Abstract
The MoaA and MoaC proteins catalyze the first step during molybdenum cofactor biosynthesis, the conversion of a guanosine derivative to precursor Z. MoaA belongs to the S-adenosylmethionine (SAM)-dependent radical enzyme superfamily, members of which catalyze the formation of protein and/or substrate radicals by reductive cleavage of SAM by a [4Fe–4S] cluster. A defined in vitro system is described, which generates precursor Z and led to the identification of 5′-GTP as the substrate. The structures of MoaA in the apo-state (2.8 Å) and in complex with SAM (2.2 Å) provide valuable insights into its mechanism and help to define the defects caused by mutations in the human ortholog of MoaA that lead to molybdenum cofactor deficiency, a usually fatal disease accompanied by severe neurological symptoms. The central core of each subunit of the MoaA dimer is an incomplete triosephosphate isomerase barrel formed by the N-terminal part of the protein, which contains the [4Fe–4S] cluster typical for SAM-dependent radical enzymes. SAM is the fourth ligand to the cluster and binds to its unique Fe as an N/O chelate. The lateral opening of the incomplete triosephosphate isomerase barrel is covered by the C-terminal part of the protein containing an additional [4Fe–4S] cluster, which is unique to MoaA proteins. Both FeS clusters are separated by ≈17 Å, with a large active site pocket between. The noncysteinyl-ligated unique Fe site of the C-terminal [4Fe–4S] cluster is proposed to be involved in the binding and activation of 5′-GTP.
In humans, genetic deficiencies of enzymes involved in molybdenum cofactor (Moco) biosynthesis lead to the pleiotropic loss of the molybdoenzymes sulfite oxidase, aldehyde oxidase, and xanthine dehydrogenase and trigger an autosomal recessive and usually fatal disease, which is characterized by severe neurological symptoms (1, 2). The first step in Moco biosynthesis, the conversion of a guanosine derivative to precursor Z, an oxygen-sensitive 6-alkyl pterin with a cyclic phosphate, is catalyzed by MoaA and MoaC (MOCS1A and MOCS1B in humans) (3, 4). As in the pathways of folate, riboflavin, and biopterin synthesis, a guanosine derivative serves as the initial biosynthetic precursor (5, 6), but in contrast to all other pathways, its C8 atom is retained and incorporated in a rearrangement reaction as the first carbon of the precursor Z side chain.
MoaA belongs to the family of S-adenosylmethionine (SAM)-dependent radical enzymes, members of which catalyze the formation of protein and/or substrate radicals by reductive cleavage of SAM by a [4Fe–4S] cluster (7–9). Members of this family are involved in various metabolic processes, but until now only two members have been structurally characterized: Biotin synthase (BioB) (10), which converts dethiobiotin to biotin and coproporphyrinogen III oxidase (HemN) (11), which catalyzes the conversion of coproporphyrinogen III to protoporphyrinogen IX during heme biosynthesis. These enzymes display related folds in their N-terminal regions, which also harbor the oxygenlabile [4Fe–4S] cluster. This cluster is ligated by only three Cys residues in the apoenzyme, whereas in the complex with SAM, the vacant coordination site on one of the Fe atoms is simultaneously occupied by N and O atoms from the methionine moiety of the cofactor.
MoaA shares 14% and 11% identity in the N-terminal region with BioB and HemN, respectively, but is completely unrelated with these proteins in the C-terminal region, which is in MoaA characterized by another Cys-rich signature motif. Recently, it could be shown that human MOCS1A in fact assembles two oxygen-sensitive [4Fe–4S] clusters, one typical for SAM-dependent radical enzymes and an additional one unique to MoaA proteins (4). The structure of MoaC has been determined earlier, and the protein was found to be present as a hexamer composed of three dimers with a putative active site located at the dimer interface (12). Because some of the SAM-dependent radical enzymes require another protein onto which the radical is transferred, it has been speculated that MoaC might act in a similar function. Here, we describe the structure of MoaA, the final player in the Moco biosynthetic pathway (13). In addition, an in vitro system for precursor Z synthesis is reported, which led to the identification of 5′-GTP as the substrate.
Methods
Cloning, Expression, and Purification. See Supporting Text, which is published as supporting information on the PNAS web site.
Crystallization of MoaA. Crystals were grown under anaerobic conditions inside a glove box (Coy Laboratory Products, Ann Arbor, MI) containing <2 ppm O2 with the hanging drop vapor diffusion technique by incubating MoaA (34 mg/ml, 100 mM Tris·HCl, pH 9.0/300 mM NaCl) with 10 mM DTT for 30 min on ice and subsequent mixing in a 1:1 ratio with precipitant solution [90 mM Na Hepes, pH 7.5/3.15 M Na formate/3% (vol/vol) DMSO]. Crystals appeared within 2 days and were cryoprotected by soaking in mother liquor containing 30% (vol/vol) glycerol. They belong to space group P212121 with cell dimensions of a = 48.1, b = 102.4, and c = 191.2 Å and contain two molecules per asymmetric unit (57% solvent content).
Data Collection and Structure Determination. Fe–multiwavelength anomalous diffraction data were collected on a single crystal on a Rigaku (Tokyo) RU-H3R rotating-anode x-ray generator equipped with double focusing mirror optics and an R axis II imaging plate detector at a wavelength of 1.5418 Å and on beamline X26C at the National Synchrotron Light Source at Brookhaven National Laboratory (Upton, NY) on a Quantum 4R ADSC charge-coupled device detector at a wavelength of 1.7406 Å (Fe peak). All data sets were indexed, integrated, and scaled with HKL software (14). For subsequent calculations, the ccp4 suite was used (15) with exceptions as indicated. Four Fe sites were located with solve (16), which was also used for phase refinement. Phases were subsequently improved with resolve (17), and a model was built with the program o (18). The second monomer was generated with the ncs operation. The structure was refined with refmac5 incorporating translation, libration, and screw-rotation displacement (TLS) refinement in all cycles (19, 20). Each monomer was refined separately, and solvent molecules were automatically added with arp (21).
The MoaA–SAM complex structure was obtained by soaking crystals cocrystallized with 5 mM SAM in precipitant solution containing 10 mM SAM and 30% (vol/vol) glycerol for 30 min. Data were collected on beamline X26C at the National Synchrotron Light Source, Brookhaven National Laboratory at a wavelength of 1.1 Å. The structure was solved by difference Fourier techniques and refined as described for apo-MoaA.
Information concerning data collection and refinement statistics is available in Tables 1 and 2, which are published as supporting information on the PNAS web site. Figures were created with molscript (22), bobscript (23), pymol (DeLano Scientific, San Carlos, CA), spock (24), and raster3d (25).
Assays of Precursor Z Synthesizing Activity and Reductive Cleavage of SAM. Nitrate reductase overlay assays and in vivo assays in moaA (KB2037) and moaC (KB2066) mutant cells were conducted as described (3). In vitro assays were performed under anaerobic conditions at room temperature in a total volume of 180 μlof100 mM Tris·HCl, pH 9.0/300 mM NaCl in the presence of 2 mM DTT and a 10-fold molar excess of SAM, 5′-GTP, MgCl2, Na2S2O4, and 50 μM MoaC. The reaction was started by adding 50 μM MoaA and terminated, at specified times, by the addition of 220 μl 100 mM Tris·HCl, pH 7.2/50 μl of acidic iodine to convert precursor Z to its stable fluorescent product compound Z. After incubation at room temperature for 14 h, compound Z was purified as described (3). SAM cleavage was performed in a total volume of 160 μl in the presence of 2 mM DTT and a 10-fold molar excess of SAM, 5′-GTP, and Na2S2O4. The reaction was started by adding 50 μM MoaA and terminated by the addition of 40 μl of 50% (wt/vol) trichloroacetic acid. SAM and 5′-deoxyadenosine (5′-dA) were analyzed by reversed-phase HPLC in 50 mM NH4H2PO4, pH 2.5, containing 10% (vol/vol) methanol.
Results and Discussion
Functional Characterization of MoaA and MoaC. MoaA and MoaC from Staphylococcus aureus were heterologously expressed as His-tagged proteins in Escherichia coli. MoaA was purified under anaerobic conditions and additionally treated with Fe and l-cysteine in the presence of the cysteine desulfurase IscS. This resulted in a protein with ≈9 mol of Fe per mol protein, indicating the assembly of two extremely oxygen-sensitive [4Fe–4S] clusters in analogy with mocs1a (4). In solution, MoaA exists as a monomer (41 kDa) and dimer (82 kDa), as determined by size exclusion chromatography (data not shown). MoaC is mainly in a hexameric form (120 kDa), which, however, easily dissociates into the dimer (40 kDa). For both proteins, the functional oligomeric states are unknown. In vitro assembly studies in the presence and absence of substrates and crosslinking experiments, as well as coexpression and subsequent copurification, failed to identify a tight MoaA–MoaC protein complex.
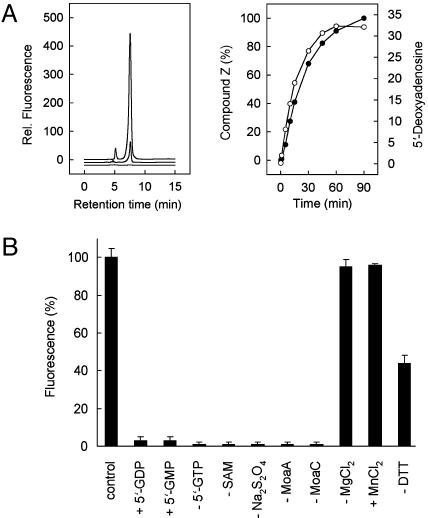
Purified MoaA and MoaC are able to catalyze the formation of precursor Z in a 5′-GTP- and SAM-dependent reaction (Fig. 1); however, neither 5′-GDP nor 5′-GMP can be used. Precursor Z was identified by coelution with precursor Z isolated from E. coli chlM cells known to accumulate this compound (26). Anaerobically purified MoaA with ≈4 mol of Fe per mol of protein shows activities of only 10–12% identifying MoaA with the two reconstituted [4Fe–4S] clusters as the catalytically competent form. Precursor Z synthesis is significantly increased in the presence of 2 mM DTT (Fig. 1B), although the specific function of DTT is unknown. Precursor Z synthesis depends strictly on the presence of MoaA and MoaC. The reduction of the FeS cluster(s) with Na2S2O4 is necessary to induce the cleavage of SAM yielding a 5′-dA radical (5′-dA•), as is the case in other SAM-dependent radical enzymes (8, 9). The reductive cleavage of SAM is increased by a factor of ≈10 in the presence of 5′-GTP. A time dependence of 5′-dA generation and precursor Z formation shows a simultaneous increase with a total yield of 0.65 mol of 5′-dA/mol of MoaA (Fig. 1 A).
Fig. 1.
In vitro synthesis of precursor Z. (A Left) Synthesis of precursor Z determined by measurement of its stable oxidized derivative, compound Z, by reversed-phase HPLC. MoaA (50 μM) and MoaC (50 μM) were incubated under anaerobic conditions in the presence of 2 mM DTT with a 10-fold molar excess of SAM, 5′-GTP, MgCl2, and Na2S2O4 for 45 min (standard assay). (Top to bottom) Reconstituted MoaA; anaerobically purified MoaA; without MoaC. (Right) Time dependence of precursor Z formation (•) and reductive cleavage of SAM in the presence of 5′-GTP generating 5′-dA (○). (B) Requirements for precursor Z synthesis. The maximal amount of compound Z formed in a standard assay (control, see A) was set to 100%. 5′-GTP was absent from the experiment with 5′-GDP and 5′-GMP.
Structure of the MoaA Monomer. MoaA was crystallized under anaerobic conditions, and the structure was solved by multiwavelength anomalous diffraction techniques using Fe as the anomalous scatterer. The final model, refined at 2.8-Å resolution (Rcryst = 18.9%, Rfree = 21.1%) contains 327 (Gln-3 to Gln-329) of 340 residues. The 3 N-terminal residues and the His-tag as well as 11 C-terminal residues are disordered. The central core of each monomer, which is formed by the N-terminal part of the protein (amino acids 3–223), is composed of an incomplete [(αβ)6] triosephosphate isomerase (TIM) barrel (27) with a lateral opening (Fig. 2A). A canonical TIM barrel [(αβ)8] consists of an inner ring of eight parallel β-strands that is wrapped by an outer wheel comprised typically of eight α-helices. However, some proteins display deviations from the classical (αβ)8-barrel fold, including variations in the number of β-strands, their orientation (typically all parallel), local structural distortions, and also lack of barrel closure (28). The C-terminal part of the protein (amino acids 224–329) covers the lateral opening of the barrel (Fig. 2 A). This part consists of a three-stranded antiparallel β-sheet, an extended loop leading to the other side of the protein and three α-helices, which complete the C-terminal part. Interestingly, a hydrophilic channel is observed in the center of the TIM barrel.
Fig. 2.
Overall structure of MoaA. (A) Ribbon diagram of the monomer. The N-terminal TIM barrel structure is shown in maroon, and the C-terminal part is shown in gold. (B) Ribbon diagram of the dimer. The two monomers are colored in blue and orange. (C) Molecular surface of the dimer colored as in B viewed along the hydrophilic channel in one of the monomers. FeS clusters are shown in cpk (Fe, green; S, yellow), and SAM is shown in stick representation.
MoaA contains two [4Fe–4S] clusters, one in the N-terminal part and one in the C-terminal part of the protein (Fig. 2 A). Both FeS clusters are on opposite sides of the hydrophilic channel. The N-terminal cluster is ligated by three Cys residues, which originate from a signature sequence motif (Cx3Cx2C) characteristic of all SAM-dependent radical enzymes (8, 9). This motif is contained in a 31-residue loop extending from β-strand 1 to α-helix 1 of the TIM barrel (Fig. 6, which is published as supporting information on the PNAS web site), and the 3 Cys coordinate 3 of the 4 Fe of the [4Fe–4S], whereas the fourth Fe has a vacant protein coordination site in the apoenzyme. In the structure of apo-MoaA, a yet-unidentified residual difference density is present at the noncysteinyl ligated Fe site that is too strong to be modeled as a single water molecule. The second [4Fe–4S] cluster is ligated by a Cx2Cx13C motif and is assembled between two loops in the C-terminal part of the protein leading to a closure of the barrel structure. This [4Fe–4S] cluster is unique to MoaA poteins but similar to the N-terminal cluster, it does not feature a Cys ligand to the fourth Fe atom.
Structure and Function of the MoaA Dimer. MoaA is present as an elongated homodimer with dimensions of 79 Å × 58 Å × 45 Å (Fig. 2 B and C). The two monomers are related by a noncrystallographic 2-fold symmetry axis, which is approximately parallel to the axis of the barrel. The contact interface covers an area of ≈1,400 Å2, corresponding to ≈9% of the surface area of each monomer. The dimer interface is mainly formed by contacts between helices α7 of the N-terminal domain and α10 of the C-terminal domain of each monomer and is composed of polar (41%) as well as hydrophobic (59%) residues. As described above, the enzyme exists in both a monomeric and dimeric form in solution, but the functional significance of the dimer is not entirely clear at present, because residues located in the dimer interface region are not conserved in MoaA proteins. The C-terminal α-helix (α10) is located in the dimer interface and points toward the active site of the second monomer (Fig. 2 B and C). The conformation of this helix is indicative of helix swapping, a known mechanism in oligomeric assembly (29).
MoaAs from eubacteria and eukaryotes contain a functionally important double Gly motif at the C terminus (3). Deletion of both Gly residues affects binding and redox properties of the FeS cluster(s), suggesting a location in the active site (Fig. 7A, which is published as supporting information on the PNAS web site). However, there is no interpretable electron density for the last 11 amino acids, indicating that these residues are highly mobile. It is possible that the C terminus adopts an ordered conformation only upon binding of 5′-GTP. Based on the crystal structure, there are two possible conformations of the C terminus: either it points back into the active site of the same monomer, or, more likely, into the active site of the other monomer. The C terminus is in both cases at a distance of ≈20–23 Å to the noncysteinyl ligated Fe site of the C-terminal FeS cluster. Maybe the last C-terminal amino acids protrude into the hydrophilic channel leading to closure of the active site after 5′-GTP-binding (Fig. 2C). In contrast to eubacteria and eukaryotes, precursor Z synthesis in archaea does not depend on a C-terminal double Gly motif and, interestingly, at least MoaAs from Methanococcus jannaschii and Archaeoglobus fulgidus are exclusively in a monomeric form (unpublished data). These data indicate differences in the oligomeric state and catalytic mechanism of archaeal MoaAs.
The Active Site of MoaA and Substrate Binding. The 2.2-Å resolution (Rcryst = 21.3, Rfree = 24.1) structure of MoaA in complex with SAM revealed no conformational changes, because the apo and SAM complex structures can be superimposed with an rms deviation of 0.35 Å. The bound SAM constitutes the fourth ligand to the [4Fe–4S] cluster located in the N-terminal domain and binds as an N/O chelate to the unique Fe position of this cluster through its amino-group nitrogen (N-Fe 2.6 Å) and carboxyl-group oxygen (O-Fe 1.9 Å) (Fig. 3A). In addition, the sulfonium sulfur of SAM is at a distance of 3.3 Å to the same Fe and 3.6 Å to the adjacent sulfur atom. The SAM-binding site comprises residues from disparate parts of the sequence and 3D structure (Figs. 3A and 6). However, amino acid residues surrounding the bound SAM within a distance of 5 Å are conserved in MoaA proteins. SAM is bound in extended conformation stretching across the top of the barrel and is stabilized by comparatively few interactions, including four hydrogen bonds. The net result of these interactions is that SAM is completely buried from solvent and appears ideally positioned for electron transfer from the [4Fe–4S] cluster. This binding mode is analogous to what has been observed in BioB and HemN (10, 11).
Fig. 3.
The active site of MoaA. (A) Stereoview of the FeS clusters and SAM. SAM is shown in a Fo–Fc omit map contoured at 3 σ. Hydrogen bonds are indicated as dashed lines. (B) Closer view of the active-site pocket created by predominantly positively charged amino acids. Conserved amino acids are labeled in red. Secondary structural elements and cofactors are rendered transparent. (C) Surface representation viewed from both sides into the active site channel. Conserved residues are mapped onto the molecular surface. (D) σA weighted 2Fo–Fc map of the C-terminal FeS cluster with a DTT molecule as the fourth ligand contoured at 1 σ. Surrounding amino acids are shown as a Cα trace colored in orange. Amino acids, FeS clusters, and SAM are shown in ball and stick.
Residual difference density close to the FeS–SAM substrate complex, which is also present in the apo-structure, could be modeled as a phosphate or sulfate (not shown). This anion is at distance of ≈5 Å to the Fe coordinating the SAM and the 3′-OH group of SAM. Because no phosphate/sulfate-containing buffers were used during purification, reconstitution, and crystallization, this anion must be already bound tightly before isolation of the protein. Accordingly, it is stabilized by several hydrogen bonds with D128 (O4-N 2.8 Å, O1-Oδ1 3.0 Å, and O3-Oδ2 2.9 Å), and Y30 (O3-OH 3.2 Å). So far, no functional role is known for the involvement of this anion.
The two FeS clusters define the general location of the active site of MoaA. The closest distance between them is ≈17 Å (Fe4–Fe4), and they create a large active site pocket interrupted by the hydrophilic channel. The active site is constructed from predominantly basic residues (Fig. 3 B and C), including five highly conserved Arg and Lys side chains, which are ideally positioned to compensate the negative charges of the 5′-GTP phosphate groups. Attempts to get a structure with bound 5′-GTP either by cocrystallization or soaking experiments have failed so far. Presumably, the 5′-GTP interaction is weak as already observed for the second substrate SAM. Additionally, a decreased binding can be expected due to the high salt concentrations necessary for crystal growth.
Although the substrate for precursor Z synthesis could be identified as 5′-GTP, the MoaA structure revealed no typical nucleotide binding motif/fold like a P-loop (30) or Rossmann fold (31) frequently found in dinucleotide binding proteins. This is not unexpected because during precursor Z synthesis the pyrophosphate moiety of 5′-GTP is only transiently attached to the protein and subsequently released. Binding and release of pyrophosphate usually requires a divalent cation like Mg2+ or Mn2+ as a cofactor. Somewhat surprisingly, precursor Z synthesis does not depend on MgCl2 or MnCl2 (Fig. 1B), and there is no obvious cation-binding site.
An interesting feature of the MoaA structure is a residual difference density associated with the unique Fe site of the C-terminal FeS cluster, which could be modeled as DTT (Fig. 3D). DTT binds with one of its sulfur atoms to the fourth Fe atom (S4–Fe4 2.2 Å), thus also resulting in a tetrahedral coordination of this Fe atom. The remainder of the DTT is not involved in any contacts with the protein. As described above, synthesis of precursor Z is significantly increased in the presence of DTT. Maybe, DTT helps in vitro to stabilize the labile nonligated Fe and maintains it in the ferrous state. During catalysis DTT is presumably replaced by the substrate (see below). Furthermore, it has been shown in case of a ferredoxin that binding of DTT to the [4Fe–4S] cluster led to a 200 mV negative shift of the redox potential of the FeS cluster (32).
In addition to their primary function in mediating electron redox processes, FeS clusters are known to control protein structure, act as environmental sensors, serve as modulators of gene regulation, and also participate in radical generation (33), as is the case for the FeS cluster common to all SAM-dependent radical enzymes. A structural role or function in electron transfer of the C-terminal FeS cluster is considered unlikely as the cluster is easily degraded and has an incomplete cysteinyl ligation. A putative function of this MoaA specific FeS cluster could be analogous to the N-terminal FeS cluster that is involved in anchoring the substrate SAM to position it in a proper conformation for the subsequent radical chemistry. One can imagine that the unique Fe site of the C-terminal FeS cluster anchors the 5′-GTP to position the guanine C8 atom close to the 5′-C atom of the 5′-dA radical (5′-dA•) generated by the N-terminal FeS cluster. The spatial proximity could propagate the radical based reaction by abstracting a hydrogen atom from the C8 atom leading to the proposed rearrangement reaction (5, 6). This function in substrate binding would be analogous to aconitase, where it could be shown that an FeS cluster participates in the enzymatic mechanism (34, 35). Aconitase assembles like MoaA a [4Fe–4S] cluster ligated by only three Cys and the noncysteinyl-ligated unique Fe site is involved in the binding and activation of substrates.
Location of Missense Mutations Identified in Moco-Deficient Patients. Mutations within the MOCS1A ORF that is homologous to MoaA account for ≈50% of all known cases of Moco deficiency (1, 2). All of the genetic lesions affect highly conserved regions (Fig. 6). Several mutations are found in close proximity to the conserved Cys motifs of the N- and C-terminal [4Fe–4S] clusters (R73W, R123W, R319Q, and G324E; S. aureus R17, R71, R268, and G273). The R319Q mutation is the most common mutation, accounting for 14% of Moco-deficient alleles and 21% of all identified mutations. Furthermore, missense mutations were found in the conserved 123RxTGGEP129 sequence (S. aureus 71RxTGGEP77) and additionally in the catalytically essential C-terminal double Gly motif (G384S; S. aureus G339) (3) that is not resolved in the structure. To obtain insights into the molecular mechanisms leading to human Moco deficiency, site-directed mutagenesis was used to generate the described missense mutations in the gene encoding MOCS1A. The activity of these variants was analyzed with a nitrate reductase overlay assay. None of the resulting variants were able to complement an E. coli moaA mutant (Fig. 7B). All identified missense mutations led to a loss of activity of MOCS1A, and the purified proteins show a decrease in Fe content by ≈20–50% and different UV-visible absorption spectra, suggesting changes in the environment of the FeS clusters (Fig. 7C).
The MoaA structure revealed that all affected residues are located in the active-site pocket (Fig. 4). G273 (G324 in MOCS1A) has main chain dihedral angles characteristic of left-handed helices (φ = 72.5°, ψ = 43.7°), which are uncommon for non-Gly residues. G273 is located in a turn connecting two β-strands that lead into the two loops ligating the C-terminal FeS cluster. Disruption of this turn by a G324E mutation presumably results in major conformational changes and subsequently in loss of the C-terminal FeS cluster leading to MOCS1A destabilization and enhanced proteolytic degradation. These structural considerations are in agreement with the fact that this mutant protein cannot be purified in a stable form (Fig. 7C), and that the loss of the C-terminal FeS cluster destabilizes the protein (4). Mutation of the three active site residues, R73/R17, R123/R71, and R319/R268, presumably affects the proposed interaction with the negatively charged phosphate groups of 5′-GTP. G75/G127 is hydrogen bonded to the SAM amino nitrogen (Fig. 3A) and ensures its correct orientation. The mutations G126D and G127R in this loop region presumably result in loss of SAM–FeS binding. The specific roles of these disease-associated amino acids in the catalytic mechanism remain to be investigated.
Fig. 4.
Location of missense mutations detected in Moco-deficient patients. Secondary structure elements and cofactors are rendered transparent. Human mutations, FeS clusters, and SAM are shown as ball and stick.
Structural Comparison of Members of SAM-Dependent Radical Enzymes. A search for structural homologs of MoaA with the program dali (36) identified, besides a large number of TIM barrel proteins, the two SAM-dependent radical enzymes BioB and HemN as the closest structural homologs. HemN was the highest match with a Z score of 11.7 and an rms deviation of 4.7 Å for 232 residues, whereas BioB was the second highest match with a Z score of 11.3 and an rms deviation of 4.1 Å for 208 residues. The structures of BioB (10), HemN (11), and MoaA show in their N-terminal parts a common structural core involved in [4Fe–4S] cluster and SAM binding (Fig. 5). Nevertheless, in each case, unique structural elements near the C terminus and additional cofactors/substrates are required for the generation of radicals on structurally divergent substrates. Common to all structures is a complete [(αβ)8, BioB] or incomplete [(αβ)6, MoaA, HemN] TIM barrel ligating the [4Fe–4S]–SAM substrate complex. BioB assembles an additional air stable [2Fe–2S] cluster, the postulated source of sulfur for biotin formation. The nonhomologous C-terminal part of BioB with two additional α-helices and β-strands completes the TIM barrel. HemN contains a second SAM molecule immediately adjacent to the first, and both SAM molecules were proposed to be involved in catalyzing the two propionate decarboxylation reactions. Besides this, a distinct C-terminal domain that is missing in the BioB and MoaA structures covers the lateral opening of the incomplete TIM barrel. Finally, MoaA ligates a second oxygen-sensitive [4Fe–4S] cluster by only three Cys residues, with the unique Fe site proposed to be involved in anchoring the substrate 5′-GTP. A search for structural homologs of the C-terminal part of MoaA with the program dali (36) revealed no structurally significant homolog.
Fig. 5.
Structural comparison of SAM-dependent radical enzymes and reactions catalyzed. α-Helices are colored in orange, β-sheets are colored in blue, and turns are colored in gray. Nonstructurally conserved secondary structural elements are rendered transparent. FeS clusters and substrates are shown in ball and stick.
Investigations of a Putative Function of Human MOCS1B in a Radical-Based Reaction Mechanism. SAM-dependent radical enzymes can be divided into two subfamilies (8, 9). Subfamily I is a two-protein system composed of an FeS protein, which is the activating protein catalyzing the reductive cleavage of SAM into methionine and the 5′-dA radical (5′-dA•), which then generates a glycyl radical on the catalytic protein. The glycyl radical in turn creates a thiyl radical. Representative members of this subfamily are anaerobic ribonucleotide reductase and pyruvate formate lyase. BioB and HemN belong to subfamily II, which is a single-protein system combining activating and catalytic activities by direct substrate radical formation. So far, it is not known to which subfamily MOCS1A/MoaA belongs. Because two proteins, MoaA/MOCS1A and MoaC/MOCS1B, are essential for precursor Z synthesis, one could speculate that MoaA/MOCS1A is the activating enzyme that leads to the generation of catalytically active glycyl- and thiyl-radicals on MoaC/MOCS1B. Based on the structure of E. coli MoaC, a putative active site has been mapped that is located at the dimer interface (12). Four sequence stretches, including the highly conserved amino acids G117, C141, and G175 in MOCS1B, are involved. Site-directed mutagenesis of these amino acids led only in the case of G175 to a complete inactivation (Fig. 8, which is published as supporting information on the PNAS web site). Additionally, the G117A variant does not show the typical accumulation of precursor Z, although wild-type levels of molybdopterin are present. Therefore, G175 and G117 are functionally and/or structurally essential and could be involved in harboring a catalytic and essential glycyl radical. However, it is unlikely that MoaC/MOCS1B belongs to subfamily I, because (i) generation of a G175A variant in the homologous E. coli MoaC resulted only in an activity decrease by 40% (12), (ii) MoaC/MOCS1B does not contain the typical radical sequence motif [consensus RV(C/S)GY] (9), (iii) MOCS1B has no catalytically essential Cys residue, and (iv) MoaC (12) does not show the conserved 3D structure with a central 10-stranded α/β barrel as known from members of this family (37). Although precursor Z synthesis depends strictly on MoaC/MOCS1B, there is so far no obvious function for MoaC/MOCS1B. Based on these considerations, it is unlikely that during the formation of precursor Z, a protein-based radical mechanism is involved and MoaA/MoaC can therefore be classified as the first member of a third subfamily (subfamily III), characterized by an accessory protein that is not involved in the radical-based mechanism.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant DK54835 (to H.S.). The National Synchrotron Light Source in Brookhaven is supported by the Department of Energy and the National Institutes of Health, and beamline X26C is supported in part by the Research Foundation of the State University of New York at Stony Brook.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: SAM, S-adenosylmethionine; 5′-dA, 5′-deoxyadenosine; TIM, triosephosphate isomerase; BioB, biotin synthase; HemN, coproporphyrinogen III oxidase; Moco, molybdenum cofactor; MOCS, molybdenum cofactor synthesis.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 1TV7 and 1TV8).
References
- 1.Reiss, J. (2000) Hum. Genet. 106, 157–163. [DOI] [PubMed] [Google Scholar]
- 2.Reiss, J. & Johnson, J. L. (2003) Hum. Mutat. 21, 569–576. [DOI] [PubMed] [Google Scholar]
- 3.Hänzelmann, P., Schwarz, G. & Mendel, R. R. (2002) J. Biol. Chem. 277, 18303–18312. [DOI] [PubMed] [Google Scholar]
- 4.Hänzelmann, P., Hernández, H. L., Menzel, C., García-Serres, R., Huynh, B. H., Johnson, M. K., Mendel, R. R. & Schindelin, H. (2004) J. Biol. Chem. 279, 34721–34732. [DOI] [PubMed] [Google Scholar]
- 5.Wuebbens, M. M. & Rajagopalan, K. V. (1995) J. Biol. Chem. 270, 1082–1087. [DOI] [PubMed] [Google Scholar]
- 6.Rieder, C., Eisenreich, W., O'Brien, J., Richter, G., Gotze, E., Boyle, P., Blanchard, S., Bacher, A. & Simon, H. (1998) Eur. J. Biochem. 255, 24–36. [DOI] [PubMed] [Google Scholar]
- 7.Sofia, H. J., Chen, G., Hetzler, B. G., Reyes-Spindola, J. F. & Miller, N. E. (2001) Nucleic. Acids Res. 29, 1097–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jarrett, J. T. (2003) Curr. Opin. Chem. Biol. 7, 174–182. [DOI] [PubMed] [Google Scholar]
- 9.Frey, P. A. & Magnusson, O. T. (2003) Chem. Rev. 103, 2129–2148. [DOI] [PubMed] [Google Scholar]
- 10.Berkovitch, F., Nicolet, Y., Wan, J. T., Jarrett, J. T. & Drennan, C. L. (2004) Science 303, 76–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Layer, G., Moser, J., Heinz, D. W., Jahn, D. & Schubert, W. D. (2003) EMBO J. 22, 6214–6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wuebbens, M. M., Liu, M. T., Rajagopalan, K. V. & Schindelin, H. (2000) Structure Fold. Des. 8, 709–718. [DOI] [PubMed] [Google Scholar]
- 13.Rizzi, M. & Schindelin, H. (2002) Curr. Opin. Struct. Biol. 12, 709–720. [DOI] [PubMed] [Google Scholar]
- 14.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 15.Collaborative Computational Project 4 (1994) Acta Crystallogr. D 50, 760–763.15299374 [Google Scholar]
- 16.Terwilliger, T. C. & Berendzen, J. (1999) Acta Crystallogr. D 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Terwilliger, T. C. (2000) Acta Crystallogr. D 56, 965–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, 110–119. [DOI] [PubMed] [Google Scholar]
- 19.Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997) Acta Crystallogr. D 53, 240–255. [DOI] [PubMed] [Google Scholar]
- 20.Winn, M. D., Isupov, M. N. & Murshudov, G. N. (2001) Acta Crystallogr. D 57, 122–133. [DOI] [PubMed] [Google Scholar]
- 21.Perrakis, A., Morris, R. & Lamzin, V. S. (1999) Nat. Struct. Biol. 6, 458–463. [DOI] [PubMed] [Google Scholar]
- 22.Kraulis, P. J. (1991) J. Appl. Crystallogr. 24, 946–950. [Google Scholar]
- 23.Esnouf, R. M. (1997) J. Mol. Graphics 15, 132–134. [DOI] [PubMed] [Google Scholar]
- 24.Christopher, J. A. & Baldwin, T. O. (1998) J. Mol. Graphics 16, 285. [Google Scholar]
- 25.Merritt, E. A. & Bacon, D. J. (1997) Methods Enzymol. 277, 505–524. [DOI] [PubMed] [Google Scholar]
- 26.Johnson, M. E. & Rajagopalan, K. V. (1987) J. Bacteriol. 169, 117–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murzin, A. G., Brenner, S. E., Hubbard, T. & Chothia, C. (1995) J. Mol. Biol. 247, 536–540. [DOI] [PubMed] [Google Scholar]
- 28.Nagano, N., Orengo, C. A. & Thornton, J. M. (2002) J. Mol. Biol. 321, 741–765. [DOI] [PubMed] [Google Scholar]
- 29.Liu, Y. & Eisenberg, D. (2002) Protein Sci. 11, 1285–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saraste, M., Sibbald, P. R. & Wittinghofer, A. (1990) Trends Biochem. Sci. 15, 430–434. [DOI] [PubMed] [Google Scholar]
- 31.Rossmann, M. G., Moras, D. & Olsen, K. W. (1974) Nature 250, 194–199. [DOI] [PubMed] [Google Scholar]
- 32.Butt, J. E., Sucheta, A., Armstrong, F. A., Breton, J., Thomson, A. J. & Hatchikiant, E. C. (1993) J. Am. Chem. Soc. 115, 1413–1421. [Google Scholar]
- 33.Johnson, M. K. (1998) Curr. Opin. Chem. Biol. 2, 173–181. [DOI] [PubMed] [Google Scholar]
- 34.Kent, T. A., Dreyer, J.-C., Kennedy, M. C., Huynh, B. H., Emptage, M. H., Beinert, H. & Münck, E. (1982) Proc. Natl. Acad. Sci. USA 79, 1096–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beinert, H., Kennedy, M. C. & Stout, C. D. (1996) Chem. Rev. 96, 2335–2373. [DOI] [PubMed] [Google Scholar]
- 36.Holm, L. & Sander, C. (1995) Trends Biochem. Sci. 20, 478–480. [DOI] [PubMed] [Google Scholar]
- 37.Eklund, H. & Fontecave, M. (1999) Structure (Cambridge, U.K.) 7, R257–262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.