Abstract
Using single-molecule fluorescence spectroscopy, time-resolved conformational changes between fluorescently labeled tRNA have been characterized within surface-immobilized ribosomes proceeding through a complete cycle of translation elongation. Fluorescence resonance energy transfer was used to observe aminoacyl-tRNA (aa-tRNA) stably accommodating into the aminoacyl site (A site) of the ribosome via a multistep, elongation factor-Tu dependent process. Subsequently, tRNA molecules, bound at the peptidyl site and A site, fluctuate between two configurations assigned as classical and hybrid states. The lifetime of classical and hybrid states, measured for complexes carrying aa-tRNA and peptidyl-tRNA at the A site, shows that peptide bond formation decreases the lifetime of the classical-state tRNA configuration by ≈6-fold. These data suggest that the growing peptide chain plays a role in modulating fluctuations between hybrid and classical states. Single-molecule fluorescence resonance energy transfer was also used to observe aa-tRNA accommodation coupled with elongation factor G-mediated translocation. Dynamic rearrangements in tRNA configuration are also observed subsequent to the translocation reaction. This work underscores the importance of dynamics in ribosome function and demonstrates single-particle enzymology in a system of more than two components.
Protein synthesis, catalyzed by the ribosome, is rapid, processive, and highly regulated. In bacteria, two RNA–protein subunits consisting of a small 30S subunit and a large 50S subunit assemble around a mRNA template into a roughly spherical 70S particle that translates the nucleotide sequence into a polypeptide chain through repetitive, codon-dependent binding of aminoacylated tRNA.
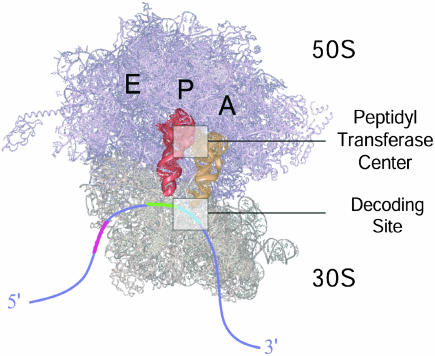
The landmark structures of the 30S (1, 2), 50S (3, 4), and 70S (5) ribosomal particles provide a molecular basis for understanding ribosome function. tRNA molecules bind to the ribosome in a solvent-accessible channel at the subunit interface. Three binding sites for tRNA, called the aminoacyl site (A site), peptidyl site (P site), and exit site (E site), have been identified on both the large and small subunit (Fig. 1). Each tRNA is separated from its neighbor at the elbow region (the site of ≈90° bending) by 25–45 Å (5, 6). The anticodon stem loops of A- and P-site tRNA form Watson–Crick base pairs with adjacent mRNA codons on the 30S subunit (5, 7), whereas the 3′ CCA terminal residues of A- and P-site tRNAs base-pair with conserved ribosomal RNA (rRNA) loops (8–10) within the 50S subunit peptidyltransferase center, the site of peptide bond formation (11). Additional interactions with rRNA and ribosomal proteins position tRNA molecules on the ribosome (5).
Fig. 1.
70S ribosome showing binding sites for A-, P-, and E-site tRNA molecules (model coordinates courtesy of M. Yusopov, Laboratoire de Biologie et Genomique Structurales de Institut de Génétique et de Biologie Moléculaire et Cellulaire, Strasbourg, France). The 50S subunit is shown in purple and the 30S subunit in tan. aa-tRNA (A), peptidyl-tRNA (P), and deacylated-tRNA (E) binding sites are labeled in black at their approximate location on the 70S ribosome. A-site tRNA is shown in gold and P-site tRNA in red; mRNA is shown in blue. The locations of the peptidyltransferase center and decoding site are highlighted.
The essential components and principal steps of translation have been delineated through genetic, biochemical, and kinetic methods (12). The process of polypeptide elongation has been most extensively characterized (13). Binding of aminoacyl-tRNA (aa-tRNA) to the A site occurs as a “ternary complex” with the GTPase elongation factor-Tu and GTP [EF-Tu(GTP)]. Ribosome interaction with the ternary complex can enhance GTP hydrolysis by EF-Tu by as much as 5 × 104, provided that there is complementary codon–anticodon pairing (14). After hydrolysis of GTP, the 3′ end of A-site aa-tRNA dissociates from EF-Tu(GDP) and accommodates at the peptidyltransferase center immediately adjacent to the tRNA in the P site. Rapid peptide bond formation transfers the peptide attached to the P-site tRNA to the A-site amino acid (15).
After peptide bond formation, two scenarios for tRNA conformation on the ribosome have been proposed. In one model, the tRNA molecules are initially in a classical state in which each tRNA is bound to the A and P sites on both the 30S and 50S subunits. Upon peptide bond formation, the tRNAs move quickly and spontaneously into a hybrid state (11) where tRNA binding at the 30S subunit remains intact while the 3′ CCA ends of the P- and A-site tRNA bind to the 50S E and P sites, respectively (16, 17). Alternatively, recent biochemical studies (18–20), cryo-electron microscopy (21, 22), and x-ray crystallographic studies (23) suggest that the tRNAs remain in the classical state after peptide bond formation and progress to the hybrid state in a separate step.
The postpeptide bond formation complex is resolved by EF-G-catalyzed translocation. EF-G catalyzes movement of the A- and P-site tRNA–mRNA complexes to the E and P sites, respectively (24). Like EF-Tu, EF-G is a ribosome-activated GTPase (25). Translocation is thought to occur through a multistep process in which the acceptor and anticodon stems of tRNA move in distinct steps separated by GTP hydrolysis (18, 20). Translocation allows further elongation of the peptide by bringing the next mRNA codon into a vacated A site for subsequent aa-tRNA selection. The cycle of tRNA movement through the ribosome is completed when deacylated tRNA exits the ribosome from the E site.
Here we report reconstitution of a highly active, surface-based translation system and direct observations of protein synthesis by using single-molecule fluorescence methods (26). This system allows us to follow multistep, asynchronous processes usually obscured in bulk studies (27). Heterogeneous populations inherent to multicomponent, multistep systems can be sorted by their activity and characterized independently. Rare, transient events can be directly observed (28).
Using single-molecule fluorescence resonance energy transfer (smFRET) we directly monitor the time-dependent distances between donor and acceptor fluorophores attached to the elbow region of tRNA molecules on the ribosome. Surface immobilization of the translation apparatus does not perturb enzymatic tRNA delivery to the A site, peptide bond formation, or translocation. Conformational fluctuations of tRNA molecules reveal continuous, dynamic exchange between discrete configurations in pretranslocation and posttranslocation states. These data shed light on the nature of tRNA–ribosome interactions and provide an essential link between static structural studies of the ribosome and the mechanism of translation.
Methods
Ribosomes, Buffers, Translation Factors, and mRNAs. Tight-coupled 70S ribosomes were purified from Escherichia coli MRE600 S30 as reported by Powers and Noller (29) and Robertson and Wintermeyer (30). 30S and 50S subunits were purified from 70S ribosomes by dialysis against Tris-polymix buffer (see below) at 1 mM Mg2+ followed by sucrose gradient ultracentrifugation in the same buffer. The composition of our buffer system has been adjusted to optimize the activity of purified ribosomes by using in vitro translation assays as described (31). The optimal buffer conditions for in vitro translation were 50 mM Tris-OAc (pH25°C 7.5), 100 mM KCl, 5 mM NH4OAc, 0.5 mM Ca(OAc)2, 5 mM Mg(OAc)2, 6 mM 2-mercaptoethanol, 5 mM putrescine, and 1 mM spermidine.
Initiation factors IF-1, IF-2, and IF-3 from E. coli were purified from overexpressing strains (a kind gift from Claudio Gualerzi, University of Camerino, Camerino, Italy) as reported (32, 33).
EF-Tu, EF-Ts, and EF-G were PCR-cloned from E. coli C600 genomic DNA into the pProEX HT-b plasmid system. The pProEX HT plasmid system introduces a six-histidine (6xHis) affinity tag at the amino terminus of the expressed protein with a highly specific Tev-protease cleavage site located between the 6xHis tag and the cloned protein. Each EF was expressed and purified over a Ni-NTA resin column following standard procedures (34). 6xHis tags were removed by Tev-protease cleavage.
The pT7gp32.1–225 plasmid, encoding a T4 gene product 32-derived mRNA, was constructed by PCR cloning of codons 1–224 of the WT T4 gene product 32 mRNA from plasmid pSY6 (a kind gift from Yousif Shamoo, Rice University, Houston). PCR cloning also introduced a T7 RNA polymerase promoter, a 25-nt spacer region, a strong (UAAGGA) Shine-Dalgarno ribosomal binding site upstream of the ORF, a UAA stop codon as codon 225, and a BamHI site for cloning and transcription purposes. Restriction digestion of pT7gp32.1–225 with BamHI was followed by in vitro run-off transcription with T7 RNA polymerase as described (35). Direct biotinylation of the 5′ end of the mRNA was achieved by doping the transcription reaction with 1:10 ratio of 5′-biotin-GG-3′ RNA primer with respect to GTP.
tRNA Aminoacylation and Formylation. Plasmids encoding E. coli methionyl tRNA synthetase and formylmethionyl-tRNA formyltransferase (a kind gift from Sylvain Blanquet, Centre National de la Recherche Scientifique-Ecole Polytechnique, Palaiseau Cedex, France) were overexpressed and purified as described (36). The formyl donor, 10-formyltetrahydrofolate, was prepared as described (37). Aminoacylation and formylation of tRNAfMet and tRNAfMet(Cy3-s4U) was achieved as reported (38), and aminoacylation of tRNAPhe was achieved following standard protocols (39).
Fluorescent Labeling of Ribosomes and tRNAs. Nonspecific Cy3 labeling of 50S subunits at a level of one to two dyes per subunit was achieved by incubating 4.3 μM 70S ribosomes and 98 μM Cy3-NHS ester in Tris-polymix buffer at 15 mM Mg(OAc)2 for 30 min at 37°C. Cy3-labeled 70S ribosomes were dissociated as described above, and Cy3–50S subunits were purified by sucrose gradient ultracentrifugation.
Labeling of tRNAfMet with Cy3-maleimide at the s4U8 position and labeling of tRNAPhe with Cy5-NHS ester at the acp3U47 position were performed following published procedures (40, 41). Cy3-Met-tRNAfMet, dye labeled at the α-amino group of methionine, was prepared by aminoacylation of tRNAfMet as described (36) and subsequent labeling with Cy3- or Cy5-NHS ester following published procedures (42). Dye-labeled tRNAs were purified from unlabeled tRNA by HPLC using a TSK-phenyl 5-PW column.
70S Complex Formation. 70S complexes were initiated on gene32-derived mRNA in vitro in Tris-polymix buffer at 5 mM Mg(OAc)2 using slight modifications of previously reported protocols (43). Initiation complexes were purified by sucrose density ultracentrifugation in Tris-polymix buffer at 20 mM Mg(OAc)2.
The EF-Tu(GTP)aa-tRNA Complex. Phe-tRNAPhe or Phe-tRNAPhe(Cy5-acp3U) was complexed with EF-Tu(GTP) in three steps (1). A 10-mM GTP charging mixture was prepared by incubating 10 mM GTP, 30 mM phosphoenolpyruvate, and 12 units/ml pyruvate kinase in Tris-polymix buffer at 15 mM Mg(OAc)2 (2). Nucleotide exchange was accomplished by addition of 10 mM GTP charging mixture to a solution of 12 μM EF-Tu and 12 μM EF-Ts in Tris-polymix buffer (excluding putrescine and spermidine) to a final concentration of 1 mM GTP (3). Phe-tRNAPhe (10 μM) was added to the reaction to achieve a final concentration of 1.5 μM in Phe-tRNAPhe.
Puromycin Assays. Puromycin assays were initiated by stopped-flow delivery of neutralized puromycin (1 mM, pH 7.5) in Tris-polymix buffer to surface-immobilized ribosome complexes carrying Cy3-Met-tRNAfMet in the P site. Cy3-Met–Phe-tRNAPhe was generated in the A site by incubating surface-immobilized ribosomes with 200 nM EF-Tu(GTP)Phe-tRNAPhe for 1 min. Cy3-Met–Phe-tRNAPhe was translocated to the P site by incubating the surface-immobilized ribosome complexes with 2 μM EF-G(GTP) for 1 min.
Total Internal Reflection Fluorescence Microscopy. Preparation of quartz microscope slides and glass coverslips for use in total internal reflection fluorescence microscopy was derived from Ha et al. (28). The flow-cell constructed from the quartz slide and glass coverslip was passivated with a solution of 10 μM BSA and 10 μM duplex DNA to prevent nonspecific binding of ribosome complexes and EF-Tu(GTP)Phe-tRNAPhe(Cy5-acp3U) ternary complex.
A lab-built, prism-based total internal reflection apparatus, based on an inverted microscope, was used for all experiments. Cy3- or Cy5-labeled molecules were excited by using a diode-pumped 532-nm laser or a 635-nm diode laser. Fluorescence emission was collected by a 1.2 numerical aperture/×60 water-immersion objective and imaged onto a cooled, back-illuminated charge-coupled device camera with nine-pixel binning at 100-ms exposure time. The dead time after stopped-flow delivery of substrates is estimated at ≈500 ms.
To extend the lifetimes and reduce the noise of Cy3 and Cy5 fluorescence for fluorescence microscopy, an oxygen scavenging system composed of 1% β-d-glucose, 25 units/ml glucose oxidase, and 250 units/ml catalase was added to all samples.
Results
Surface Immobilization of Fluorescent Ribosome Complexes. 70S complexes containing fluorescently labeled components are assembled with mRNA and tRNA in vitro by using purified E. coli ribosomes, initiation factors, formylated aminoacyl-initiator tRNA (fMet-tRNAfMet), and a 5′-biotinylated gene32-derived mRNA. The activity of ribosome complexes used in our experiments was demonstrated in vitro by using standard toeprinting assays (44). Ternary complexes consisting of Phe-tRNAPhe bound to EF-Tu(GTP) were delivered to 70S complexes with fMet-tRNAfMet in the P site with or without EF-G in the presence of the nucleotide exchange factor EF-Ts and nucleotide charging enzymes. Approximately 90% of ribosomes bind Phe-tRNAPhe at the A site and translocate in the presence of EF-G (Fig. 5, which is published as supporting information on the PNAS web site).
Assembled and purified ribosome complexes were immobilized via biotinylated mRNA at the surface of a quartz microscope slide coated with a combination of polyethylene glycol (PEG) and streptavidin-biotin-PEG (28). Quartz surfaces were further passivated by incubation with a 10 μM BSA/10 μM of double-stranded DNA solution. Surface immobilization of 70S complexes is highly specific; the ratio of specific to nonspecific binding is ≈500:1 by comparison to a nonbiotinylated control complex. Typically, 150–200 ribosome complexes, in an area of ≈60 × 120 μm, are illuminated by total internal reflection using either a 532- or 635-nm laser. Images were collected at 10 frames per s by a cooled, back-illuminated charge-coupled device camera. Single ribosomes are characterized by single-step photobleaching of fluorescence. The rate of dye photobleaching is retarded, and the fluorescence signal of Cy3 and Cy5 dyes is increased by using an enzymatic oxygen scavenger system.
Ribosome complexes immobilized at the quartz surface can be observed for extended periods. The stability of immobilized complexes was confirmed by observing 70S particles in which the 50S subunit was nonspecifically labeled with Cy3 dyes through the surface lysine residues of ribosomal proteins. 70S complexes remain stably associated with the surface at [Mg2+] ≥5 mM. Cy3 fluorescence originating from the 50S subunit of surface-bound complexes is rapidly released upon solvent exchange into buffer doped with 50 mM EDTA. This observation is consistent with dissociation of the 50S subunit from the 30S subunit in the absence of Mg2+ (45). The kinetic stability of the immobilized 70S complex highly depends on tRNA occupancy and buffer conditions (unpublished data).
Surface-Immobilized Ribosomes Are Highly Active in Peptide Bond Formation. The peptidyltransferase activity of surface-immobilized ribosomes was assayed with the antibiotic puromycin, an analogue of the 3′ terminal residue of aa-tRNA that acts as an acceptor in the peptidyltransferase reaction when the A site is vacant (46). A single-molecule version of the puromycin assay was developed by using ribosomes initiated with Cy3-Met-tRNAfMet in the P site, dye-labeled with Cy3-NHS dye at the α-amino group of the amino acid. Upon reaction with Cy3-labeled amino acid, the peptidyl-puromycin product rapidly dissociates from the ribosome and the surface-localized fluorescence disappears. The reaction of puromycin with Cy3-Met-tRNAfMet at the P site (1 mM puromycin, pH 7.5) is best described by a double exponential process with fast and slow components that decay at rates of 0.08 s–1 and 0.004 s–1, respectively (Fig. 2). The rate of the faster reaction component agrees with previous literature values (19, 47, 48). The origin of the slow reacting component has yet to be delineated.
Fig. 2.
Single-molecule puromycin assay. Surface-immobilized ribosome complexes carrying Cy3-Met-tRNAfMet in the P site (dye-labeled at the α-amino group) react with puromycin. Stopped-flow addition of puromycin (1 mM, pH 7.5) to the immobilized complexes results in peptide bond formation and rapid loss of Cy3 signal caused by dissociation of the puromycin-Met-Cy3 product from the surface. Before A-site tRNA accommodation, the loss of fluorescence was fit to a double exponential, A1*exp(–t/τ1) + A2* exp(–t/τ2) + y0, where A1 and A2 represent the population (%) of fast and slow decay reactions (solid lines). The fitting parameters from a double-exponential fit were: A1 = 79, τ1 = 12.6 ± 0.3 s, A2 = 18, τ2 = 236 ± 26 s, and y0 = 3. After enzymatic delivery of Phe-tRNAPhe, the rate of puromycin reaction is dramatically reduced, consistent with tRNA accommodation at the A site and peptide bond formation. Less that 5% of the population shows a fast decay after Phe-tRNAPhe delivery. However, some residual reaction with puromycin remains. For comparison, an independent measurement of the intrinsic Cy3 signal decay caused by peptidyl-tRNA dissociation from the A site is shown (dotted line). Recovery of the rapid puromycin reaction was achieved by incubation with EF-G in the presence of GTP. The fitting parameters were A1 = 58, τ1 = 6.4 ± 0.3 s, A2 = 27, τ2 = 154 ± 8 s, and y0 = 15. Thus, ≈73% (58/79) of ribosomes return to reacting rapidly with puromycin.
Surface-Immobilized Ribosomes Are Competent in tRNA Accommodation and Translocation. Preincubation of surface-immobilized ribosomes with EF-Tu(GTP)Phe-tRNAPhe results in dramatic reduction in the apparent rate of puromycin reaction. The data, fit to a single exponential process, yield an estimated reaction rate of 0.001 s–1 (Fig. 2). These data are consistent with >95% tRNA accommodation and formation of peptidyl-tRNA at the A site, which impedes puromycin reactivity (16). The observed rate, corrected for peptidyl-tRNA dissociation from the A site (49), agrees with previous studies that cite slow reaction of peptidyl-tRNA with puromycin before translocation (19, 50). The residual reactivity of peptidyl-tRNA suggests that the 50S A site remains at least partially accessible to puromycin.
Incubation of pretranslocation complexes with EF-G in the presence of GTP restores rapid puromycin reactivity to ≈70% of the population (Fig. 2). These data are consistent with translocation of Cy3-Met-Phe-tRNAPhe to the P site (19). The cycle of peptidyltransferase activities reported by the single-molecule puromycin assay correlate with our toeprinting results (Fig. 5) and strongly suggest that surface immobilization does not perturb ribosome function in tRNA delivery and translocation.
tRNA Accommodation at the A Site Can Be Observed by smFRET. To monitor tRNA accommodation and dynamics on the ribosome by using smFRET, tRNAfMet and tRNAPhe were labeled with fluorescent dye molecules at specific positions by virtue of naturally occurring modified nucleotides. tRNAfMet was labeled with Cy3 at the 4-thiouridine (s4U) residue present at position 8 (40), and tRNAPhe was labeled with Cy5 at the 3-(3-amino-3-carboxypropyl)uridine (acp3U) residue at position 47 (41). Positions 8 and 47 are located at the elbow region of tRNA, and their labeling does not affect tRNA function. Elbow-labeled tRNA molecules retain full activity in aminoacylation, formylation, complex formation with EF-Tu(GTP) (data not shown), and in the efficiencies of accommodation or translocation (Fig. 5).
In the smFRET experiment, Cy3 is illuminated, and Cy3 and Cy5 emission intensities (I) are simultaneously monitored with the FRET value defined as ICy5/(ICy3 + ICy5). Cy5 emission arising through smFRET ≥0.25 (our experimental noise threshold) indicates that Cy3 and Cy5 molecules are colocalized within 60 Å. Low concentrations of dye-labeled ternary complex [EF-Tu(GTP)Phe-tRNAPhe(Cy5-acp3U)] (10–100 nM) were delivered to avoid high background fluorescence caused by weak, direct excitation of the complex in solution.
Stopped-f low delivery of ternary complex to surface-immobilized ribosome complexes carrying fMet-tRNAfMet(Cy3-s4U) in the P site generates a rapidly evolving smFRET signal for ≈70% of surface-immobilized ribosomes (Fig. 3 and Fig. 6, which is published as supporting information on the PNAS web site). The time evolution of the FRET signal for the ensemble of individual ribosomes under observation is displayed as a time-resolved population FRET histogram generated from the superimposition of many smFRET signals. To eliminate the blurring of data caused by initial asynchronous binding of ternary complex to the ribosome, we introduce a postsynchronization technique in which all smFRET time traces are synchronized to the first FRET signal above a minimum threshold (FRET ≥0.25) (Fig. 4A). Single-molecule traces show an increase in smFRET consistent with an accommodation process in which at least one intermediate exists with a lifetime of 100–200 ms (Fig. 7, which is published as supporting information on the PNAS web site). A detailed examination of this multistep accommodation process is discussed elsewhere (56).
Fig. 3.
Single-molecule Cy3 and Cy5 fluorescence and FRET. tRNA delivery to ribosome complexes carrying fMet-tRNAfMet(Cy3-s4U) in the P site was monitored under Cy3 excitation. Cy5 fluorescence arises from Phe-tRNAPhe(Cy5-acp3U) binding to the ribosome and colocalization of Cy3 and Cy5 fluorophores within the same complex. (Upper) Cy3 and Cy5 emission intensity are shown in green and red, respectively. (Lower) The corresponding FRET value, ICy5/(ICy3 + ICy5), is shown in blue.
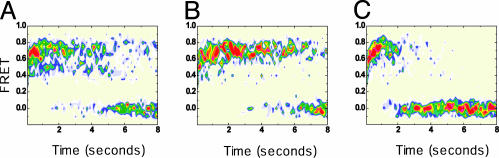
Fig. 4.
Postsynchronized, time-resolved population FRET histograms reporting on intermolecular tRNA–tRNA distances within individual ribosomes of an ensemble of particles. Histograms show the evolution of FRET for an ensemble of surface-immobilized 70S complexes carrying tRNAfMet(Cy3-s4U) in the P site after delivery of EF-Tu(GTP)Phe-tRNAPhe(Cy5-acp3U) where individual FRET time traces have been synchronized to the first FRET value ≥ 0.25. Population information is color-coded from tan (lowest occupancy) to red (highest occupancy). The histogram plots generated by stopped-flow delivery of EF-Tu(GTP)Phe-tRNAPhe(Cy5-acp3U) depend on the acylation state of tRNA in the P site and in the presence of EF-G. (A) Complexes carrying fMet-tRNAfMet(Cy3-s4U) in the P site. (B) Complexes carrying OH-tRNAfMet(Cy3-s4U) in the P site. (C) Exactly as in A but in the presence of EF-G. Within experimental uncertainty, the high FRET peaks observed have the same mean FRET value, 0.74 ± 0.05. Similarly, the lower FRET peaks observed in A and B have a mean FRET value of 0.45 ± 0.05. Peaks centered at zero FRET arise from Cy5 blinking, photobleaching, and tRNA dissociation.
The smFRET signal resulting from elbow-labeled tRNA accommodation is observed to undergo time-dependent fluctuations. Three peaks are observed in the time-resolved population FRET histogram presented in Fig. 4A with mean FRET values centered at 0, 0.45, and 0.74. Fluctuations between all three FRET states are observed. Transition to the 0 FRET state is caused by known photo-physical properties of the Cy5 fluorophore and peptidyl-tRNA dissociation from the A site. Dye molecules can reversibly enter a nonfluorescing, “blinking” state and irreversibly photobleach, where the time to enter either state is approximately inversely proportional to the excitation intensity (Fig. 8, which is published as supporting information on the PNAS web site). Blinking appears to be a general property of the Cy5 fluorophore that is exacerbated by its attachment to the elbow region of tRNAPhe. Fluctuations between FRET = 0.74 and 0.45 represent dynamic exchange between two distinct tRNA configurations on the ribosome.
The increase in the number of molecules with FRET ≥0.25, plotted as a function of time (i.e., a FRET arrival time plot), is best described by a double exponential function (Fig. 6). At 10 nM concentration, ≈70% of the ternary complex bind with a rate constant of 0.7 s–1 and ≈30% bind with a rate constant of 0.03 s–1. The faster rate (0.7 s–1) depends linearly on ternary complex concentration, consistent with a bimolecular reaction with a second-order rate constant of 7 × 107 M–1·s–1. This value is in good agreement with previous determinations of the bimolecular rate of ternary complex binding to ribosome (15). The slower rate (0.03 s–1) may arise from nonenzymatic delivery of Phe-tRNAPhe(Cy5-acp3U) to the A site or a subpopulation of less active ribosomes.
tRNA Molecules Are Dynamic on the Ribosome. Our single-molecule puromycin studies show that ternary complex delivery to ribosomes containing amino acid-labeled tRNA in the P site leads to efficient accommodation and peptide bond formation. We assume that accommodation of elbow-labeled aa-tRNA also leads to peptide bond formation. The high-FRET state centered at 0.74 (Fig. 4) is identified as the classical state and is the point of closest approach between the two tRNAs. The rate of arrival to the 0.74 FRET state agrees with the reported rate of full accommodation (15) (Fig. 6).
The assignment of the 0.74 FRET state is also supported by the structure of the 70S particle (51). Based on that structure, the inter-dye distance between position 47 on the A-site tRNA and position 8 of the P-site tRNA is estimated to be ≈45 Å. To use FRET values as a measure of distance, the distance parameter R0 of the Cy3–Cy5 pair (known to be ≈50 Å) and the average orientation factor must be known (52).
Single-molecule fluorescence anisotropy measurements show there is evidence for some rotational hindrance during the lifetime of the excited Cy3 dye (<300 ps) when attached to tRNA. The rotational relaxation of Cy5 dye is not hindered when attached to tRNA; however, the anisotropy R factor increases when Cy5-labeled tRNA is in the ribosome (Table 1, which is published as supporting information on the PNAS web site). Thus, we cannot claim that our FRET values are a reliable measure of absolute distance. However, if free rotation is assumed to occur in 100-ms integration time of each video frame, the observed FRET value of 0.74 corresponds to a distance of ≈42 Å and is in good agreement with the crystal structure. Thus, the distances derived from FRET further supports the assignment of 0.74 FRET to the classical state.
The transition of FRET = 0.74 to 0.45 (see Fig. 3) implies that the distance between tRNA elbows increases by ≈10 Å. The most likely assignment of this alternate configuration is the hybrid state. Previous literature states that tRNA molecules adopt a hybrid-state configuration when the A site is occupied by peptidyl-tRNA, whereas they remain in a classical state when the A site is occupied by aa-tRNA (16). Indeed, when ternary complex is delivered to ribosomes carrying deacylated tRNA (OH-tRNAfMet) in the P site so that peptide bond formation is not permitted, we observe a strongly favored classical-state tRNA configuration (Fig. 4B). With aa-tRNA at the A site, the lifetime of the classical state is 1.3 s. However, both the classical- and hybrid-state tRNA configurations remain accessible and are in dynamic exchange. When ternary complex is delivered to ribosomes with aa-tRNA in the P site, the lifetime of the classical state after peptide bond formation is 0.2 s. The lifetime in the hybrid state before returning to the classical state is independent of peptide bond formation (Fig. 9, which is published as supporting information on the PNAS web site). These lifetime changes manifest themselves as changes in the relative populations of classical and hybrid states: the hybrid state is occupied 14% of the time with aa-tRNA at the A site and 51% of the time when peptide bond formation creates peptidyl-tRNA at the A site (Fig. 9).
Movement of tRNAs on the ribosome necessarily involves the breaking and forming of numerous RNA–RNA and RNA–protein interactions. In agreement, the observed dynamic exchange between the two tRNA configurations is Mg2+ dependent. At a lower Mg2+ concentration (5 mM) the 0.45 FRET state is poorly defined and shifts toward a slightly higher FRET value (Fig. 10, which is published as supporting information on the PNAS web site). This result is consistent with averaging that arises from interconversion of the high- and low-FRET states at a rate that is comparable to or faster than the video frame collection rate (10 s–1). In support of this view, data acquired at 5-ms integration time using a confocal microscope reveal that many FRET fluctuations take place on time scales much faster than 100 ms (data not shown).
The single-molecule puromycin assay shows that translocation will occur if ternary complex and EF-G(GTP) are delivered to immobilized ribosomes carrying Cy3-Met-tRNAfMet in the P site. Similarly, toeprinting assays demonstrate that elbow-labeled tRNA delivered to the A site with EF-G(GTP) efficiently translocate (Fig. 5). The time-resolved population FRET histogram of accommodation, peptide bond formation, and translocation is shown in Fig. 4C. In the presence of EF-G(GTP), the time to achieve a FRET signal of 0.75 is comparable to the time observed in its absence. However, two significant differences are observed. First, there is an indication of a transient ≈0.45–0.5 FRET state in the first 500 ms. Second, the system evolves to a predominantly low-FRET tRNA configuration within 2–3safter aa-tRNA arrival.
If the transition of the population to lower FRET values is the signature for complete translocation, the apparent rate of translocation is ≈0.5 s–1. This value is consistent with previous reports (24, 53). Interestingly, single-molecule time traces reveal both high- and low-FRET-state tRNA configurations after translocation. Because single-molecule fluorescence anisotropy measurements of the posttranslocation complex indicate no observable changes in dye anisotropy (Table 1), we interpret the observed changes in FRET to reflect conformational dynamics of the tRNA molecules in the posttranslocated state(s). Translocation significantly increases the rate of Cy3 and Cy5 blinking and photobleaching, as can be seen by comparing the population of molecules in the 0 FRET state in Fig. 4. Because the local environmental conditions affect the fluorescence properties of the dyes, the interpretation of posttranslocation tRNA dynamics is significantly more difficult.
Discussion
Current models of translation propose that tRNA molecules are positioned statically on the ribosome in either classical- or hybrid-state configurations. Our smFRET experiments show that after accommodation of aa-tRNA at the A site tRNA molecules are in dynamic exchange between the classical and hybrid states on the ribosome. The relative populations of the two states show that classical- and hybrid-state tRNA configurations are roughly similar in free energy. Assuming a simple two-state exchange, peptide bond formation appears to decrease the free energy of activation for the transition from classical to hybrid states by 4.2 kJ·mol–1, whereas the free energy of activation for the reverse transition appears unaffected (Fig. 9). Peptide bond formation thus destabilizes the classical state.
The ≈6-fold increase in transition rate to the hybrid state when aa-tRNA in the A site is converted to peptidyl-tRNA is consistent with the finding that peptidyl-tRNA binds more weakly to the A site than aa-tRNA (49, 54). Both the transition of A-site peptidyl-tRNA from classical to hybrid state and dissociation require disruption of RNA–RNA interactions linking the A loop and the peptidyl-tRNA. In support of this finding, we observe that the rate of transitions between the hybrid and classical states increase at lower concentrations of Mg2+ ion (5 vs. 15 mM; Fig. 10). A more complete understanding of how the growing peptide chain promotes formation of the hybrid state will require the lifetime of tRNA configurations to be measured with A-site peptidyl-tRNAs of various amino acid compositions and lengths.
The exchange between hybrid- and classical-state tRNA configurations may play an important role in the translation cycle. EF-G-catalyzed translocation is faster with peptidyl-tRNA in the A site relative to aa-tRNA (49). Our data show that peptidyl-tRNA at the A site spends 3.5-fold more time in the hybrid state than aa-tRNA. We suggest that the increased rate of translocation would be explained if the hybrid-state tRNA configuration favors EF-G binding and translocation.
tRNA conformational dynamics are also observed in the ribosomal complexes after translocation of tRNA–mRNA complexes to the P and E sites. Although interpretation of these data are compromised by poor dye photophysics, our data suggest tRNAs dynamics take place in the posttranslocation complex. Large-scale movement of the L1 stalk was recently suggested from electron microscopic reconstructions of pretranslocation and posttranslocation complexes of the ribosome and may be related to E-site tRNA positioning and dynamics (22, 55).
In summary, this work demonstrates that single-molecule fluorescence and smFRET provide a powerful link between static high-resolution structures of the ribosome and biochemical studies of translation. We have temporally resolved several steps of a complete cycle of translation by monitoring individual, surface-immobilized ribosomes. These measurements reveal mechanistic features of translation previously unavailable, such as rapid tRNA movements between classical- and hybrid-state configurations and additional fluctuations after translocation.
The detailed interplay between tRNA dynamics and ribosome conformation must await further experiments in which direct dye labeling of ribosomal RNA and/or ribosomal proteins can be used to monitor simultaneously tRNA position, tRNA–ribosome motions, and changes in ribosome structure. Single-molecule approaches will also allow rare events linked to gene regulation such as translational pausing, frameshifting, and stop codon read-through to be investigated. These studies will be especially important when extended to eukaryotic translation systems. In the future it should also be possible to generalize the methods described here to the study of transcription, splicing, and signaling.
Supplementary Material
Acknowledgments
We thank Eric Lau for technical support and enzyme purification and Elisabetta Viani Puglisi for critical discussions and scientific input. S.C.B. is supported by the Giannini Family Foundation, and R.L.G. is supported by the American Cancer Society. This work was supported, in part, by National Institutes of Health Grant GM51266; a grant from the David and Lucile Packard Foundation (to J.D.P.); grants from the National Science Foundation, the National Aeronautics and Space Administration, and the Air Force Office of Scientific Research (to S.C.); and David and Lucille Packard Foundation Interdisciplinary Science Program Grant 2000-01671 (to S.C. and J.D.P.).
Abbreviations: aa-tRNA, aminoacyl-tRNA; A site, aminoacyl site; P site, peptidyl site; E site, exit site; EF, elongation factor; FRET, fluorescence resonance energy transfer; smFRET, single-molecule FRET.
References
- 1.Wimberly, B. T., Brodersen, D. E., Clemons, W. M., Jr., Morgan-Warren, R. J., Carter, A. P., Vonrhein, C., Hartsch, T. & Ramakrishnan, V. (2000) Nature 407, 327–339. [DOI] [PubMed] [Google Scholar]
- 2.Schluenzen, F., Tocilj, A., Zarivach, R., Harms, J., Gluehmann, M., Janell, D., Bashan, A., Bartels, H., Agmon, I., Franceschi, F. & Yonath, A. (2000) Cell 102, 615–623. [DOI] [PubMed] [Google Scholar]
- 3.Ban, N., Nissen, P., Hansen, J., Moore, P. B. & Steitz, T. A. (2000) Science 289, 905–920. [DOI] [PubMed] [Google Scholar]
- 4.Harms, J., Schluenzen, F., Zarivach, R., Bashan, A., Gat, S., Agmon, I., Bartels, H., Franceschi, F. & Yonath, A. (2001) Cell 107, 679–688. [DOI] [PubMed] [Google Scholar]
- 5.Yusupov, M. M., Yusupova, G. Z., Baucom, A., Lieberman, K., Earnest, T. N., Cate, J. H. & Noller, H. F. (2001) Science 292, 883–896. [DOI] [PubMed] [Google Scholar]
- 6.Fairclough, R. H., Cantor, C. R., Wintermeyer, W. & Zachau, H. G. (1979) J. Mol. Biol. 132, 557–573. [DOI] [PubMed] [Google Scholar]
- 7.Agrawal, R. K., Penczek, P., Grassucci, R. A., Li, Y., Leith, A., Nierhaus, K. H. & Frank, J. (1996) Science 271, 1000–1002. [DOI] [PubMed] [Google Scholar]
- 8.Samaha, R. R., Green, R. & Noller, H. F. (1995) Nature 377, 309–314. [DOI] [PubMed] [Google Scholar]
- 9.Green, R., Switzer, C. & Noller, H. F. (1998) Science 280, 286–289. [DOI] [PubMed] [Google Scholar]
- 10.Kim, D. F. & Green, R. (1999) Mol. Cell 4, 859–864. [DOI] [PubMed] [Google Scholar]
- 11.Green, R. & Noller, H. F. (1997) Annu. Rev. Biochem. 66, 679–716. [DOI] [PubMed] [Google Scholar]
- 12.Wilson, D. N., Blaha, G., Connell, S. R., Ivanov, P. V., Jenke, H., Stelzl, U., Teraoka, Y. & Nierhaus, K. H. (2002) Curr. Protein Pept. Sci. 3, 1–53. [DOI] [PubMed] [Google Scholar]
- 13.Rodnina, M. V., Stark, H., Savelsbergh, A., Wieden, H. J., Mohr, D., Matassova, N. B., Peske, F., Daviter, T., Gualerzi, C. O. & Wintermeyer, W. (2000) Biol. Chem. 381, 377–387. [DOI] [PubMed] [Google Scholar]
- 14.Rodnina, M. V. & Wintermeyer, W. (2001) Annu. Rev. Biochem. 70, 415–435. [DOI] [PubMed] [Google Scholar]
- 15.Pape, T., Wintermeyer, W. & Rodnina, M. V. (1998) EMBO J. 17, 7490–7497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moazed, D. & Noller, H. F. (1989) Nature 342, 142–148. [DOI] [PubMed] [Google Scholar]
- 17.Odom, O. W., Picking, W. D. & Hardesty, B. (1990) Biochemistry 29, 10734–10744. [DOI] [PubMed] [Google Scholar]
- 18.Borowski, C., Rodnina, M. V. & Wintermeyer, W. (1996) Proc. Natl. Acad. Sci. USA 93, 4202–4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Semenkov, Y., Shapkina, T., Makhno, V. & Kirillov, S. (1992) FEBS Lett. 296, 207–210. [DOI] [PubMed] [Google Scholar]
- 20.Zavialov, A. V. & Ehrenberg, M. (2003) Cell 114, 113–122. [DOI] [PubMed] [Google Scholar]
- 21.Agrawal, R. K., Penczek, P., Grassucci, R. A., Burkhardt, N., Nierhaus, K. H. & Frank, J. (1999) J. Biol. Chem. 274, 8723–8729. [DOI] [PubMed] [Google Scholar]
- 22.Valle, M., Zavialov, A. V., Sengupta, J., Rawat, U., Ehrenberg, M. & Frank, J. (2003) Cell 114, 123–134. [DOI] [PubMed] [Google Scholar]
- 23.Schmeing, T. M., Seila, A. C., Hansen, J. L., Freeborn, B., Soukup, J. K., Scaringe, S. A., Strobel, S. A., Moore, P. B. & Steitz, T. A. (2002) Nat. Struct. Biol. 9, 225–230. [DOI] [PubMed] [Google Scholar]
- 24.Wintermeyer, W. & Rodnina, M. V. (2000) Essays Biochem. 35, 117–129. [DOI] [PubMed] [Google Scholar]
- 25.Hamel, E. & Nakamoto, T. (1972) J. Biol. Chem. 247, 6810–6817. [PubMed] [Google Scholar]
- 26.Weiss, S. (1999) Science 283, 1676–1683. [DOI] [PubMed] [Google Scholar]
- 27.Zhuang, X., Kim, H., Pereira, M. J., Babcock, H. P., Walter, N. G. & Chu, S. (2002) Science 296, 1473–1476. [DOI] [PubMed] [Google Scholar]
- 28.Ha, T., Rasnik, I., Cheng, W., Babcock, H. P., Gauss, G. H., Lohman, T. M. & Chu, S. (2002) Nature 419, 638–641. [DOI] [PubMed] [Google Scholar]
- 29.Powers, T. & Noller, H. F. (1991) EMBO J. 10, 2203–2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robertson, J. M. & Wintermeyer, W. (1981) J. Mol. Biol. 151, 57–79. [DOI] [PubMed] [Google Scholar]
- 31.Chambliss, G. H., Henkin, T. M. & Leventhal, J. M. (1983) Methods Enzymol. 101, 598–605. [DOI] [PubMed] [Google Scholar]
- 32.Dahlquist, K. D. & Puglisi, J. D. (2000) J. Mol. Biol. 299, 1–15. [DOI] [PubMed] [Google Scholar]
- 33.Soffientini, A., Lorenzetti, R., Gastaldo, L., Parlett, J. H., Spurio, R., LaTeana, A. & Islam, L. (1994) Protein Expression Purif. 5, 118–124. [DOI] [PubMed] [Google Scholar]
- 34.Hoffmann, A. & Roeder, R. G. (1991) Nucleic Acids Res. 19, 6337–6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wyatt, J. R., Chastain, M. & Tinoco, I. J. (1991) BioTechniques 11, 764–769. [PubMed] [Google Scholar]
- 36.Fourmy, D., Meinnel, T., Mechulam, Y. & Blanquet, S. (1993) J. Mol. Biol. 231, 1068–1077. [DOI] [PubMed] [Google Scholar]
- 37.Kahn, D., Fromant, M., Fayat, G., Dessen, P. & Blanquet, S. (1980) Eur. J. Biochem. 105, 489–497. [DOI] [PubMed] [Google Scholar]
- 38.Schmitt, E., Blanquet, S. & Mechulam, Y. (1999) Acta Crystallogr. D 55, 332–334. [DOI] [PubMed] [Google Scholar]
- 39.Carbon, J. & David, H. (1968) Biochemistry 7, 3851–3858. [DOI] [PubMed] [Google Scholar]
- 40.Watson, B. S., Hazlett, T. L., Eccleston, J. F., Davis, C., Jameson, D. M. & Johnson, A. E. (1995) Biochemistry 34, 7904–7912. [DOI] [PubMed] [Google Scholar]
- 41.Plumbridge, J. A., Baumert, H. G., Ehrenberg, M. & Rigler, R. (1980) Nucleic Acids Res. 8, 827–843. [PMC free article] [PubMed] [Google Scholar]
- 42.Reuben, M. A., Kusnezov, I. J. & Wickstrom, E. (1979) Biochim. Biophys. Acta 565, 219–223. [DOI] [PubMed] [Google Scholar]
- 43.Pavlov, M. Y. & Ehrenberg, M. (1996) Arch. Biochem. Biophys. 328, 9–16. [DOI] [PubMed] [Google Scholar]
- 44.Hartz, D., McPheeters, D. S., Traut, R. & Gold, L. (1988) Methods Enzymol. 164, 419–425. [DOI] [PubMed] [Google Scholar]
- 45.van Duin, J., Dieijen, G., van Knippenberg, P. H. & Bosch, L. (1970) Eur. J. Biochem. 17, 433–440. [DOI] [PubMed] [Google Scholar]
- 46.Traut, R. R. & Monro, R. E. (1964) J. Mol. Biol. 10, 63–72. [DOI] [PubMed] [Google Scholar]
- 47.Katunin, V. I., Muth, G. W., Strobel, S. A., Wintermeyer, W. & Rodnina, M. V. (2002) Mol. Cell 10, 339–346. [DOI] [PubMed] [Google Scholar]
- 48.Synetos, D. & Coutsogeorgopoulos, C. (1987) Biochim. Biophys. Acta 923, 275–285. [DOI] [PubMed] [Google Scholar]
- 49.Semenkov, Y. P., Rodnina, M. V. & Wintermeyer, W. (2000) Nat. Struct. Biol. 7, 1027–1031. [DOI] [PubMed] [Google Scholar]
- 50.Sharma, D., Southworth, D. R. & Green, R. (2004) RNA 10, 102–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cate, J. H., Yusupov, M. M., Yusupova, G. Z., Earnest, T. N. & Noller, H. F. (1999) Science 285, 2095–2104. [DOI] [PubMed] [Google Scholar]
- 52.Dale, R. E. & Eisinger, J. (1976) Proc. Natl. Acad. Sci. USA 73, 271–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Studer, S. M., Feinberg, J. S. & Joseph, S. (2003) J. Mol. Biol. 327, 369–381. [DOI] [PubMed] [Google Scholar]
- 54.Karimi, R. & Ehrenberg, M. (1994) Eur. J. Biochem. 226, 355–360. [DOI] [PubMed] [Google Scholar]
- 55.Agrawal, R. K., Lata, R. K. & Frank, J. (1999) Int. J. Biochem. Cell Biol. 31, 243–254. [DOI] [PubMed] [Google Scholar]
- 56.Blanchard, S. C., Gonzalez, R. L., Jr., Kim, H. D., Chu, S. & Puglisi, J. D. (2004) Nat. Struct. Mol. Biol., in press. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.