Abstract
Structural and molecular studies have shown that the vitamin D receptor (VDR) mediates 1α,25(OH)2-vitamin D3 gene transactivation. Recent evidence indicates that both VDR and the estrogen receptor are localized to plasma membrane caveolae and are required for initiation of nongenomic (NG) responses. Computer docking of the NG-specific 1α,25(OH)2-lumisterol to the VDR resulted in identification of an alternative ligand-binding pocket that partially overlaps the genomic pocket described in the experimentally determined x-ray structure. Data obtained from docking five different vitamin D sterols in the genomic and alternative pockets were used to generate a receptor conformational ensemble model, providing an explanation for how VDR and possibly the estrogen receptor can have genomic and NG functionality. The VDR model is compatible with the following: (i) NG chloride channel agonism and antagonism; (ii) variable ligand-stabilized trypsin digest banding patterns; and (iii) differential transcriptional activity, employing different VDR point mutants and 1α,25(OH)2-vitamin D3 analogs.
The steroid hormone 1α,25(OH)2-vitamin D3 (1,25D) (Fig. 1A), other steroid hormones, retinoids, and thyroid hormones form the family of ligands for the nuclear receptor (NR) superfamily (1), the members of which produce genomic responses through selective interaction of the liganded receptor with promoters of appropriate genes and basal transcription machinery. Many of these hormones also activate rapid, nongenomic (NG), cellular signaling cascades (2, 3) (except retinoids) that range from activation of ion channels (4, 5) to promoting kinase and other cytosolic signaling cascades (6–9). Defining the structure-function requirements for 1,25D and 17β-estradiol (E2) rapid actions has been aided by the synthesis of analogs that are NG agonists like 1α,25(OH)2-lumisterol (JN) (ref. 10 and Fig. 1A) and 4-estren-3α, 17β-diol (EST) (11) or antagonists like 1β,25(OH)2-vitamin D3 (HL) (ref. 12 and Fig. 1A), but that are only weak genomic transactivators (13). Thus, important structural attributes of the sterol dictate its agonistic properties and subsequent genomic vs. NG signaling profile (14, 15).
Fig. 1.
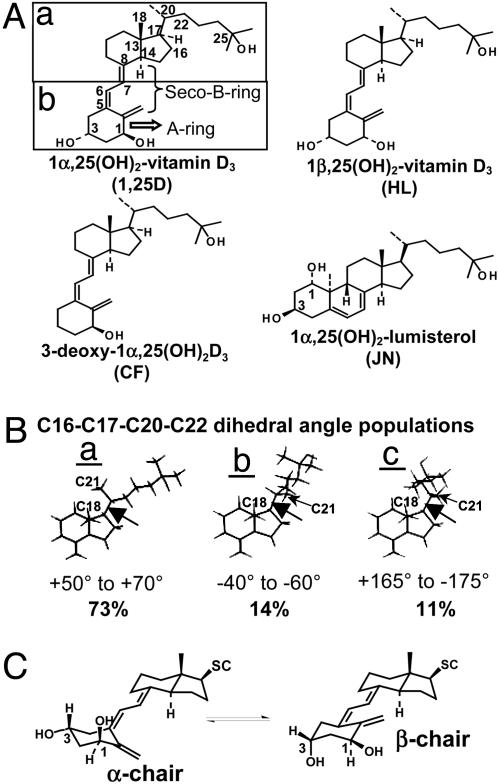
VDR natural and synthetic ligands. (A) The structures of 1,25D, metabolites, and analogs used in this study are shown. The carbon atoms of 1,25D with a hydroxyl moiety (C1, C3, and C25) or a flexible dihedral (360° rotation) are labeled. (Aa and b) Highlighted are the different chemical and physical properties of the two functional halves of the 1,25D molecule (40). (B) Dot maps of 1,25D (27) in a 4.0-kcal energy window were generated by using GMMX (pcmodel V9.0). (B) For side-chain population a, the lowest-energy conformer of the 262 1,25D CD ring fragments is depicted. The C16,17,20,22 dihedral (black arrow) of 98% of these conformers can be grouped into three dihedral population categories defining a window of 20°. For population a, the dihedral range is between +50° and +70° (73%); for population b, between –40° and –60° (14%); and for population c, between +165° and –175° (11%). (C) The α- and β-chair 1,25D A rings are depicted.
When 1,25D rapid signaling cascades were first discovered, it was hypothesized that the observed activities were propagated by a novel membrane protein(s) (16), because analogs JN and HL did not compete well with [3H]1,25D for binding to the nuclear vitamin D receptor (VDR) (17). Recently, in studies using a VDR knockout (KO) mouse (18) and a naturally occurring human VDR mutation (19), 1,25D-mediated rapid responses were shown to require a functional VDR. Both the VDR and estrogen receptor (ER) have been found localized to the plasma membrane in caveolae (7, 20); therefore, it has been proposed that the VDR and ER propagate some NG signaling (6, 9, 18, 21, 22). However, given the poor affinity of JN and HL for the VDR, it is difficult to understand how these sterols can facilitate their activities through the VDR.
Results obtained from the modeling (insight 2000.1) of JN, 1,25D, and HL in the VDR ligand-binding domain (LBD) showed that the VDR could possibly accept and form favorable nonbonding interactions with vitamin D sterols in a distinct ligand-binding pocket [an alternative ligand-binding pocket (A pocket)] from the genomic pocket (G pocket) that was previously defined by x-ray crystallography (23, 24). Our proposed A-pocket accepts ligands that differ in shape from those in the classical G pocket (3, 23, 24).
The data from these models has led to the proposal that the VDR can function as a rapid response receptor through a conformational ensemble mechanism (3, 25) whereby the flexible 1,25D steroid hormone samples an ensemble of energetically similar protein conformations (26). In addition, the ensemble model and existence of an A pocket may provide an explanation for the observed sex-nonspecific, nongenotropic signaling through the ERα receptor by EST (3, 9, 11). The physiological relevance of the ensemble model and functional importance of an A pocket within the VDR is further substantiated by applying the model to the following observations: (i) variable ligand-induced partial trypsin digest electrophoresis-banding patterns, and (ii) differential transcriptional activity employing different VDR point mutants and vitamin D sterols.
Materials and Methods
Reagents. JN was provided by W. H. Okamura (University of California, Riverside). The gifts of 1,25D, 25(OH)-vitamin D3 (25D), HL, and 3-deoxy-1α,25(OH)2-vitamin D3 (CF) were from M. Uskokovic (Hoffmann La Roche, Nutley, NJ). The VDR mutant construct S278A was a gift from E. Collins (San Jose State University, San Jose, CA).
pcmodel V9.0 Dot Map Calculation. Dot maps, tracking the position of the 25-OH group in each conformer, were produced by using only the CD ring fragment (Fig. 1 Aa) (ref. 27). Each dot represents the position of the 25-OH group. Conformational search calculations were performed by using the gmmx driver (pcmodel V9.0; Serena Software, Bloomington, IN) default settings.
Docking JN and 1,25D in the A Pocket. The unoptimized form [builder module, insight 2000.1; Accelrys (San Diego) force field-based simulations] of 6-s-cis-locked JN, was overlaid on 1,25D in the G pocket of the VDR LBD (amino acids 118–427, Δ165–215; Protein Data Bank ID code 1DB1) by superimposing the C1, C3, and C5 atoms of JN on the C3, C1, and C5 atoms of 1,25D, respectively (Fig. 1 A). The 1,25D was then deleted from the assembly, JN was merged into the assembly, and minimization was performed (discover_3, cff91 force field; insight 2000.1). The α/β-chair forms of 1,25D were docked in the A pocket by manually rotating the C5,6,7,8 dihedral to ±20° (i.e., a high-energy 6-s-cis conformation) to begin the calculations. Starting calculations in this manner and using both the α- and β-chair forms (Fig. 1C) in the computations tests the possible 1,25D A ring (Fig. 1 A) α/β-chair orientations in the A pocket (a total of four). Favorable complexes are determined in the molecular modeling protocol by convergence on a derivative ≤0.5 kcal/mol–1·Å–1 and subsequent dynamic simulations showing energy conservation (Fig. 5, which is published as supporting information on the PNAS web site; Accelrys force field-based simulations). See Supporting Text, which is published as supporting information on the PNAS web site, for detailed modeling protocol and procedures for calculating interaction energies (IEs).
Modeling CD Ring Fragments Generated with pcmodel V9.0. From the conformer populations (see Fig. 1 Ba–c) generated in the gmmx calculation (see above), CD ring fragments (Fig. 1 Aa) were selected and superimposed on the C18, C13, and C14 atoms of 1,25D docked in either the G or A pocket. Carbon 7 was then fused with C6 of the A ring fragment (Fig. 1 A). All of the data in Table 1 were produced by using this method. The final JN/VDR (A pocket) and starting 1,25D/VDR (G pocket) structures served as the starting structures for all of these models.
Table 1. Summary of VDR LBD–ligand interaction energies.
| Vitamin D sterol | A-ring chair/seco-B-ring rotomer* | VDR pocket† | IE kcal/mol‡ | Average A-pocket IE kcal/mol§ | VDR ΔIE kcal/mol¶ |
|---|---|---|---|---|---|
| 1,25D | β/trans | G | -108 | ||
| α/trans | G | -100 | -91 | 17 | |
| β/cis | A | -93 | |||
| α/trans | A | -88 | |||
| JN | β/cis | G | -104 | -89 | 15 |
| β/cis | A | -89 | |||
| HL | β/trans | G | -100 | ||
| α/trans | G | -93 | -81 | 19 | |
| β/cis | A | -80 | |||
| α/trans | A | -95 | |||
| 25D | α/trans | A | -86 | ||
| β/cis | A | -78 | -84 | 8 | |
| β/trans | G | -91 | |||
| CF | α/trans | A | -76 | ||
| β/cis | A | -85 | -82 | 16 | |
| β/trans | G | -98 |
A-ring chair form (α or β) used to start the calculation and the final orientation of the seco-B ring (cis or trans).
G, G pocket; A, A pocket. For all experiments, the H305 is the H-bond acceptor and H397 the donor to the 25-OH group (23).
The IEs were calculated by using a distance-dependent dielectric (1*r2, cff91 force field) and are presented in units of kcal/mol. The calculated IE represents the van der Waals and Coulombicstabilizing forces between the ligand and the rest of the assembly. Complexes where no A ring H-bonds are formed are indicated by italic IE values.
The average A pocket IEs of were calculated by applying their known A ring α/β-chair favorabilities in CDCl3 (dielectric = 4.8). For example, HL has an α/β chair favorability of 10:90 (ref. 36 and Fig. 1C), thus the average A pocket IE = (-80 × 0.90) + (-95 × 0.10). The 1,25D, CF, and 25D are known to have an α/β-chair favorability of 45:55 (41), 35:65 (calculated), and 60:40 (40), respectively.
The ΔIE value was calculated by subtracting the average A pocket value from the β-chair G pocket IE for the given ligand (bold values). The resulting ΔIE denotes the inferred ligand preference for the G pocket, in kcal/mol, under steady-state conditions.
Electrophysiology. Patch-clamping of ROS 17/2.8 cells was used to determine the change in outward Cl– currents in response to administration of 1,25D and/or its analogs (see ref. 5 for a complete description of the methodology).
Protease Sensitivity. The 35S-VDR was generated from pcDNA3.1(–)NheI(–)VDR by using the Promega TNT reticulocyte lysate kit. The reaction tubes were incubated for 2 h at 30°C, then placed on ice. The 1,25D or analog (10–5 M) was added, the tubes were incubated at room temperature for 20 min (28), and then 15 μg/ml trypsin was added to the tubes and incubated for 20 min at room temperature, followed by a 5-min incubation at 80°C with SDS. After SDS/PAGE, bands were visualized by autoradiography.
Relative Competitive Index (RCI) and EC50 Determination. Analog RCIs were determined in COS-1 cells transfected with pcDNA3.1(–)NheI(–)VDR. Transactivation (genomic response) of the osteocalcin promoter linked to a secreted alkaline phosphatase was determined in CV1 cells (see Supporting Text).
Results
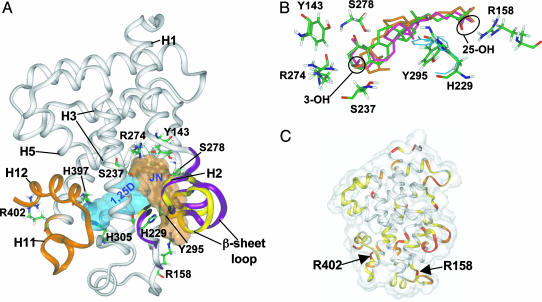
Identification of an A Pocket in the VDR LBD: A-Ring and Side Chain Selectivity. Whereas the 6-s-cis-locked JN (Fig. 1 A) is a full agonist for 1,25D-mediated rapid responses, it is only a weak genomic agonist (10). These results parallel the structural and molecular evidence that 1,25D genomic responses are mediated by its 6-s-trans form. To investigate whether the VDR could bind 6-s-cis (NG conformer) and 6-s-trans (genomic conformer) vitamin D sterols differently, we developed an in silico molecular modeling technique incorporating ligand attributes known to be required for the rapid activation of Cl– channels (5, 18): the 3β-OH and 25-OH groups and a 6-s-cis sterol. When analog JN was manually docked in the VDR LBD, keeping these structural criteria in mind, it was observed that when the 3β-OH contacts and forms a H-bond with R274, the molecule extends toward the Helix 2 (H2) hinge domain/β-sheet region of the LBD (A pocket, Fig. 2A and see Docking JN and 1,25D in the A Pocket in Materials and Methods) rather than toward the H11/H12 region (G pocket, Fig. 2 A).
Fig. 2.
VDR molecular models depicting the G and A pockets. (A) Ribbon diagrams of the assemblies resulting from molecular modeling of the VDR LBD (x-ray coordinates; Protein Data Bank ID code 1DB1) with 6-s-trans 1,25D in the G pocket (cyan Connolly surface) and JN docked in the A pocket (light-brown Connolly surface). The orange ribbon, H11 and H12, is shown with H12 in the closed, transcriptionally active conformation. The yellow ribbons indicate the orientation of the H2/β-sheet region in VDRwt, whereas the purple ribbons indicate their final position after JN was docked in the A pocket. Important amino acid residues discussed in the text are labeled. (B) Superimposition of the VDR amide backbone atoms of the 1,25D α/β-chair and JN A pocket models. JN's oxygen atoms are red and carbon atoms are green (Colat colors). The α-chair form of 1,25D is orange and the β-chair is pink. The starting orientations of Y295 and H229 are indicated by the thin, cyan wireframe and their final positions are indicated by the thicker, Colat-colored wireframe. (C) Superimposition of the 1,25D/VDR G pocket and the 6-s-cis 1,25D/VDR A pocket (backbone rms = 1.18 Å) models rendered to show the VDR LBD Connolly surface (transparent). The ribbon diagram is color-coded to represent the degree of movement observed when the two models are compared. The red regions indicate >3.0 Å, orange between 2.0 and 3.0 Å, yellow between 1.0 and 2.0 Å, and white <1.0 Å movement when the atomic rms are compared. The location of important, flexible Arg residues discussed in the text are labeled for reference.
The JN/VDR in silico model and subsequent dynamics experiments showed that JN can plausibly form a favorable complex with the VDR LBD in a pocket other than the G pocket (Figs. 2 A and 5). The two strongest H-bonds stabilizing JN in this orientation are formed by 3β-OH (Fig. 1 A) with S237 (2.83 Å; H3) and R274 (2.90 Å; C terminus H5; Fig. 2 A and B). The 1α-OH of JN forms an indirect H-bond with R274 by means of a H2O bridge and the 25-OH H-bonds with R158 (Fig. 2B). Importantly, S237 and R274 form H-bonds with the 1α-OH group of 1,25D in the VDR LBD x-ray structures (23, 24), where the steroid is in the G pocket (Fig. 2 A).
In the complex with JN in the A pocket, the first dihedral angle of the side chain (carbons C16,17,20,22) adopts a gauche+ conformation. This rotamer belongs to population a (Fig. 1B), which represents 73% of the side-chain conformations generated in the dot map calculation (262 conformers; see Materials and Methods). Interestingly, side-chain conformers in population b (14%), which adopt a gauche–conformation, fit the G pocket, whereas population c (Fig. 1B) (11%; trans conformation) fits the ligand-binding crevice of the serum vitamin D-binding protein (ref. 29 and Fig. 1B).
Docking the β-chair (Fig. 1C), 6-s-cis configuration of 1,25D in the A pocket indicates 1,25D can form the same protein– ligand contacts with the A pocket as JN (locked β-chair). When compared with JN, the β-chair of 1,25D maintains a 6-s-cis orientation (C5,6,7,8 dihedral = 53°); however, with additional H-bond contacts to the 1α-OH group (S278, loop and Y143, H1; see Fig. 2B). The α-chair of 1,25D (Fig. 1C) can also form a favorable complex with the A pocket (Fig. 2B). Here the secosteroid region adopts a 6-s-trans conformation (C5,6,7,8 dihedral = 152°). For both conformers, α- or β-chair, the side chain adopts a gauche+ conformation (+60° and +50°, respectively; population a in Fig. 1B).
The final 6-s-trans conformation (α-chair) is favored by ≈7 kcal/mol over the final 6-s-cis conformer (β-chair), as deduced from calculating their total potential energies (Supporting Text). This finding is important because the α-chair lacks the S278 and Y143 H-bonds in the A pocket formed by the β-chair (Fig. 2B). Thus, we conclude that the A pocket shows no preference for closed, 6-s-cis, or opened, 6-s-trans, vitamin D sterols, because the loss in H-bond stabilization of the α-chair conformer is compensated for by its increased intramolecular stability when compared with the β-chair (Fig. 2B and Table 1, IEs). This finding contrasts with the β-chair and 6-s-trans ligand selectivity observed for the G pocket (Fig. 2 A and Table 1, IEs).
When a 1,25D side-chain conformer from population b (Fig. 1B) was docked in the G pocket, the five H-bond contacts made by 1,25D paralleled those observed in the x-ray structures (23, 24): the 1α-OH H-bonds with R274 (H5) and S237 (H3), the 3β-OH H-bonds with S278 (loop) and is proximal to Y143 (H1), and the 25-OH H-bonds with H305 (acceptor and loop) and H397 (donor, H11; Fig. 2 A). Thus, using CD ring fragments generated in pcmodel as the starting side-chain conformations in docking experiments allows for the side-chain statistical distribution of conformers to be factored into the ligands selectivity for a given pocket.
When JN occupies the A pocket, the donor/acceptor relationship of the Y295-H229 H-bond reverses, causing a >3.5-Å movement of Y295 (Fig. 2B). This H-bond reversal, movement of R158, and flexibility in the β-sheet loop (Fig. 2 A and C) allow JN to form favorable van der Waals and Coulombic nonbonding contacts with residues lining the A pocket. In contrast, ligand access to the G pocket and the static S237 and R274 residues (Fig. 6, which is published as supporting information on the PNAS web site) requires movement of the C-terminal end of H11 and the whole of H12. Thus, the in silico models suggest that the ligand may enter the A pocket through the H2/β-sheet region by a mechanism analogous to a flickering gate (3, 30) (Fig. 2B) where the enthalpy of activation for entry is small compared with the theoretical enthalpy of activation required for H11/H12 repositioning (31, 32). A portal to the NR LBD through the region between H1 and H3 has been proposed by Wagner et al. (33). This region has also been proposed to be the sole entrance portal to the LBD in peroxisome proliferator-activated receptor γ (PPARγ) (34). It remains a possibility that an A pocket exists in other NR LBDs, based on the fact that the structure in the H1/H3 region is highly divergent across the family.
The Conformational Ensemble Model: Theoretical A and G Pocket Accessibility. Based on the modeling and crystallographic results, it is proposed that both the apo-VDR LBD and free 1,25D exist in multiple, statistically averaged configurations that can create an ensemble of stable ligand/receptor assemblies, capable of driving different physiological signaling cascades (3). In this model, a vitamin D sterol can bind with different affinity to either the G or A pocket. Therefore, the occupancy of the two LBD cavities and ligand/receptor IEs (Table 1) are influenced by the following: (i) ligand chemistry, which dictates pocket stability and selectivity; (ii) the concentration of ligand and protein (law of mass action); and (iii) the gating properties of the two receptor portals, which are influenced by the receptor's local environment.
We postulate that ligand occupancy of the A pocket is kinetically favored, whereas occupancy of the G pocket is thermodynamically favored for most vitamin D sterols, for the following reasons: (i) the A vs. G pocket accessibility and side-chain selectivity discussed above, (ii) the calculated IEs (Table 1) suggest that all vitamin D sterols prefer occupying the G pocket, and (iii) an increased hydrophobic effect is imparted by G pocket occupancy, given that the G pocket completely encapsulates the ligand, whereas in the A pocket, the terminal end of the side chain is solvent-exposed (Fig. 2 A).
Below, we describe the application of the VDR receptor ensemble model in describing (i) Cl– channel agonism and antagonism, (ii) differential ligand-induced banding patterns observed in partial trypsin digest experiments, (iii) vitamin D sterol/CV1 cell transient transfection experiments incorporating VDR wild–type (VDRwt) and point mutants, and (iv) ERα-dependent EST NG signaling (11).
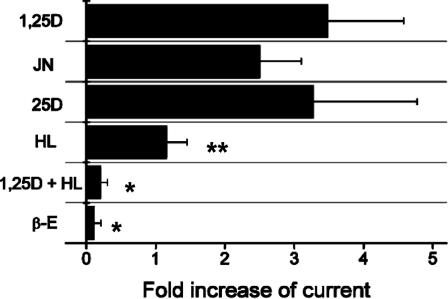
Activation of Chloride Channels by 1,25D, JN, and 25D and Antagonism by HL. The use of 1,25D activates chloride channels in ROS 17/2.8 cells at nanomolar concentrations (Fig. 3). We postulate that this NG effect is mediated by VDR A pocket occupancy by 1,25D (3), and is transient and therefore occurs before reaching a steady state. JN (1.0 nM) showed ≥70% the channel activity observed with 1,25D (0.5 nM) (Fig. 3). This result could be explained by the slightly lower IE for JN in the A pocket compared with 1,25D's averaged A pocket IE (Table 1). The natural metabolite 25D showed activity that paralleled 1,25D at 0.5 nM (Fig. 3), but surprisingly, an ≈7 kcal/mol lower A pocket IE compared with 1,25D [Table 1; –84 (see §) vs. –91 kcal/mol]. This energy loss may be compensated by a higher population of 25D molecules in the agonistic, α-chair, 6-s-trans conformation local to the hydrophobic membrane [25D favors α-chair 100% in SDS micelles (35)]. In addition, 25D has a much weaker G pocket IE (Table 1) than 1,25D; therefore, the difference in IE between the G and A pockets (ΔIE) is low compared with JN and 1,25D (Table 1), suggesting that it should show an increased selectivity for the A pocket.
Fig. 3.
Effect of 1,25D and its analogs on chloride channel opening in ROS 17/2.8 cells. The chloride currents were elicited by a depolarizing step to 80 mV, in the presence of the indicated sterols. Currents were obtained with glutamate as the permanent anion because seals were more stable and long-lasting than in the presence of Cl–. Anion currents were isolated from inward Ba+2 currents after blockage of Ca+2 channels with 100 μM Cd+2. The ligand concentrations used were 0.5 nM 1,25D, 0.5 nM 25D, 1.0 nM HL, 1.0 nM HL plus 0.5 nM 1,25D, 1.0 nM JN, and 10 nM E2 (β-E). The mean effect of each analog was statistically compared with the effect attained by 0.5 nM 1,25D (*, P < 0.05; **, P < 0.01, n = 4–9).
If only the G pocket existed in the VDR LBD, then HL (Fig. 1 A) would be expected to be a better Cl– channel agonist than 25D, given their calculated G pocket IEs (Table 1). However, HL (1.0 nM) blocks 1,25D (0.5 nM)-induced Cl– channel activity when they are coincubated, whereas 1.0 nM HL alone shows only 20–30% the activity of 1,25D (Fig. 3). Importantly, the HL α/β-chair equilibrium is 10:90 (36) in a hydrophobic environment, resulting in HL preferential occupancy of the G pocket (ΔIE = 19 kcal/mol) and incompetent interaction with the A pocket (Table 1). Thus, when 1,25D and HL are assayed together, HL preferentially binds the G pocket, sterically blocking 1,25D's ability to efficiently occupy the A pocket (Fig. 2 A) and trigger the response. The mild, NG, agonistic activity of HL by itself could be indicative of the α-chair's capability (only 10%; ref. 36) of forming a favorable, competent complex with the A pocket (Table 1).
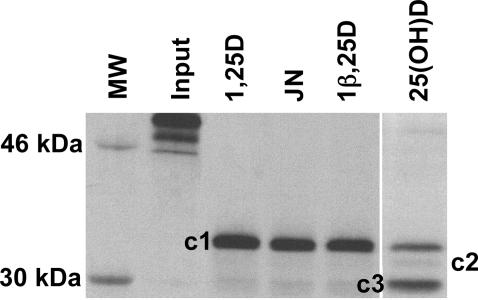
Importance of the A Pocket in Explaining Protease Sensitivity Results. When 35S-VDRwt is incubated with 1,25D and subjected to a partial trypsin digest, three ligand-stabilized VDR LBD conformations (fragments) are observed: a dense ≈34-kDa band [Fig. 4, cleavage 1 (c1)], a very faint ≈32-kDa band (c2), and a faint ≈30-kDa band (c3) (28, 37). The partial trypsin digest results for 1,25D, 25D, HL, and JN show that under saturating ligand conditions (10–5 M), only 25D shows an increase in the c3 band density, suggesting an increased cleavage frequency of the LBD C-terminal region (Fig. 2 A, orange ribbon). However, 1,25D, JN, and HL show qualitatively identical banding patterns (Fig. 4), which is consistent with a noncleaved C terminus (28, 37). The altered banding pattern for 25D can be understood by its preference to bind to the A pocket as suggested by its poor G pocket IE (–91 kcal/mol) and lower ΔIE (8 kcal/mol) compared with 1,25D, HL, and JN (15–19 kcal/mol, Table 1). Ligands occupying the A pocket are not local to or sterically contacting H12; therefore, loss of ligand-mediated H12 stability by a preferential binding to the A pocket is consistent with the mousetrap model (31), where the terminal end of H11 uncoils, allowing trypsin access to R402 (Fig. 2 A and C). JN has a 1,25D-like banding pattern (Fig. 4) because it shows G pocket selectivity under the experimental steady state conditions (Table 1). The same argument can be used to explain the dense c1 LBD conformer stabilized by HL (Fig. 4 and Table 1).
Fig. 4.
Protease sensitivity of 35S-VDRwt in the presence of 1,25D or select analogs. SDS/PAGE gel of partial trypsin digests of 35S-VDR (residues 1–427) incubated with 10–5 M of the indicated vitamin D sterol. The dense band in the 1,25D lane is the ≈34-kDa fragment (c1) and represents the closed H12 conformer (Fig. 2 A), whereas the dense bands in the 25(OH)D lane are c1 and the ≈30-kDa band (c3). Input represents 35S-VDR not subjected to trypsin treatment.
VDR Mutational Analysis Supporting the A Pocket Ensemble Concept. We made S278A and Y143F/S278A mutant VDRs to test the ensemble model with 1,25D and CF (Fig. 1 A) (38). These two mutations knock out one and both, respectively, of the H-bonds formed by the 3β-OH of 1,25D in the G pocket, but they presumably would have no effect on CF's G pocket stability. In the A pocket, the point mutants should affect the β-chair stability of both 1,25D and CF. Importantly, CF only forms favorable contacts with the A pocket in the high-energy, β-chair conformation where it does not contact the static S237 and R274 residues (Fig. 6); however, CF's 1α-OH does contact S237 and R274 in the G pocket. Thus, if the models are valid, CF should be directed to the G pocket by the mutations.
In VDRwt, CF showed an ≈8-fold decrease in affinity, as determined in a steroid competition assay (RCI), but only a 4-fold decrease in transactivation efficiency (EC50) compared with 1,25D (Table 2). In the RCI experiment, the CF and [3H]1,25D are assayed together; therefore, the RCI results are consistent with the models because CF and 1,25D have parallel G pocket selectivity (ΔIE, Table 1), but 1,25D is 10 kcal/mol more stable in the G pocket (Table 1). The models suggest that CF is an efficient transactivator in VDRwt because it has the same G pocket selectivity and forms the same hydrophobic contacts as 1,25D in the G pocket. Said differently, the decreased stability of CF in the A pocket compared with 1,25D (Table 1) allows CF to overcome the 10 kcal/mol loss in G pocket IE when present by itself and to be a potent transactivator. The S278A mutation has no significant effect on CF's RCI, but causes a 5-fold decrease in CF's genomic EC50 (Table 2); however, S278A did not affect 1,25D's EC50 (Table 2). These results are supported by the slight increase in CF's ΔIE (18 kcal/mol), but not 1,25D's (17 kcal/mol; Table 3, which is published as supporting information on the PNAS web site), when compared with their VDRwt ΔIE (Table 1). The 1,25D's unaltered transactivation efficacy is likely due to the ability of Y143 to continue to provide stability to the 3β-OH in the G pocket and 1α-OH in the A pocket (Fig. 2 A and B). This result leads to no change in 1,25D's G pocket selectivity.
Table 2. CF RCI and genomic transactivation (EC50 values) in VDRwt and targeted VDR hydroxyl mutants.
| Genomic transactivation, EC50
|
|||
|---|---|---|---|
| VDR construct | RCI for CF, % | 1,25D, nM | CF, nM |
| VDRwt | 13 | 1.6 ± 1.0 (n = 23) | 5.4 ± 1.3 (n = 3) |
| S278A | 20 | 1.9 ± 0.3 (n = 5) | 0.88 ± 0.29 (n = 3) |
| Y143F/S278A | 130 ± 41 (n = 2) | 94 ± 34 (n = 3) | 1.3 ± 0.3 (n = 3) |
RCI values for CF with the indicated VDR constructs in COS-1 cells. The RCI of 1,25D is, by definition, set to 100%. The concentration of steroid producing 50% of the maximal transactivation of the osteocalcin promoter is defined as the EC50. All EC50 values shown are significantly different between 1,25D and CF with P < 0.05.
The Y143F/S278A mutation had the same effect on CF's EC50 observed in S278A; however, there was an ≈10-fold increase in CF's RCI (Table 2). This result can be explained by the Y143F/S278A mutation affecting 1,25D's affinity for the G pocket more than its stability in the A pocket (loss of ≈7 kcal/mol and ≈3 kcal/mol in the calculated IE, respectively; Table 3). Because of CF's chemistry, the double mutation knocks out CF's 1-OH stabilizing contacts in the A pocket, but its affinity for the G pocket (IE = –96 kcal/mol) is maintained. Thus, the ≈10-fold increase in CF's RCI compared with S278A and VDRwt can be explained by CF's complete loss of A pocket stability, which strengthens its G pocket selectivity (ΔIE = 21 kcal/mol), whereas 1,25D's G pocket selectivity is reduced by the double mutation (ΔIE = 13 kcal/mol), supporting its low EC50 (Table 2). Importantly, if no A pocket existed, then, based on CF's Y143F/S278A RCI being ≥100, CF and 1,25D should transactivate with nearly the same efficacy in the double mutant; however, this result is not supported by the data (Table 2).
ERα-Dependent NG Signaling. The ERα receptor with an opened H12 conformation (Protein Data Bank ID code 1A52) was docked with E2 and EST by manually placing them in the putative ERα A pocket so the 3-OH remained in close proximity (≤ 3.5 Å) to R394 and E353, whereas the 17β-OH oriented toward R335. In silico minimization experiments showed that these two sterols' different physiological profiles (9, 11) could possibly be related to how they selectively interact with the receptor ERα LBD. E2 showed a more favorable IE with the G pocket (-66) vs. the A pocket (–61 kcal/mol), whereas EST showed reciprocal activity (G pocket = –61 vs. A pocket –66 kcal/mol). These de novo results, suggesting EST stabilizes a different ratio of ERα ensembles, provide a plausible explanation for EST's poor transactivation capability, yet potent bone fortification properties (11). It is noted this is not the first A pocket proposed to exist for ERα (39). The van Hoorn proposal identifies the A pocket in the same relative region of the ERα LBD as proposed in this communication, but the molecular contacts made by the ligands differ in the two models.
Conclusions
Based on the existence of an A pocket and the known literature, the two regions of the VDR LBD that seem to be integral to ligand stability and subsequent VDR cellular signaling are the H11/H12 and H2/β-sheet regions (Fig. 2 A). Importantly, the H2/β-sheet region is structurally diverse across the NR superfamily (see Protein Data Bank files •••), and is the most solvated region in the VDR x-ray construct, implying that it is a highly flexible region of the molecule (Fig. 2C). Both the 6-s-cis and 6-s-trans forms of the conformationally flexible 1,25D molecule formed viable complexes with the VDR A pocket. Interestingly, the 1,25D cis conformer utilizes the β-chair conformer, whereas its trans conformer utilizes the α-chair.
The steroid receptor ensemble model was shown here to be compatible with the following experimental observations: (i) NG signaling of specific ligands known to require a functional VDR or ERα LBD; (ii) potent HL antagonism of 1,25D activation of Cl– channels; (iii) VDR/vitamin D sterol proteolysis results; and (iv) differential effects of VDR point mutations on 1,25D and CF transactivational efficiencies and CF RCIs. Our data do not definitively define the structural attributes of the A pocket, nor does it suggest that no other membrane/cytosolic receptor(s) exist, and that the NR is the only receptor that sterols bind to activate NG events (3). The data do provide a plausible molecular and kinetic model, termed the conformational ensemble model (3), that shows how the broad and divergent 1,25D and E2 cellular signaling could be facilitated by their cognate nuclear hormone receptors.
The ensemble hypothesis coupled with the law of mass action describes a very sensitive and dynamic system that would be influenced by the local environment where the binding event is occurring (i.e., pH, protein milieu, alternative splicing, etc.). For the VDR system, this means that the concentration of metabolites and therefore expression level of processing enzymes like CYP24 play an integral role in controlling the intracellular signaling cascades. Perhaps the most important attribute of this model is that it may provide a new platform for drug development. Future work involving docking known vitamin D therapeutics to both the G and A pockets of the VDR to determine whether drug activity can be linked to differential stability in the two pockets is warranted. In addition, we have used modeling, molecular biology, biochemistry, and the ensemble hypothesis to develop a mechanism describing superagonism and antagonism of a VDR reporter construct in CV1 cells (M.T.M., C.M.B., and A.W.N., unpublished data).
Supplementary Material
Acknowledgments
We thank Professors Helen L. Henry and William H. Okamura for useful discussions and comments. This work was supported by National Institutes of Health Grant DK-09012 (to A.W.N.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: VDR, vitamin D receptor; VDRwt, VDR wild-type; NR, nuclear receptor; NG, nongenomic; G pocket, genomic pocket; A pocket, alternative ligand-binding pocket; LBD, ligand-binding domain; JN, 1α,25(OH)2-lumisterol; IE, interaction energy; RCI, relative competitive index; 1,25D, 1α,25(OH)2-vitamin D3; 25D, 25(OH)-vitamin D3; HL, 1β,25(OH)2-vitamin D3; CF, 3-deoxy-1α,25(OH)2-vitamin D3; ER, estrogen receptor; cn, cleavage n; Hn, helix n; E2, 17β-estradiol; EST, 4α-estren-3α, 17β-diol.
References
- 1.Mangelsdorf, D. J., Thummel, C., Beato, M., Herrlich, P., Schütz, G., Umesono, K., Blumberg, B., Kastner, P., Mark, M., Chambon, P., et al. (1995) Cell 83, 835–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Losel, R. & Wehling, M. (2003) Nat. Rev. Mol. Cell Biol. 4, 46–56. [DOI] [PubMed] [Google Scholar]
- 3.Norman, A. W., Mizwicki, M. T. & Norman, D. P. G. (2004) Nat. Rev. Drug Discov. 3, 27–41. [DOI] [PubMed] [Google Scholar]
- 4.Caffrey, J. M. & Farach-Carson, M. C. (1989) J. Biol. Chem. 264, 20265–20274. [PubMed] [Google Scholar]
- 5.Zanello, L. P. & Norman, A. W. (1997) J. Biol. Chem. 272, 22617–22622. [DOI] [PubMed] [Google Scholar]
- 6.Rebsamen, M. C., Sun, J., Norman, A. W. & Liao, J. K. (2002) Circ. Res. 91, 17–24. [DOI] [PubMed] [Google Scholar]
- 7.Chambliss, K. L. & Shaul, P. W. (2002) Steroids 67, 413–419. [DOI] [PubMed] [Google Scholar]
- 8.Pedram, A., Razandi, M., Aitkenhead, M., Hughes, C. C. & Levin, E. R. (2002) J. Biol. Chem. 277, 50768–50775. [DOI] [PubMed] [Google Scholar]
- 9.Kousteni, S., Bellido, T., Plotkin, L. I., O'Brien, C. A., Bodenner, D. L., Han, L., Han, K., DiGregorio, G. B., Katzenellenbogen, J. A., Katzenellenbogen, B. S., et al. (2001) Cell 104, 719–730. [PubMed] [Google Scholar]
- 10.Norman, A. W., Okamura, W. H., Hammond, M. W., Bishop, J. E., Dormanen, M. C., Bouillon, R., Van Baelen, H., Ridall, A. L., Daane, E., Khoury, R., et al. (1997) Mol. Endocrinol. 11, 1518–1531. [DOI] [PubMed] [Google Scholar]
- 11.Kousteni, S., Chen, J. R., Bellido, T., Han, L., Ali, A. A., O'Brien, C. A., Plotkin, L., Fu, Q., Mancino, A. T., Wen, Y., et al. (2002) Science 298, 843–846. [DOI] [PubMed] [Google Scholar]
- 12.Norman, A. W., Bouillon, R., Farach-Carson, M. C., Bishop, J. E., Zhou, L.-X., Nemere, I., Zhao, J., Muralidharan, K. R. & Okamura, W. H. (1993) J. Biol. Chem. 268, 20022–20030. [PubMed] [Google Scholar]
- 13.Song, X., Bishop, J. E., Okamura, W. H. & Norman, A. W. (1998) Endocrinology 139, 457–465. [DOI] [PubMed] [Google Scholar]
- 14.Norman, A. W., Mizwicki, M. T. & Okamura, W. H. (2003) in Proceedings of the Vitamin D and Cancer Meeting, eds. Reichrath, J., Friedrich, M. & Tilgen, W. (Springer, Homburg, Germany), pp. 55–82.
- 15.Norman, A. W., Song, X.-D., Zanello, L. P., Bula, C. M. & Okamura, W. H. (1999) Steroids 64, 120–128. [DOI] [PubMed] [Google Scholar]
- 16.Nemere, I., Dormanen, M. C., Hammond, M. W., Okamura, W. H. & Norman, A. W. (1994) J. Biol. Chem. 269, 23750–23756. [PubMed] [Google Scholar]
- 17.Bouillon, R., Okamura, W. H. & Norman, A. W. (1995) Endocr. Rev. 16, 200–257. [DOI] [PubMed] [Google Scholar]
- 18.Zanello, L. P. & Norman, A. W. (2004) Proc. Natl. Acad. Sci. USA 101, 1589–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nguyen, T. M., Lieberherr, M., Fritsch, J., Guillozo, H., Alvarez, M. L., Fitouri, Z., Jehan, F. & Garabedian, M. (2003) J. Biol. Chem. 279, 7591–7597. [DOI] [PubMed] [Google Scholar]
- 20.Norman, A. W., Olivera, C. J., Barreto Silva, F. R. & Bishop, J. E. (2002) Biochem. Biophys. Res. Commun. 298, 414–419. [DOI] [PubMed] [Google Scholar]
- 21.Razandi, M., Oh, P., Pedram, A., Schnitzer, J. & Levin, E. R. (2002) Mol. Endocrinol. 16, 100–115. [DOI] [PubMed] [Google Scholar]
- 22.Kim, Y. S., MacDonald, P. N., Dedhar, S. & Hruska, K. A. (1996) Endocrinology 137, 3649–3658. [DOI] [PubMed] [Google Scholar]
- 23.Rochel, N., Wurtz, J. M., Mitschler, A., Klaholz, B. & Moras, D. (2000) Mol. Cell 5, 173–179. [DOI] [PubMed] [Google Scholar]
- 24.Tocchini-Valentini, G., Rochel, N., Wurtz, J. M., Mitschler, A. & Moras, D. (2001) Proc. Natl. Acad. Sci. USA 98, 5491–5496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bursavich, M. G. & Rich, D. H. (2002) J. Med. Chem. 45, 541–558. [DOI] [PubMed] [Google Scholar]
- 26.Kenakin, T. (2002) Annu. Rev. Pharmacol. Toxicol. 42, 349–379. [DOI] [PubMed] [Google Scholar]
- 27.Midland, M. M., Plumet, J. & Okamura, W. H. (1993) Bioorg. Med. Chem. Lett. 3, 1799–1804. [Google Scholar]
- 28.Peleg, S., Sastry, M., Collins, E. D., Bishop, J. E. & Norman, A. W. (1995) J. Biol. Chem. 270, 10551–10558. [DOI] [PubMed] [Google Scholar]
- 29.Verboven, C., Rabijns, A., De Maeyer, M., Van Baelen, H., Bouillon, R. & De Ranter, C. (2002) Nat. Struct. Biol. 9, 131–136. [DOI] [PubMed] [Google Scholar]
- 30.Gutfreund, H. (1995) in Kinetics of the Life Sciences: Receptors, Transmitters, and Cataylsts, ed. Gutfreund, H. (Cambridge Univ. Press, Cambridge, U.K.), pp. 231–275.
- 31.Egea, P. F., Mitschler, A., Rochel, N., Ruff, M., Chambon, P. & Moras, D. (2000) EMBO J. 19, 2592–2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Egea, P. F., Klaholz, B. P. & Moras, D. (2000) FEBS Lett. 476, 62–67. [DOI] [PubMed] [Google Scholar]
- 33.Wagner, R. L., Apriletti, J. W., McGrath, M. E., West, B. L., Baxter, J. D. & Fletterick, R. J. (1995) Nature 378, 690–697. [DOI] [PubMed] [Google Scholar]
- 34.Nolte, R. T., Wisely, G. B., Westin, S., Cobb, J. E., Lambert, M. H., Kurokawa, R., Rosenfeld, M. G., Willson, T. M., Glass, C. K. & Milburn, M. V. (1998) Nature 395, 137–143. [DOI] [PubMed] [Google Scholar]
- 35.Okamura, W. H., Zhu, G. D., Hill, D. K., Thomas, R. J., Ringe, K., Borchardt, D. B., Norman, A. W. & Mueller, L. J. (2002) J. Org. Chem. 67, 1637–1650. [DOI] [PubMed] [Google Scholar]
- 36.Sheves, M., Friedman, N. & Mazur, Y. (1977) J. Org. Chem. 42, 3597–3599. [DOI] [PubMed] [Google Scholar]
- 37.Liu, Y. Y., Collins, E. D., Norman, A. W. & Peleg, S. (1997) J. Biol. Chem. 272, 3336–3345. [DOI] [PubMed] [Google Scholar]
- 38.Okamura, W. H., Mitra, M. N., Procsal, D. A. & Norman, A. W. (1975) Biochem. Biophys. Res. Commun. 65, 24–30. [DOI] [PubMed] [Google Scholar]
- 39.van Hoorn, W. P. (2002) J. Med. Chem. 45, 584–589. [DOI] [PubMed] [Google Scholar]
- 40.Okamura, W. H. & Zhu, G.-D. (1997) in Vitamin D, eds. Feldman, D., Glorieux, F. H. & Pike, J. W. (Academic, San Diego), pp. 939–971.
- 41.Helmer, B., Schnoes, H. K. & DeLuca, H. F. (1985) Arch. Biochem. Biophys. 241, 608–615. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.