Abstract
To better understand the pathways involved in the pathogenesis of small cell lung carcinoma (SCLC), we compared the patterns of molecular changes present in these tumors and their accompanying bronchial epithelium with those present in the other two major types of lung cancer [squamous cell carcinoma (SQC) and adenocarcinoma (ADC)]. We obtained DNA from 68 microdissected invasive lung tumors (22 SCLCs, 21 ADCs, and, 25 SQCs) and 119 noncontiguous foci of histologically normal or hyperplastic epithelia from 10 tumors of each histological type. We determined loss of heterozygosity and microsatellite alterations at 12 chromosomal regions frequently deleted in lung cancers using 19 polymorphic microsatellite markers. Our major findings are as follows: (a) the mean index of allelic loss in SCLC (0.85) and SQC (0.71) tumors was higher than that in ADC (0.39) tumors; (b) although there was considerable overlap, each tumor type had a characteristic pattern of allelic loss; (c) most samples of bronchial epithelium accompanying SCLC (90%) had allelic loss at one or more loci compared with samples accompanying SQC (54%) or ADC (10%); (d) the mean index of allelic loss was much higher in bronchial epithelial samples from SCLC (0.27) than in those from SQC (0.08) or ADC (0.01); and (e) although the mean indices of microsatellite alterations in the tumor types were similar, the bronchial epithelial samples accompanying SCLC had a 10-fold higher mean index (0.063) than those accompanying SQC (0.006) or ADC (0.006). Our findings indicate that extensive genetic damage in the accompanying normal and hyperplastic bronchial epithelium is characteristic of SCLC tumors and suggest major differences in the pathogenesis of the three major lung cancer types.
Introduction
Lung cancer is classified into two major groups, SCLC3 and NSCLC (1). SQC, ADC, and large cell carcinoma are the major histological types of NSCLC (2). As with other epithelial malignancies, lung cancers are believed to arise after a series of progressive pathological changes (preneoplastic lesions) in the bronchial epithelium (3). In patients with SQC, the changes include (from earliest to most advanced) hyperplasia, squamous metaplasia, dysplasia of various stages, and CIS (3). The early abnormal epithelial changes such as hyperplasia and squamous metaplasia (without dysplasia) may reflect reactive changes and may not be truly preneoplastic. These changes frequently regress after smoking cessation (4), and the molecular changes present in them are similar to those present in histologically normal epithelium (5). In contrast, advanced dysplastic changes and CIS persist after smoking cessation (6) and contain more advanced molecular changes (5). The reactive and preneoplastic changes are frequently extensive and multifocal, and they occur throughout the respiratory tree, indicating a field effect (3). ADC may be accompanied by hyperplastic or dysplastic changes in the peripheral airway including atypical adenomatous hyperplasia (3), although the neoplastic potential of these lesions has not been demonstrated. Specific preneoplastic changes have not been described for SCLC (3), although smoking-related changes similar to those accompanying SQC may be present (7).
Many mutations, especially those involving recessive oncogenes, have been described in clinically evident lung cancer (8). Whereas some of these are common to all lung cancer types, some are more frequent in specific tumor types (8). For risk assessment and very early lung cancer detection, it would be helpful to know the respiratory epithelial molecular events preceding the development of lung carcinoma. LOH and MA analyses using polymorphic microsatellite DNA markers are frequently used to identify allelic losses at specific chromosomal loci. Recently, we have established that allele losses at chromosomal regions on 3p usually followed by 9p21, 8p21–23, and 17p13 (TP53) losses occur relatively early during the multistage development of SQC of the lung, commencing in histologically normal and mildly abnormal (squamous metaplasia/hyperplasia) stages (5, 9). K-ras mutations and 3p and 9p allele losses have been detected in atypical adenomatous hyperplasia lesions in the peripheral airway (10). However, there is no information about the genetic changes occurring in the bronchial epithelium accompanying lung ADC and SCLC.
In previous studies, we (9, 11) and others (12) have reported a very high incidence of LOH and MAs in the normal and abnormal bronchial mucosa of cancer patients and in current and former smokers. Of interest, approximately half of histologically normal and mildly abnormal bronchial biopsies from smokers show allelic loss, and a subset of these specimens shows allele loss at multiple chromosomal sites, a phenomenon frequently observed in CIS and invasive cancer. To further understand the pathways involved in the pathogenesis of lung cancers, we compared the patterns of the molecular changes found in normal and hyperplastic epithelium accompanying SCLC with those present during the development of the other two major types of lung cancer, namely, SQC and ADC. We compared these findings with those present in the invasive component of the corresponding histological type of lung cancer.
Materials and Methods
Tumor Specimens
Paraffin-embedded materials from 68 surgically resected primary lung carcinomas were analyzed. They consisted of 22 SCLCs, 25 SQCs, and 21 ADCs. They were obtained from cases resected between 1980 and 1997 at Parkland Hospital (Dallas, TX) and M. D. Anderson Cancer Center (Houston, TX). Most lung cancer cases were relatively recent in origin (1990–1997). However, because of the rarity of SCLC resections, five cases were resected between 1980 and 1990. The clinicopathological stage was determined after surgery using standard criteria. Because curative intent surgeries were performed, staging of all cancers, including SCLC cases, was based on the tumor-node-metastasis (TNM) system (13). The patients consisted of 30 women and 38 men ranging in age from 30 – 84 years (mean age, 62 years). Most of the tumors were early-stage tumors (stage I, 72%; stage II, 15%; stage IIIA, 10%). On postoperative staging, two cases (3%; one SCLC and one ADC) were determined to be stage IV. Although they varied from stage I to stage IV, all of the patients were smokers, and most were heavy smokers (mean, 39 pack-years; range, 15–120 pack-years). No significant differences in age, gender, and smoking history were detected between the patients with the three different lung tumor types. The mean age for SCLC patients was 68 years, and the mean age for SQC and ADCA patients was 60 years.
Normal and Hyperplastic Epithelia Accompanying Lung Cancers
We selected 30 cases representing the three major histological types of lung cancer (10 SCLCs, 10 ADCs, and 10 SQCs) that contained multiple noncontiguous foci of histologically normal or hyperplastic bronchial mucosa. From these cases, we identified a total of 119 histologically discrete foci of normal (n = 44) and hyperplastic (n = 75) epithelia, each consisting of at least 800 cells (mean, 3.9 foci/cancer case; range, 2–5 foci/cancer case). All epithelial specimens were obtained from centrally located large bronchi (lobar, segmental, and subsegmental). There were no significant differences in smoking exposure or demographic factors between the different tumor types. Most of the bronchial epithelial samples were taken from a different histological block than the tumor; however, the exact spatial relationship between the epithelial samples and their corresponding tumors could not be determined with precision.
Microdissection and DNA Extraction
Microdissection from archival paraffin-embedded tissues was performed either by laser capture microdissection or manually using a micromanipulator from multiple microslides of each sample (5, 9, 11, 14, 15). DNA extraction was performed as described previously (5). Dissected lymphocytes or stromal cells from the same slides were used as a source of constitutional DNA from each case. After DNA extraction, 5 µl of the proteinase K-digested samples containing DNA from at least 200 cells were used for each multiplex PCR reaction.
Polymorphic DNA Markers and PCR-based Microsatellite Analysis
To evaluate LOH and MAs, we used primers flanking 19 dinucleotide and multinucleotide microsatellite repeat polymorphisms spanning 12 chromosomal regions frequently deleted in lung cancer (8, 9, 16). The microsatellite markers and the chromosomal regions analyzed were as follows: (a) 3p12 (D3S1274); (b) 3p14.2 at the FHIT gene (D3S4103); (c) 3p21 (D3S44623/Luca 2.2, D3S1478, and D3S1029); (d) 3p22–24 (D3S2432 and D3S1537); (e) 4p (D4S404); (f) 4q25–32 (D4S194 and D4S408); (g) 5q21–22 (L5.71 in the APC-MCC region); (h) 6q16 –21 (D6S1021); (i) 8p21–23 (D8S1130 and D8S11106); (j) 9p21 (IFNA, D9S1748); (k) 13q14 (dinucleotide repeat at the RB gene); and (l) 17p13 (TP53 dinucleotide and pentanucleotide repeats). Primer sequences can be obtained from the Genome Database, with five exceptions that have been published and referenced previously (5). For all samples, multiplex PCR (up to six markers) was performed in the first amplification, followed by uniplex PCR for individual microsatellite markers as described previously (17).
Data Analysis
Because heterozygosity at the different loci varied between subjects, the number of chromosomal regions tested varied in subjects. Thus, indices were calculated to compare molecular changes between microdissected invasive lung carcinomas and epithelial samples and between subjects (5, 9, 17). The FRL-sample index indicates LOH for all informative chromosomal regions per microdissected specimen (maximum, 12 regions/specimen). The FRL-subject index indicates the LOH for all informative chromosomal regions per subject in their bronchial epithelium specimens. The indices were calculated as shown below.
| (1) |
| (2) |
To compare the incidence of MAs, we used the MA index (17). The MA index indicates the MA for all microsatellite markers per microdissected specimen (maximum, 19 microsatellite markers/specimen) and was calculated as shown below.
| (3) |
Statistical analyses were performed using the nonparametric Wilcoxon and Fisher exact tests. The cumulative binomial test was used to examine the likelihood that a particular event (for example, loss of the same allele in the invasive carcinoma and an associated epithelial sample) occurs at a particular probability when observed in repeated trials. When the results are compared with a chance occurrence or nonoccurrence, the particular probability of comparison is 0.5.
Results
Patterns of LOH in Invasive Lung Carcinoma Types
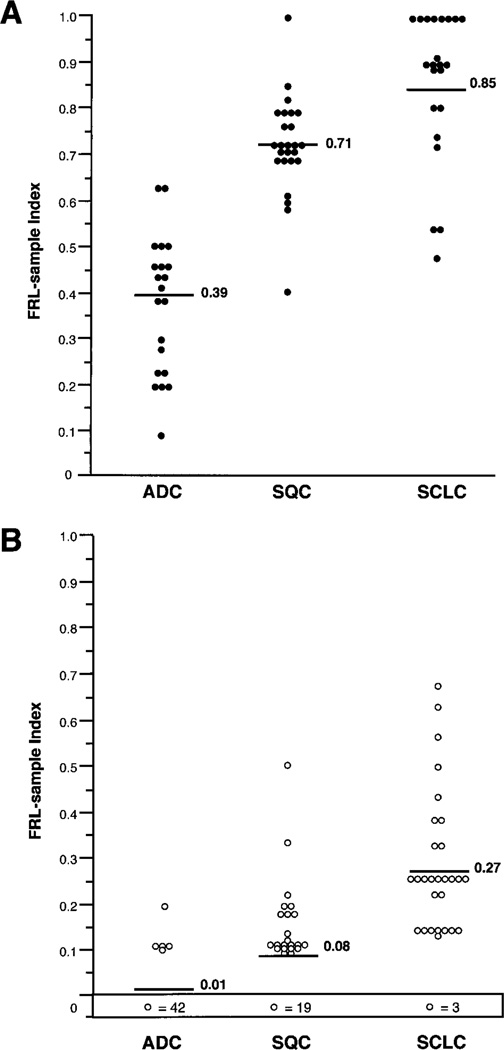
Sixty-eight resected lung cancers were microdissected and tested for allele loss at 12 chromosomal regions, and examples of these analyses are illustrated in Fig. 1. The FRL-sample index was used to compare allelic loss at all chromosomal regions analyzed between lung tumor types. Whereas similar high FRL-sample index means were observed between SCLC (mean, 0.85) and SQC (mean, 0.71), ADC demonstrated a significantly lower FRL-sample index mean (mean, 0.39; P < 0.001) in the chromosomal regions analyzed, indicating less extensive allelic losses compared with SQC and SCLC (Fig. 2A). Although there was considerable overlap, each tumor type had a characteristic pattern of allelic loss. SCLC and SQC demonstrated similar patterns of allele loss for the various chromosomal regions, with three exceptions (Table 1). SCLCs demonstrated higher incidences of LOH at the 5q21–22 (69% versus 27%; P = 0.042) and 13q14 (RB gene loci; 67% versus 29%; P = 0.047) regions, whereas SQCs had a higher incidence of LOH at the 8p21–23 region (92% versus 29%; P < 0.001). In contrast, ADCs had a distinct pattern, with lower frequencies of allelic loss than SCLCs or SQCs at all but two chromosomal regions analyzed (Table 1).
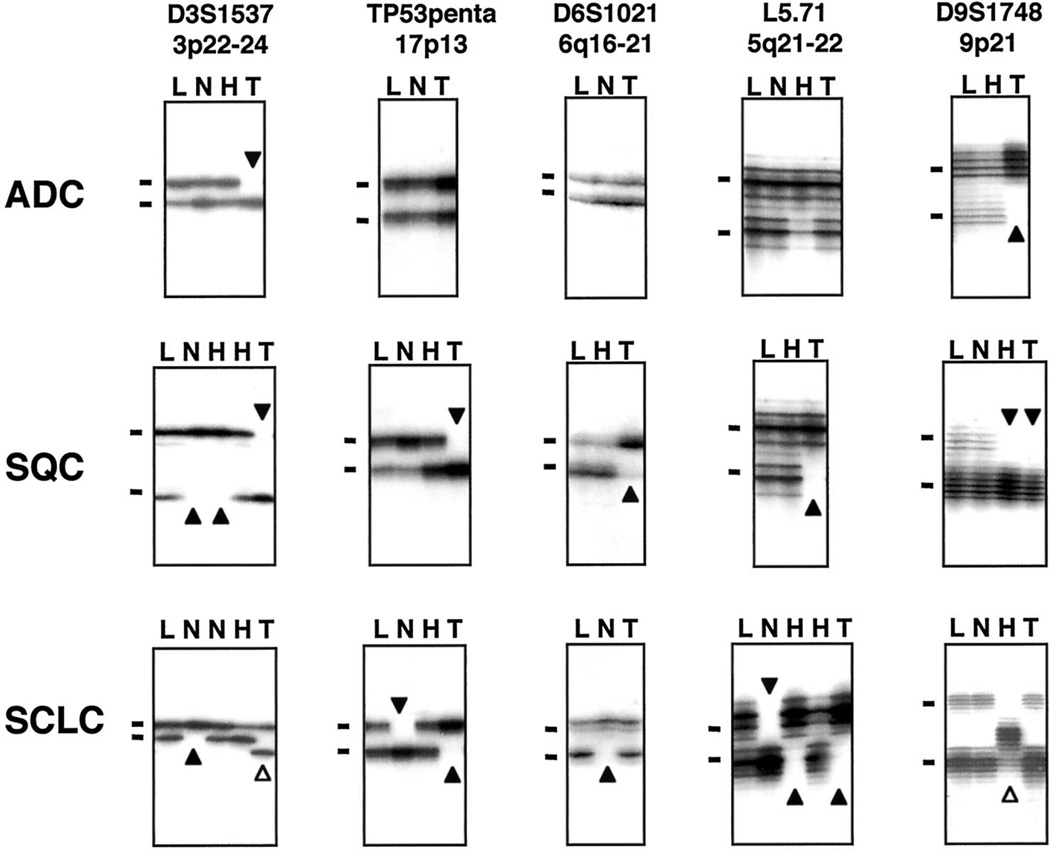
Fig. 1.
Representative autoradiograph of gels demonstrating LOH (closed arrowheads) and MAs (open arrowheads) at five chromosomal regions (3p22–24, 17p13-TP53, 6q16 –21, 5q21–22, and 9p21) in bronchial epithelium and the corresponding lung tumor specimens. There are two examples of MAs [marker D3S1537 (3p22–24) in the SCLC tumor sample and marker D9S1748 (9p21) in the hyperplastic bronchial epithelium]. L, lymphocytes; N, histologically normal bronchial epithelium; H, hyperplastic bronchial epithelium; T, invasive lung tumor. Bars, the main allele bands.
Fig. 2.
Overall incidence of allelic loss represented by the FRL-sample index of each (A) invasive lung cancer specimen (n = 68) and (B) histologically normal and hyperplastic bronchial epithelium specimens (n = 119) by histological tumor type. The box at the bottom of the chart in B indicates the number of bronchial epithelial samples with no allelic loss (FRL-sample = 0). Bars and numbers indicate the mean FRL-sample indices for each category.
Table 1.
LOH frequencies in invasive lung cancers (n = 68) and accompanying bronchial epithelia (n = 119)
| Invasive tumors | Bronchial epithelial samples | |||||
|---|---|---|---|---|---|---|
| Chromosomal regions | ADC n = 21 | SQC n = 25 | SCLC n = 22 | ADC n = 47 | SQC n = 41 | SCLC n = 31 |
| 3p12 | 2/14 (14%) | 11/12 (92%) | 7/9 (78%) | 0/32a | 1/30 (3%) | 2/10 (20%) |
| 3p14 FHIT | 5/16 (31%) | 19/21 (91%) | 13/16 (81%) | 0/43 | 1/40 (3%) | 3/27 (11%) |
| 3p21 | 7/20 (35%) | 22/24 (92%) | 16/19 (84%) | 0/47a | 15/41 (37%)b | 8/25 (32%) |
| 3p22–24 | 4/18 (22%) | 18/23 (78%) | 17/22 (77%) | 1/47 (2%)a | 3/41 (7%) | 6/31 (19%) |
| Any 3p | 10/21 (48%) | 23/25 (92%) | 19/22 (86%) | 1/47 (2%)a | 14/41 (34%)b | 14/31 (45%) |
| 4p | 0/10 | 2/6 (33%) | 6/11 (55%) | 0/23 | 2/21 (10%) | 4/23 (18%) |
| 4q25–32 | 0/6 | 4/8 (50%) | 6/9 (67%) | 0/27a | 2/31 (7%) | 7/31 (23%) |
| 5q21–22 | 0/14 | 3/11 (27%) | 11/16 (69%) | 0/36a | 0/30 | 7/22 (32%)c |
| 6q16–21 | 4/16 (25%) | 4/9 (44%) | 7/13 (54%) | 0/36 | 0/35 | 2/13 (15%) |
| 8p21–23 | 6/19 (32%) | 22/24 (92%) | 6/21 (29%) | 0/39a | 3/41 (7%) | 8/30 (27%)c |
| 9p21 | 10/16 (63%) | 9/13 (69%) | 10/19 (53%) | 2/43 (5%)a | 4/38 (11%) | 10/31 (32%)c |
| 13q14 RB | 5/15 (33%) | 4/14 (29%) | 10/15 (67%) | 1/31 (3%) | 0/25 | 2/16 (13%) |
| 17p13 TP53 | 8/18 (44%) | 18/22 (82%) | 18/20 (90%) | 1/38 (3%)a | 0/31 | 12/19 (63%)c |
| Any region | 19/21 (90%) | 25/25 (100%) | 22/22 (100%) | 5/47 (10%)a | 22/41 (54%)b | 28/31 (90%)c |
Significant differences between bronchial epithelium accompanying ADC and SCLC were detected at 3p12 (P = 0.041), 3p21 (P < 0.001), 3p22–24 (P = 0.014), any 3p (P < 0.001), 4p (P = 0.006), 4q25–32 (P = 0.013), 5q21 (P < 0.001), 8p21–23 (P = 0.001), 9p21 (P = 0.002), 17p13 (P < 0.001), and any region (P < 0.001).
Significant differences between bronchial epithelium accompanying ADC and SQC were detected at 3p21 (P < 0.001), any 3p (P < 0.001), and any chromosomal region (P < 0.001).
Significant differences between bronchial epithelium accompanying SQC and SCLC were detected at 5q21 (P = 0.005), 8p21–23 (P = 0.029), 9p21 (P = 0.026), and 17p13 (P < 0.001), and any chromosomal regions (P < 0.001).
Frequency of Allelic Loss in Bronchial Epithelium Accompanying Lung Cancers
A total of 119 foci of normal (n= 44) or hyperplastic (n = 75) respiratory epithelia containing sufficient cells for multiple assays were microdissected from 30 of the lung cancer pulmonary resection samples (10 samples of each type). These cases were selected because multiple noncontiguous foci of nonneoplastic epithelium were available. Several examples of allelic loss in the bronchial epithelium accompanying the corresponding invasive tumors are shown in Fig. 1.
Most samples of bronchial epithelium accompanying SCLC (90%) had allelic loss at one or more loci as compared with SQC (54%) or ADC (10%). We detected a very high incidence of LOH in normal and hyperplastic epithelium accompanying SCLCs, as expressed by a high FRL-sample mean (mean, 0.27) and by high frequencies of LOH at any chromosomal region (90% of the samples) compared with the other two lung cancer types (P < 0.001; Table 1; Fig. 2B). Bronchial epithelial specimens obtained from SQC demonstrated an intermediate LOH frequency (mean FRL-sample index, 0.08; LOH at any region seen in 54% of the samples) compared with epithelial samples from ADCs (mean FRL-sample index, 0.01; LOH at any region seen in 10% of the samples), which exhibited a significantly (P < 0.001) lower frequency of allele loss. In fact, several bronchial epithelial samples from SCLCs exhibited FRL-sample indices similar to those of their corresponding tumor specimens (Fig. 2). The mean FRL-sample indices for tumors and epithelial specimens were higher in SCLC than in the other cancer types. However, even after adjusting for the relatively high tumor index, the epithelial sample index in SCLC was relatively high, as demonstrated by the epithelial sample: tumor ratio, which was calculated on a case-by-case basis (0.32 for SCLC, 0.11 for SQC, and 0.03 for ADC; Table 2). No significant differences in the incidences of allelic loss were detected between histologically normal and hyperplastic specimens for any of the tumor types. Higher frequencies of allelic loss were present in the bronchial epithelium of SCLC and ADC of male patients compared with female patients, but these gender differences were not significant.
Table 2.
Summary of the major differences in the genetic changes detected between lung cancer types
| Invasive tumor | Bronchial epithelium | |||||
|---|---|---|---|---|---|---|
| Genetic marker | ADC | SQC | SCLC | ADC | SQC | SCLC |
| 5q21–22 LOH | 0 | 27% | 69% | 0 | 0 | 32%a |
| 8p21–23 LOH | 32% | 92% | 29% | 0 | 7% | 27%a |
| 9p21 LOH | 63% | 69% | 53% | 5% | 11% | 32%a |
| 17p13-TP53 LOH | 44% | 82% | 80% | 3% | 0 | 63%a |
| Any chromosomal region LOH | 90% | 100% | 100% | 10% | 54% | 90%a |
| Mean FRL-subject indexb | 0.01 | 0.09 | 0.31a | |||
| Mean FRL-sample index | 0.39 | 0.71 | 0.85 | 0.01 | 0.08 | 0.27a |
| Epithelium FRL-sample index/tumor FRL-sample index | 0.03 | 0.11 | 0.32 | |||
| MA index | 0.019 | 0.024 | 0.078 | 0.006 | 0.006 | 0.063c |
| MA frequency | 24% | 32% | 50% | 13% | 10% | 68%a |
P < 0.001 (significant difference between bronchial samples obtained from SCLC and the other histological types of lung cancer.
Mean FRL-sample index was calculated using only the epithelial samples from each subject.
P < 0.001 (significant difference between bronchial samples obtained from SCLC and the other histological types of lung cancer.
Pattern of Allelic Loss in Bronchial Epithelium Accompanying Lung Cancers (Table 1)
Bronchial epithelial samples obtained from ADCs had a very low frequency of LOH at all chromosomal regions examined. Epithelial samples accompanying SCLCs demonstrated a significantly higher frequency of LOH at 5q21–22, 8p21–23, 9p21, and 17p13 (TP53 gene) than those present in samples obtained from SQC specimens (Table 1). Of interest, deletions at the 5q21–22 and 17p13 (TP53 gene) regions were never detected or occasionally detected in epithelial samples obtained from SQCs and ADCs. However, epithelial samples from SCLC patients had allelic loss frequencies of 32% and 63%, respectively, at these regions. In fact, the frequency of LOH at 17p13 (63%) was the highest rate of loss for any chromosomal region in epithelial samples from any type of cancer. SCLC-associated epithelial samples even had higher LOH frequencies at regions demonstrating relatively low LOH frequencies in SCLC tumors (9p21 and 8p21–23).
ASL
Our previous studies have demonstrated that at any individual locus, the allele loss patterns in bronchial preneoplastic lesions were not random but had a strong tendency to be present in the same chromosomal loci, and the identical parental allele is lost in the corresponding invasive tumor (5, 9, 11). We refer to this phenomenon as ASL. In cases in which nonneoplastic and tumor samples demonstrated loss at a particular locus, we determined whether the losses involved the same or both parental alleles. For all comparisons, allelic loss at the same chromosomal loci was present in 54 of 94 (57%) comparisons. We also compared the specific parental allele lost in the corresponding nonneoplastic and neoplastic samples. For all comparisons, the same parental allele was lost in 38 of 54 (70%) comparisons (P = 0.001). No significant differences in the ASL phenomenon were detected between specimens obtained from the three major types of lung cancer analyzed.
MAs in Invasive Carcinomas and Accompanying Bronchial Epithelium
A relatively high incidence of MAs was detected in all of the three types of lung carcinomas (24 of 68 cases; 35%). However, there were no significant differences in MA frequency between the histological tumor types (Table 2). MAs at one or more loci analyzed were also detected in histologically normal and hyperplastic respiratory epithelium accompanying lung cancers (30 of 119 cases overall; 25%). Because artifacts resulting from PCR amplification may be mistaken for MAs, especially when minute amounts of input DNA are used, all examples of MAs occurring in nontumor samples were confirmed by a replicate PCR analysis. Of interest, in the bronchial epithelium, we found a significantly higher incidence of MAs in samples accompanying SCLCs compared with those accompanying SQCs and ADCs (Table 2). No differences in the MA frequencies were detected between normal and hyperplastic specimens. None of the MAs detected in bronchial epithelia were identical to the alterations observed in the corresponding invasive tumors. No gender differences in the frequency of MAs in the bronchial epithelium accompanying lung cancers were detected. Examples of MAs in the bronchial epithelium and invasive lung tumors are shown in Fig. 1.
Discussion
Many mutations, especially those involving recessive oncogenes, have been described in invasive lung cancer (8). Whereas some of these are common to all lung cancer types, some appear to be tumor type related. Studies of a large number of lung cancers have shown different patterns of mutation between the two major types of lung cancer, namely, SCLC and NSCLC (8, 18, 19). SCLC has been better characterized as a specific entity because most of the studies compare their findings with those of NSCLC (mostly ADC and SQC) as a group (8). However, some studies have demonstrated differences in the genetic abnormality patterns between ADC and SQC (20–22), especially in the K-ras gene mutation frequency (22–25). Our results indicate that more genetic changes accumulate during tumorigenesis of SCLC and SQC than that of ADC. The incidence and pattern of allelic loss detected in our microdissected SCLC cases are consistent with previous reports in cell lines and fresh tumors (8, 9, 16). Whereas SCLC demonstrated a significantly higher incidence of allelic loss at 5q21–22 and 13q14 (RB gene) than SQC, the latter showed a significantly greater frequency of 8p21–23 losses. Although the overall incidences of allelic loss in the NSCLC groups are similar to those reported previously, many differences between SQC and ADC tumors were detected. Our findings of different patterns of LOH among all three of the major histological types of lung carcinoma link genetic changes in tumors with histological characteristics.
One of our most striking findings was the relative difference in allelic loss in bronchial epithelium accompanying lung cancers. The vast majority of samples of bronchial epithelium accompanying SCLC (90%) had allelic loss at one or more loci as compared with SQC (54%) or ADC (10%). We have previously described allelic losses in normal and mildly abnormal epithelium associated with SQC and other tumors, and higher incidences were present in more advanced preneoplastic changes [dysplasia and CIS (5, 9, 26)]. The bronchial epithelium accompanying SCLC also showed a more extensive pattern of allele loss than did the other lung cancers, in some cases involving most of the chromosomal regions analyzed and demonstrating a similar or greater incidence of genetic changes than some invasive lung carcinoma cases. The finding of a very low frequency of allelic loss in proximal airway epithelium accompanying ADCs is consistent with the origin of most of these tumors from the smaller peripheral airways. The development of epithelial cancers requires multiple mutations, the stepwise accumulation of which may represent a mutator phenotype (27). Thus, it is possible that those epithelial cells that have accumulated multiple mutations are at higher risk for progression to invasive cancer. Our findings suggest that in SCLC, in which no characteristic preneoplastic sequence of morphological changes has been described, the tumors may arise directly from either normal or hyperplastic epithelia, without passing through recognizable intermediate pathological stages (parallel theory of lung cancer development). In contrast, SQC and perhaps ADC would appear to develop after a sequence of morphological intermediate steps (sequential theory).
The multiple mutations required for cancer development that accumulate during the preneoplastic process are not random but usually follow a pattern (27). Our present findings indicate that 5q21–22 and 17p13 (TP53 gene) deletions are frequent and early changes in the bronchial epithelium accompanying SCLC, as compared with the other lung cancer types. Recently, we have reported that allele losses at those loci are infrequent and relatively late events during the multistage development of SQC and in bronchial biopsies from smokers without cancer (5). Mutational analysis of the TP53 gene in our SCLC cases would be desirable, but because of the very limited size of the epithelial samples, this was not feasible. We have recently demonstrated that allelic loss at three or more chromosome 4 regions is frequent in lung cancers, especially SCLC (16). Our findings demonstrate that deletions at chromosome 4 regions may also be detected as an early event in the pathogenesis of lung carcinomas. The finding in epithelial samples from SCLC of certain genetic changes (9p21 and 8p21–23 LOH) infrequently found in the associated tumors may reflect a field effect, rather than specific changes associated with the SCLC pathway.
In previous studies, we have noted that specific chromosomal loci and specific parental alleles lost in preneoplastic lesions of the respiratory tract are usually identical to those lost in the corresponding invasive cancer (mostly SQCs), a phenomenon referred to as ASL (5, 9, 11, 14, 15). However, in another recent study (26), we failed to demonstrate ASL in histologically normal and hyperplastic epithelia. In our present study, which focused on histologically normal or hyperplastic epithelium, ASL was noted, but at a lower rate than reported previously in histologically and molecularly more advanced lesions.
MA (also referred to as microsatellite instability) represents changes in the size of simple nucelotide repeat polymorphic microsatellite markers, resulting in altered electrophoretic mobility of one or both alleles (17). In lung cancers, MAs have been reported to occur at frequencies ranging from 0–45% (17). Whereas the mechanism underlying MAs is currently unknown, they appear to represent a manifestation of genomic instability. Although SCLCs demonstrated a higher incidence of MAs than other lung cancer histological type, the differences were not statistically significant. However, the bronchial epithelia accompanying SCLC showed a significant 10-fold higher incidence of MAs compared with the corresponding epithelial specimens associated with SQC and ADC specimens. These findings are a further indication that more extensive genetic damage is present in the respiratory epithelium of patients with SCLC than in those with the other major types of lung cancer. The findings of more extensive genetic damage in the epithelial field of SCLC patients may explain the higher frequencies of second primary lung tumor in cases with this tumor type (28).
In summary, our findings indicate that several genetic changes are common to all lung cancer histological types, whereas others appear to be tumor type specific. More extensive genetic damage accumulates during the pathogenesis of centrally located SCLC and SQC than during that of peripherally arising ADC. Our results also indicate that the most widespread and most extensive genetic damage (allelic loss and MAs) is present in the normal and mildly abnormal bronchial epithelium of patients with SCLC.
Acknowledgments
Supported by Specialized Program of Research Excellence Grant P50-CA70907 from the National Cancer Institute, NIH (Bethesda, MD) and a grant from the Bristol-Myers-Squibb Foundation Inc. (New York, NY).
Footnotes
The abbreviations used are: SCLC, small cell lung carcinoma; NSCLC, non-SCLC; SQC, squamous cell carcinoma; ADC, adenocarcinoma; CIS, carcinoma in situ; LOH, loss of heterozygosity; MA, microsatellite alteration; FRL, fractional regional loss; ASL, allele-specific loss.
References
- 1.Minna JD, Sekido Y, Fong K, Gazdar AF. Molecular biology of lung cancer. In: DeVita VT Jr, Hellman S, Rosenberg SA, editors. Cancer: Principles and Practice of Oncology. 5th. Philadelphia: Lippincott; 1997. pp. 849–857. [Google Scholar]
- 2.Travis WD, Colby TV, Corrin B, Shimosato Y, Brambilla E, Countries CF. WHO Classification of Lung and Pleural Tumors. 3rd. Berlin: Springer-Verlag; 1999. [Google Scholar]
- 3.Colby TV, Wistuba II, Gazdar A. Precursors to pulmonary neoplasia. Adv. Anat. Pathol. 1998;5:205–215. doi: 10.1097/00125480-199807000-00001. [DOI] [PubMed] [Google Scholar]
- 4.Auerbach O, Stout AP, Hammond EC, Garfinkel L. Changes in bronchial epithelium in relation to smoking and cancer of the lung. N. Engl. J. Med. 1961;265:253–267. doi: 10.1056/NEJM196108102650601. [DOI] [PubMed] [Google Scholar]
- 5.Wistuba II, Behrens C, Milchgrub S, Bryant D, Hung J, Minna JD, Gazdar AF. Sequential molecular abnormalities are involved in the multistage development of squamous cell lung carcinoma. Oncogene. 1999;18:643–650. doi: 10.1038/sj.onc.1202349. [DOI] [PubMed] [Google Scholar]
- 6.Lam S, leRiche JC, Zheng Y, Coldman A, MacAulay C, Hawk E, Kelloff G, Gazdar AF. Sex-related differences in bronchial epithelial changes associated with tobacco smoking. J. Natl. Cancer Inst. 1999;91:691–696. doi: 10.1093/jnci/91.8.691. [DOI] [PubMed] [Google Scholar]
- 7.Gazdar AF, Cohen MH, Ihde DC, Minna JD, Matthews MJ. Bronchial epithelial changes in association with small cell carcinoma of the lung. In: Muggia F, Rozencweig M, editors. Proceedings of the Second National Cancer Institute Conference on Lung Cancer Treatment; Raven Press; New York. 1979. pp. 167–174. [Google Scholar]
- 8.Sekido Y, Fong KM, Minna JD. Progress in understanding the molecular pathogenesis of human lung cancer. Biochim. Biophys Acta. 1998;1378:F21–F59. doi: 10.1016/s0304-419x(98)00010-9. [DOI] [PubMed] [Google Scholar]
- 9.Wistuba II, Behrens C, Virmani AK, Milchgrub S, Syed S, Lam S, Mackay B, Minna JD, Gazdar AF. Allelic losses at chromosome 8p21–23 are early and frequent events in the pathogenesis of lung cancer. Cancer Res. 1999;59:1973–1979. [PubMed] [Google Scholar]
- 10.Kitamura H, Kameda Y, Ito T, Hayashi H. Atypical adenomatous hyperplasia of the lung. Implications for the pathogenesis of peripheral lung adenocarcinoma. Am. J. Clin. Pathol. 1999;111:610–622. doi: 10.1093/ajcp/111.5.610. [DOI] [PubMed] [Google Scholar]
- 11.Wistuba II, Lam S, Behrens C, Virmani AK, Fong KM, LeRiche J, Samet JM, Srivastava S, Minna JD, Gazdar AF. Molecular damage in the bronchial epithelium of current and former smokers. J. Natl. Cancer Inst. 1997;89:1366–1373. doi: 10.1093/jnci/89.18.1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mao L, Lee JS, Kurie JM, Fan YH, Lippman SM, Lee JJ, Ro JY, Broxson A, Yu R, Morice RC, Kemp BL, Khuri FR, Walsh GL, Hittelman WN, Hong WK. Clonal genetic alterations in the lungs of current and former smokers. J. Natl. Cancer Inst. 1997;89:857–862. doi: 10.1093/jnci/89.12.857. [DOI] [PubMed] [Google Scholar]
- 13.Mountain CF. Revisions in the International System for Staging Lung Cancer. Chest. 1997;111:1710–1717. doi: 10.1378/chest.111.6.1710. [DOI] [PubMed] [Google Scholar]
- 14.Hung J, Kishimoto Y, Sugio K, Virmani A, McIntire DD, Minna JD, Gazdar AF. Allele-specific chromosome 3p deletions occur at an early stage in the pathogenesis of lung carcinoma. J. Am. Med. Assoc. 1995;273:558–563. [PubMed] [Google Scholar]
- 15.Kishimoto Y, Sugio K, Mitsudomi T, Oyama T, Virmani A, McIntire DD, Gazdar AF. Allele specific loss of chromosome 9p in preneoplastic lesions accompanying non-small cell lung cancers. J. Natl. Cancer Inst. 1995;87:1224–1229. doi: 10.1093/jnci/87.16.1224. [DOI] [PubMed] [Google Scholar]
- 16.Shivapurkar N, Virmani AK, Wistuba II, Milchgrub S, Mackay B, Minna JD, Gazdar AF. Deletions of chromosome 4 at multiple sites are frequent in malignant mesothelioma and small cell lung carcinoma. Clin. Cancer Res. 1999;5:17–23. [PubMed] [Google Scholar]
- 17.Wistuba II, Behrens C, Milchgrub S, Virmani AK, Jagirdar J, Thomas B, Ioachim HL, Litzky LA, Brambilla EM, Minna JD, Gazdar AF. Comparison of molecular changes in lung cancers in HIV-positive and HIV-indeterminate subjects. J. Am. Med. Assoc. 1998;279:1554–1559. doi: 10.1001/jama.279.19.1554. [DOI] [PubMed] [Google Scholar]
- 18.Virmani AK, Fong KM, Kodagoda D, McIntire D, Hung J, Tonk V, Minna JD, Gazdar AF. Allelotyping demonstrates common and distinct patterns of chromosomal loss in human lung cancer types. Genes Chromosomes Cancer. 1998;21:308–319. doi: 10.1002/(sici)1098-2264(199804)21:4<308::aid-gcc4>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 19.Mitsudomi T, Viallet J, Mulshine JL, Linnoila RI, Minna JD, Gazdar AF. Mutations of ras genes distinguish a subset of non-small-cell lung cancer cell lines from small-cell lung cancer cell lines. Oncogene. 1991;6:1353–1362. [PubMed] [Google Scholar]
- 20.O’Briant KC, Bepler G. Delineation of the centromeric and telomeric chromosome segment 11p15.5 lung cancer suppressor regions LOH11A and LOH11B. Genes Chromosomes Cancer. 1997;18:111–114. doi: 10.1002/(sici)1098-2264(199702)18:2<111::aid-gcc5>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 21.Bepler G, Fong KM, Johnson BE, O’Briant KC, Daly LA, Zimmerman PV, Garcia-Blanco MA, Peterson B. Association of chromosome 11 locus D11S12 with histology, stage, and metastases in lung cancer. Cancer Detect. Prev. 1998;22:14–19. [PubMed] [Google Scholar]
- 22.Sato S, Nakamura Y, Tsuchiya E. Difference of allelotype between squamous cell carcinoma and adenocarcinoma of the lung. Cancer Res. 1994;54:5652–5655. [PubMed] [Google Scholar]
- 23.Petersen I, Bujard M, Petersen S, Wolf G, Goeze A, Schwendel A, Langreck H, Gellert K, Reichel M, Just K, du Manoir S, Cremer T, Dietel M, Ried T. Patterns of chromosomal imbalances in adenocarcinoma and squamous cell carcinoma of the lung. Cancer Res. 1997;57:2331–2335. [PubMed] [Google Scholar]
- 24.Yokoyama S, Yamakawa K, Tsuchiya E, Murata M, Sakiyama S, Nakamura Y. Deletion mapping on the short arm of chromosome 3 in squamous cell carcinoma and adenocarcinoma of the lung. Cancer Res. 1992;52:873–877. [PubMed] [Google Scholar]
- 25.Suzuki Y, Orita M, Shiraishi M, Hayashi K, Sekiya T. Detection of ras gene mutations in human lung cancers by single-strand conformation polymorphism analysis of polymerase chain reaction products. Oncogene. 1990;5:1037–1043. [PubMed] [Google Scholar]
- 26.Park I-W, Wistuba II, Maitra A, Milchgrub S, Virmani AK, Minna JD, Gazdar AF. Multiple clonal abnormalities in bronchial epithelium of lung cancer patients. J. Natl. Cancer Inst. 1999;91:1863–1868. doi: 10.1093/jnci/91.21.1863. [DOI] [PubMed] [Google Scholar]
- 27.Loeb LA. Cancer cells exhibit a mutator phenotype. Adv. Cancer. Res. 1998;72:25–56. doi: 10.1016/s0065-230x(08)60699-5. [DOI] [PubMed] [Google Scholar]
- 28.Heyne KH, Lippman SM, Lee JJ, Lee JS, Hong WK. The incidence of second primary tumors in long-term survivors of small-cell lung cancer. J. Clin. Oncol. 1992;10:1519–1524. doi: 10.1200/JCO.1992.10.10.1519. [DOI] [PubMed] [Google Scholar]