATP-binding site is necessary for ATP-hydrolyzing activity.
Dynein is a minus-end-directed microtubule motor protein, and its isoforms are classified into cytoplasmic dyneins and axonemal dyneins. Axonemal dyneins generate specific flagellar and ciliary waveforms to move the cell through the solution. In contrast, cytoplasmic dyneins are involved in numerous essential processes within eukaryotic cells, including nuclear segregation, mitosis, maintenance of the Golgi apparatus, and trafficking of membranous vesicles, virus, and other intracellular particles. The dynein complex is very large (1–2 MDa) and comprises at least four components: heavy, intermediate, light-intermediate, and light chains (1–3).
The heavy chain, composed of two distinct parts, the N-terminal domain of 160 kDa and the C-terminal domain of 380 kDa, is the main component involved in dynein motor activity. The N-terminal domain forms the stem of the structure and interacts with associated polypeptides, including heavy chains and other components. The C-terminal domain forms the globular motor domain.
The motor domain comprises six tandemly linked AAA (ATPases associated with diverse cellular activities) modules (AAA1–AAA6). These modules are arranged in a ring with a projection, a microtubule-binding stalk, between the AAA4 and AAA5 modules (4, 5). The first four modules (AAA1–AAA4) include P-loop motifs, which are predicted to be nucleotide-binding sites. In the case of axonemal dyneins, it has been shown that at least two and perhaps all four P-loops bind ATP with physiologically significant binding constants (6, 7). The first P-loop, P1, is absolutely conserved among cytoplasmic and axonemal dyneins. P1 is considered the principal site of ATP hydrolysis based on photocleavage experiments of axonemal dyneins in vitro (8) and mutational experiments of cytoplasmic dyneins in vivo (4). Among cytoplasmic dyneins, the third P-loop, P3, is also well conserved. It has been shown that a mutation of cytoplasmic dynein in P3 brings about the same phenomena as a mutation in P1 (9). Therefore, not only P1 but also P3 is thought to be involved in cytoplasmic dynein ATP-dependent microtubule binding. Moreover, Reck-Peterson and Vale (10) have shown that dynein function requires nucleotide binding only at P1 and P3 and nucleotide hydrolysis only at P1. However, the ATPase activities of these mutants have not been measured in vitro because of the difficulty of purifying them. Thus, whether or not P3 has the ATPase activity remains unclear.
Here, we attempted to examine whether or not P3 is a catalytic site. We expressed several truncated and mutated recombinant cytoplasmic dyneins in Escherichia coli and then measured their ATPase activities. Our results suggest that there are multiple ATP-hydrolyzing sites that are potentially functional in cytoplasmic dynein.
Materials and Methods
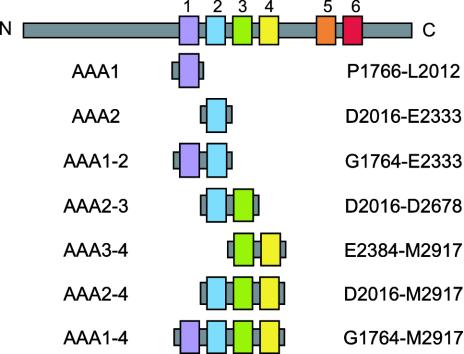
Creation of Truncated and Mutated Dyneins. The diagrams of all constructs used here are shown in Fig. 1. To construct plasmids for the expression of truncated dyneins in E. coli, gene fragments of AAA1 (P1766-L2012), AAA2 (D2016-E2333), AAA1–2 (G1764-E2333), AAA2–3 (D2016-D2678), AAA3–4 (E2384-M2917), AAA2–4 (D2016-M2917), and AAA1–4 (G1764-M2917) of yeast cytoplasmic dynein were amplified by PCR from the genomic DNA of Saccharomyces cerevisiae. Taking into consideration a report showing that a motor domain of Ustilago maydis cytoplasmic dynein consists of two polypeptides, Dyn1 and Dyn2, which include the putative ATPase region and the predicted microtubule-binding site, respectively (11), we ascertained the C-terminal end of AAA1–4. The fragments were then inserted into the SmaI site of pGEX-BCCP (biotin carboxyl carrier protein)-His6. The expression vector was constructed by inserting the BCCP-His6 gene into the EcoRI site of pGEX-2T (Amersham Pharmacia), and the gene was amplified by PCR from PinPoint Xa-1 vector (Promega).
Fig. 1.
Schematic representation of the constructs used in this study. The names are shown on the left. The first amino acid and the last amino acid of the constructs are shown on the right. The color of each module in this figure corresponds to the color of each module of protein in the other figures that follow.
Site-directed mutagenesis in the SRC motif of the AAA2 and AAA4 modules was performed by using Mutan-K (Takara Shuzo, Kyoto). The mutagenic primers 5′-CCT GCT ACT ATA ACA GCA TGC GGT TTG CTG TGG TTT-3′ and 5′-CCA GCC CTT TTC AAT GCA TGC ATA ATT AAT TGG-3′ were used to alter the wild-type AAA2-SRC sequence from TITRCGL to TITACGL and the wild-type AAA4-SRC sequence from LFNRCII to LFNACII, respectively.
The DNA sequence of all constructs was confirmed by using a Thermo Sequence fluorescent-labeled primer-cycle sequencing kit (Amersham Pharmacia).
Purification of Recombinant Dynein Fragments. All constructs were transformed into E. coli strain BL21-Codon Plus (DE3)-RIL (TOYOBO, Tokyo). Transformants were grown in 500 ml of a medium with ampicillin (50 μg/ml) at 37°C until reaching an OD600 value of ≈0.6. Protein expression was induced with 0.1 mM isopropyl-β-d-thiogalactopyranoside for 3 h at 20°C. Cells were then harvested by centrifugation and frozen in liquid N2.
All proteins expressed in E. coli were purified as follows. Frozen cells were thawed in buffer A (50 mM Tris·HCl, pH 8.0/500 mM NaCl/2 mM MgCl2/0.1 mM ATP/1 mM DTT) containing protease inhibitors (1 mM PMSF and 10 μg/ml each of pepstatin A, leupeptin, and p-toluenesulfonyl-l-arginine methyl ester) and 40 mM imidazole-HCl (pH 8.0) and lysed by sonication. The lysates were then ultracentrifuged to remove cellular debris. The following adsorption and elution of proteins were performed in a batch procedure. Ni-NTA (Ni-nitrilotriacetic acid) agarose (Qiagen, Valencia, CA) and glutathione agarose (Sigma) were equilibrated with buffer A containing 40 mM imidazole-HCl (pH 8.0) and buffer B (50 mM Tris·HCl, pH 8.0/20 mM NaCl/2 mM MgCl2/0.1 mM ATP/1 mM DTT), respectively. Proteins in the supernatant were adsorbed to Ni-NTA agarose resins, which were then washed with buffer A, and proteins nonspecifically bound to resins were removed with buffer A containing 80 mM imidazole-HCl (pH 8.0). These resins were then washed with buffer B to reduce the salt concentration, and the target protein was eluted with buffer B containing 300 mM imidazole-HCl (pH 8.0). The target protein in an eluted solution containing 1 mM each of EDTA and EGTA was adsorbed to glutathione agarose resins. The resins were washed with buffer B containing 1 mM each of EDTA and EGTA, and the target protein was then eluted with buffer C [50 mM Tris·HCl, pH 8.0/20 mM glutathione/20 mM NaCl/30% (wt/vol) sucrose/1 mM EDTA/1 mM EGTA/1 mM DTT]. Purified proteins were confirmed by SDS/PAGE according to the method of Laemmli (12). After electrophoresis, protein bands were visualized by Coomassie brilliant blue R-250 staining. Western blotting was carried out by a standard method (13). Antibodies used here were as follows. Polyclonal antibodies against His6 (Clontech) and GST (Amersham Pharmacia) were used as primary antibodies. Secondary antibodies (goat antimouse IgG and rabbit anti-goat IgG, respectively) were conjugated with alkaline phosphatase. Bands were detected by using a kit [BCIP/NBT Membrane Phosphatase Substrate System (3-C), Kirkegaard & Perry Laboratories]. Protein concentration was estimated by the method of Read and Northcote (14) by using BSA as a standard. The final yield of each truncated dynein per liter of cell culture was approximately as follows: AAA1, 400 μg; AAA2, 200 μg; AAA1–2, 60 μg; AAA3–4, 120 μg; AAA2–4, 70 μg; and AAA1–4, 30 μg.
ATPase Assays. An ATPase assay was performed by using EnzChek (Molecular Probes) in the presence of 1 mM ATP in an assay buffer [40 mM Tris·HCl, pH 8.0/15 mM NaCl/10 mM MgCl2/23% (wt/vol) sucrose] at 25°C by monitoring the absorbance at 360 nm for 5 or 10 min. In each measurement, 77 μl of a final product was used. To determine the error bars, more than three measurements were performed for each recombinant dynein fragment. For comparison, we also measured the activity of native cytoplasmic dynein purified from porcine brain (15).
The ATPase activities were estimated as follows. Raw data were fitted by linear regression, and the increase in the concentration of released Pi per second was obtained from the slope of the regression line. This value was then normalized by the molarity of the protein. Therefore, the ATPase activities were presented in units of s–1, which refers to the turnover rate.
To compare the activities of our dynein fragments with those reported for native cytoplasmic dynein (16, 17), we calculated the reported values to transform the units from nmol/min per mg to s–1. Namely, we multiplied the reported value by 0.012 on the assumption that the molecular mass of cytoplasmic dynein per head is 700 kDa.
Results and Discussion
Purification of Recombinant Dynein Fragments. First, we tried expressing truncated dyneins fused only with GST, which is known to be a highly soluble protein, at their N termini. However, in this case, GST did not help to solubilize the truncated dyneins.
To increase their solubility, the truncated dyneins were fused with BCCP, which is also known to be a highly soluble protein, at their C termini. As a result of this attempt, parts of them were expressed in E. coli as soluble forms, indicating that the expression of truncated dyneins in E. coli as soluble forms requires two highly soluble tags: GST and BCCP. As a tag for purification, His6 was conjugated with the C terminus of BCCP.
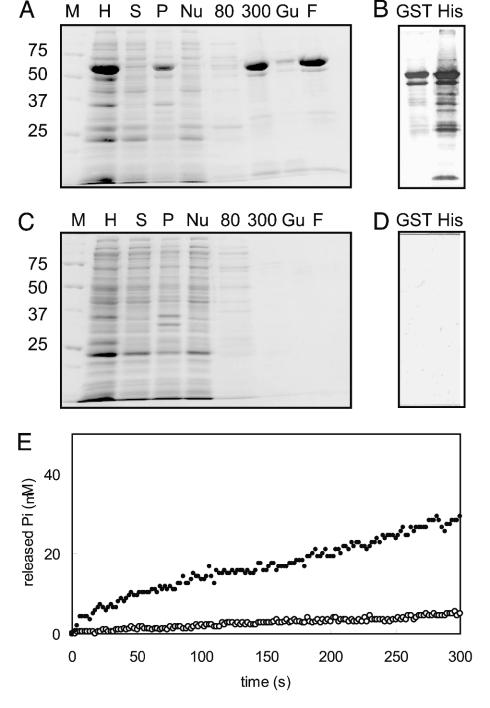
We expressed AAA1 in E. coli and purified it. Fig. 2A shows all of the stages of purification of AAA1 from the initial bacterial homogenate through to the final product. We also prepared a control sample by the same method from E. coli, which was not transformed, and all of the stages of this preparation are shown in Fig. 2C. AAA1 was obtained as the main band. Below the main band of the final product, Western blotting detected some contaminating bands (Fig. 2B). The control sample did not contain any visible bands in Coomassie brilliant blue staining, and no band was detected by Western blotting (Fig. 2D). Moreover, the nontransformed bacterial homogenates had no protein that reacted with anti-His6 antibody and/or anti-GST antibody (Fig. 6, which is published as supporting information on the PNAS web site). Thus, the contaminating proteins contained in the final product of AAA1 were thought to be degradation products; further, there was no E. coli protein in the final product.
Fig. 2.
Comparison of AAA1 and a control sample. (A and C) Purification of AAA1 and preparation of control sample on 10% SDS/PAGE gel. M, molecular mass markers; H, bacterial homogenate; S and P, supernatant and precipitant after centrifugation, respectively; Nu, unbound fraction to Ni-nitrilotriacetic acid (NTA) agarose; 80 and 300, eluted fraction with buffer B containing 80 or 300 mM imidazole-HCl (pH 8.0), respectively; Gu, unbound fraction to glutathione agarose; F, final product eluted with 20 mM glutathione. (B and D) Western blotting of the final product from the purification of AAA1 and the preparation of the control sample. Lanes at left (GST) show samples probed with the anti-GST antibody. Lanes at right (His) show samples probed with the anti-His6 antibody. (E) Time course of Pi liberation monitored by an EnzChek system. AAA1 (closed circles) and control samples (open circles).
Most degradation products reacted with anti-His6 antibody; those with molecular masses smaller than 35 kDa do not include P1 and are thought not to have ATPase activity, whereas those larger than 35 kDa include P1 and might have ATPase activity. Especially, the main degradation product, whose molecular mass was ≈10 kDa smaller than that of AAA1, reacted with both anti-His6 and anti-GST antibodies. Therefore, this degradation product is thought to cut in the region of GST (at 10 kDa from the N terminus of 27-kDa GST) and thus contains a whole AAA1 module so that its ATPase activity is thought to be the same as AAA1.
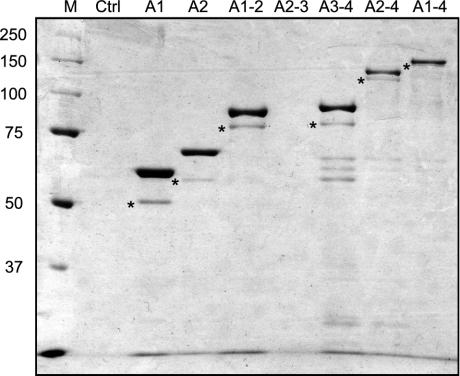
We expressed other recombinant dynein fragments in E. coli and purified them by the same method. All truncated dyneins, except for AAA2–3, were obtained as main bands (Fig. 3). The main degradation product in each lane, with a molecular mass 10 kDa smaller than that of the full-length product, contained the whole truncated dynein and was thought to have the same activity. Because AAA2–3 almost fully degraded in E. coli (Fig. 7, which is published as supporting information on the PNAS web site), AAA2–3 was not obtained. As the degradation products of 60–70 kDa in lane AAA3–4 were purified by two sequential steps by using N- and C-terminal tags and reacted with anti-His6 antibody, they were thought to be associated with smaller paired fragments. Although there were some contaminating proteins in the frontal line, they were not E. coli proteins but degradation products as discussed above. Thus, we determined the ATPase activities of dynein fragments using their corresponding final products.
Fig. 3.
Expressed proteins purified from E. coli in SDS/PAGE on 7.5% gel. M, the molecular mass markers; Ctrl, the control sample prepared from E. coli that was not transformed. The main band of each lane is the target protein. Because target proteins are fused with GST (27 kDa) and His6-conjugated BCCP (13 kDa), their molecular masses are ≈40 kDa larger than that of the included dynein fragments: AAA1, 67 kDa; AAA2, 75 kDa; AAA1–2, 103 kDa; AAA3–4, 101 kDa; AAA2–4, 142 kDa; and AAA1–4, 170 kDa. Asterisks indicate the main degradation products whose molecular masses are ≈10 kDa smaller than those of major bands. In the front line, there are some degradation products whose molecular masses are smaller than 25 kDa. The lane of AAA1 is labeled as A1, and others similarly.
There was no essential difference between wild type and mutants in respect to their purity and yield. We then performed ATPase assays using these purified recombinant dynein fragments in the following experiments.
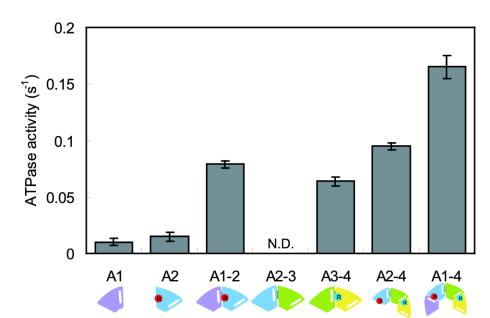
AAA2 Module Is Necessary for the ATPase Activity of P1. It is presently thought that the elements involved in nucleotide binding and hydrolysis are Walker A (or P-loop) and Walker B motifs. Moreover, the first P-loop (P1) is known to be a catalytic site (4, 8). Thus, AAA1, which contains P1 and the first Walker B, is expected to have considerable ATPase activity. Therefore, we measured the ATPase activity of AAA1, which although low (0.010 ± 0.003 s–1) was significant compared with the control sample (Fig. 2E). As the P-loop was predicted to face the cleft between the two modules (18) and P1 faces the cleft between the AAA1 and AAA2 modules, the activity of P1 might be influenced by the AAA2 module. Thus, we measured the ATPase activities of AAA2 and AAA1–2. Whereas AAA2 also showed low activity (0.015 ± 0.004 s–1), AAA1–2, which has the cleft, showed higher activity (0.079 ± 0.003 s–1) than the sum of those of AAA1 and AAA2 (Fig. 4). These results suggest that there are elements in the AAA2 module to hydrolyze ATP relating to P1.
Fig. 4.
ATPase activities of truncated dyneins. Bars indicate the SD. A schematic representation of each truncated dynein is shown under each name. White ellipsoids and red and light blue circles correspond to P-loops and catalytic arginines R2209 and R2911, respectively. AAA1 has only a P-loop (P1). AAA2 has both a P-loop (P2) and catalytic arginine, but the catalytic arginine is located far away from P2; thus, no functional ATPase site is formed. On the other hand, catalytic arginine is in close proximity to P1 in AAA1–2, and a functional ATPase site is formed.
Many AAA+ proteins are homohexamers and form ring-like structures that are thought to be important for their activity; that is, the adjacent subunit is involved in ATP hydrolysis (19–22). The SRC motif, a sequence of serine, arginine, and cysteine, is widely conserved in clamp-loading subunits of T4 phage, eubacteria, archaea, and eukaryotes. A recent study of the E. coli γ complex, a clamp loader, has shown that the arginine residue within one subunit SRC motif functions with ATP bound to an adjacent subunit. Further, the mutation of this arginine to alanine prevents ATP hydrolysis without disrupting ATP binding (20).
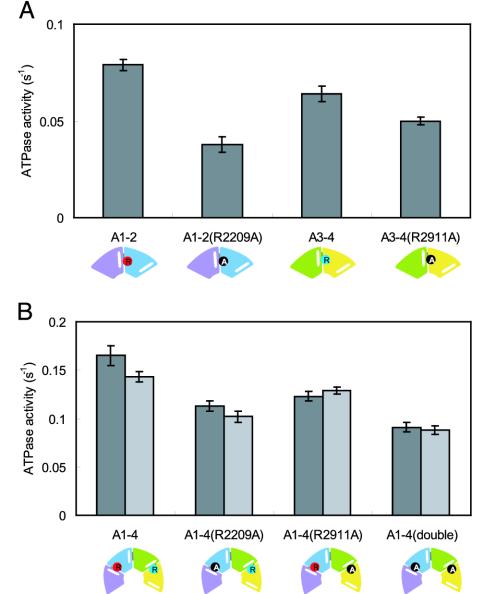
Among dyneins, a similar sequence is located in BoxVII (or at the C end of helix H5) in some AAA modules (23). Moreover, this region is predicted to participate in the formation of the ATP-binding site (18). In the case of an AAA2 module of S. cerevisiae cytoplasmic dynein, the sequence of this region is TITRCGL, and the arginine residue corresponds to R2209. (The bold letters indicate the residues corresponding to the SRC motif.) To examine whether or not R2209 is involved in ATP hydrolysis relating to P1, we mutated it to alanine and measured the ATPase activity of this mutant (Fig. 5A). AAA1–2(R2209A) showed lower activity (0.038 ± 0.004 s–1, 48% of the activity of AAA1–2), indicating that R2209 participates in the ATP hydrolysis of AAA1–2.
Fig. 5.
ATPase activities of mutants. Bars indicate the SD. A schematic representation of each recombinant dynein is shown under each name. Red and light blue circles correspond to catalytic arginines R2209 and R2911, respectively. Substituted alanines are shown in black circles. (A) SRC mutants of AAA1–2 and AAA3–4. (B) SRC mutants of AAA1–4. Dark gray bars show the experimental values. Light gray bars show the values estimated from those of wild type and/or mutants of AAA1–2 and AAA3–4. For example, the value of AAA1–4(R2209A) represents the sum of those of AAA1–2(R2209A) and AAA3–4.
P3 also Has ATPase Activity in AAA3–4. The third P-loop, P3, is also well conserved among cytoplasmic dyneins. Silvanovich et al. (9) showed that not only P1 but also P3 was involved in cytoplasmic dynein ATP-sensitive microtubule binding. There is an SRC motif in BoxVII of the AAA4 module, and the sequence around this SRC motif is LFNRCII. Therefore, P3 is expected to have ATPase activity although P1 is presently thought to be the only ATPase site in dynein. We measured the ATPase activity of AAA3–4 and found that the activity of AAA3–4 (0.064 ± 0.004 s–1) was at the same level as that of AAA1–2 (Fig. 4).
To examine the importance of the arginine residue in AAA4 (R2911) for ATP hydrolysis relating to P3, we mutated it to alanine and measured the ATPase activity of this mutant (Fig. 5A). This mutation significantly reduced the activity of AAA3–4 although this mutation was less effective than the mutation of R2209. Namely, AAA3–4(R2911A) showed lower activity (0.050 ± 0.002 s–1, 78% of the activity of AAA3–4). These results suggest that the ATPase site exists in AAA3–4 and that the arginine residue (R2911) serves to hydrolyze ATP, as R2209 does.
Multiple ATP-Hydrolyzing Sites in AAA1–4. To examine whether or not P3 has ATPase activity in AAA1–4, we measured the ATPase activity of AAA1–4. The activity of AAA1–4 (0.165 ± 0.010 s–1) was equal to the sum of those of AAA1–2 and AAA3–4. This result supports the idea that there are at least two ATPase sites, P1 and P3, in AAA1–4.
Next, we expressed three mutants [AAA1–4(R2209A), AAA1–4(R2911A), and AAA1–4(double)] whose arginine residues of the AAA2 and/or AAA4 module were mutated to alanines and measured their ATPase activities (Fig. 5B). The activity of AAA1–4(R2911A) (0.123 ± 0.005 s–1, 74% of the activity of AAA1–4) was lower than that of AAA1–4, similar to that of AAA1–4(R2209A) (0.113 ± 0.005 s–1, 68% of the activity of AAA1–4), and higher than that of AAA1–4(double) (0.091 ± 0.005 s–1, 55% of the activity of AAA1–4). Moreover, the activity of AAA1–4(R2911A) was almost equal to the sum of those of AAA1–2 and AAA3–4(R2911A). These results indicate that (i) P3 has ATPase activity in AAA1–4, and (ii) the activities of both P1 and P3 are independent and of similar levels.
The Activities of Other P-Loops. We attempted to examine the possibility that other P-loops might have ATPase activity. However, we could not examine the ATPase activity of P4 because we could not express any truncated dynein fragments larger than AAA1–4. We then examined the ATPase activity of P2. Because we could not obtain AAA2–3, as described before, we measured the ATPase activity of AAA2–4. The activity of AAA2–4 (0.095 ± 0.003 s–1) was slightly higher than that of AAA3–4 but similar to that of AAA1–2 and AAA3–4 (Fig. 4). This result is consistent with a report showing that nucleotide binding at P2 plays a minor role (10) and the fact that the arginine residue of SRC motif is not conserved in AAA3. Therefore, this result suggests that P2 does not have significant activity compared with those of P1 and P3.
Physiological Activities of Multiple ATP-Hydrolyzing Sites. There is a major difference between dynein and other motor proteins such as kinesin and myosin: the former binds four ATP whereas the latter bind only one ATP. Thus, the determination of the catalytic P-loops is important for understanding the dynein motor mechanism. So far, the abilities of several mutants to bind to and release from microtubules have been examined in vitro (9, 10); however, their ATPase activities have not been measured. In this study, we expressed and purified single and paired AAA modules of dynein, some of which have physiologically significant activities (AAA1–2, 0.079 s–1; AAA3–4, 0.069 s–1; AAA2–4, 0.095 s–1; AAA1–4, 0.165 s–1) compared with that of native cytoplasmic dynein (0.102 ± 0.014 s–1) measured by the same method. These values were also comparable with other reported values: 0.06–0.12 s–1 by Shimizu et al. (16) and 0.55 s–1 by King and Schroer (17) (see Materials and Methods). Because it is known that the catalytic activity of one P-loop is regulated by the nucleotide binding to other P-loops (24–26), we cannot directly evaluate the activity of each P-loop by the mutation at each P-loop in the motor domain. Therefore, to ascertain the activity of each P-loop, the use of these recombinant proteins is essential.
Our analysis reveals that P3 has ATPase activity not only in AAA3–4 but also in AAA1–4. This finding suggests the possibility that there are at least two ATPase sites, P1 and P3, in cytoplasmic dynein. This finding does not mean that both of the two sites are always active within the complete motor domain that forms a ring-like structure but indicates that each of the two sites has the ability to hydrolyze ATP. Recently, Mallik et al. (27) proposed that cytoplasmic dynein functions as a gear in response to a load. Then, ATP hydrolysis at P3 might trigger the downshift of a dynein gear under a high-load condition. So far, investigators have tried to elucidate the mechanism on the assumption that there is only one ATPase site in dynein as with kinesin and myosin. However, our study shows the existence of other potential ATPase sites, such as P3. Therefore, we have to take into consideration of these potential ATPase sites. From this point of view, the examination of ATPase and a motility assay by using mutated dyneins, which include the entire motor domain, will give us more precise information of the relationship between ATP hydrolysis and dynein motile activity.
Supplementary Material
Acknowledgments
We thank Junko Watai-Nishii, Shiori Toba, and Shirou Hirayama for technical help. This work was supported by Special Coordination Funds for Promoting Science and Technology of the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) and by the Core Research for Evolutional Science and Technology (CREST) program of the Japan Science and Technology Agency (JST).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: AAA, ATPases associated with diverse cellular activities; BCCP, biotin carboxyl carrier protein.
References
- 1.King, S. M. (2000) Biochim. Biophys. Acta 1496, 60–75. [DOI] [PubMed] [Google Scholar]
- 2.Asai, D. J. & Koonce, M. P. (2001) Trends Cell Biol. 11, 196–202. [DOI] [PubMed] [Google Scholar]
- 3.Vale, R. D. (2003) Cell 112, 467–480. [DOI] [PubMed] [Google Scholar]
- 4.Gee, M. A., Heuser, J. E. & Vallee, R. B. (1997) Nature 390, 636–639. [DOI] [PubMed] [Google Scholar]
- 5.Samso, M., Radermacher, M., Frank, J. & Koonce, M. P. (1998) J. Mol. Biol. 276, 927–937. [DOI] [PubMed] [Google Scholar]
- 6.Mocz, G. & Gibbons, I. R. (1996) Biochemistry 35, 9204–9211. [DOI] [PubMed] [Google Scholar]
- 7.Mocz, G., Helms, M. K., Jameson, D. M. & Gibbons, I. R. (1998) Biochemistry 37, 9862–9869. [DOI] [PubMed] [Google Scholar]
- 8.Lee-Eiford, A., Ow, R. A. & Gibbons, I. R. (1986) J. Biol. Chem. 261, 2337–2342. [PubMed] [Google Scholar]
- 9.Silvanovich, A., Li, M. G., Serr, M., Mische, S. & Hays, T. S. (2003) Mol. Biol. Cell 14, 1355–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reck-Peterson, S. L. & Vale, R. D. (2004) Proc. Natl. Acad. Sci. USA 101, 1491–1495. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Straube, A., Enard, W., Berner, A., Wedlich-Soldner, R., Kahmann, R. & Steinberg, G. (2001) EMBO J. 20, 5091–5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laemmli, U. K. (1970) Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- 13.Towbin, H., Staehelin, T. & Gordon, J. (1979) Proc. Natl. Acad. Sci. USA 76, 4350–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Read, S. M. & Northcote, D. H. (1981) Anal. Biochem. 116, 53–64. [DOI] [PubMed] [Google Scholar]
- 15.Toba, S. & Toyoshima, Y. Y. (2004) Cell Motil. Cytoskeleton 58, 281–289. [DOI] [PubMed] [Google Scholar]
- 16.Shimizu, T., Toyoshima, Y. Y., Edamatsu, M. & Vale, R. D. (1995) Biochemistry 34, 1575–1582. [DOI] [PubMed] [Google Scholar]
- 17.King, S. J. & Schroer, T. A. (2000) Nat. Cell Biol. 2, 20–24. [DOI] [PubMed] [Google Scholar]
- 18.Mocz, G. & Gibbons, I. R. (2001) Structure (Cambridge) 9, 93–103. [DOI] [PubMed] [Google Scholar]
- 19.Karata, K., Inagawa, T., Wilkinson, A. J., Tatsuta, T. & Ogura, T. (1999) J. Biol. Chem. 274, 26225–26232. [DOI] [PubMed] [Google Scholar]
- 20.Johnson, A. & O'Donnell, M. (2003) J. Biol. Chem. 278, 14406–14413. [DOI] [PubMed] [Google Scholar]
- 21.Yao, N., Coryell, L., Zhang, D., Georgescu, R. E., Finkelstein, J., Coman, M. M., Hingorani, M. M. & O'Donnell, M. (2003) J. Biol. Chem. 278, 50744–50753. [DOI] [PubMed] [Google Scholar]
- 22.Lee, S. Y., De La Torre, A., Yan, D., Kustu, S., Nixon, B. T. & Wemmer, D. E. (2003) Genes Dev. 17, 2552–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neuwald, A. F. (1999) Structure Fold. Des. 7, R19–R23. [DOI] [PubMed] [Google Scholar]
- 24.Yagi, T. (2000) Cell Struct. Funct. 25, 263–267. [DOI] [PubMed] [Google Scholar]
- 25.Shiroguchi, K. & Toyoshima, Y. Y. (2001) Cell Motil. Cytoskeleton 49, 189–199. [DOI] [PubMed] [Google Scholar]
- 26.Kikushima, K., Yagi, T. & Kamiya, R. (2004) FEBS Lett. 563, 119–122. [DOI] [PubMed] [Google Scholar]
- 27.Mallik, R., Carter, B. C., Lex, S. A., King, S. J. & Gross, S. P. (2004) Nature 427, 649–652. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.