Abstract
In model organisms, classical genetic screening via random mutagenesis provides key insights into the molecular bases of genetic interactions, helping defining synthetic-lethality, synthetic-viability and drug-resistance mechanisms. The limited genetic tractability of diploid mammalian cells, however, precludes this approach. Here, we demonstrate the feasibility of classical genetic screening in mammalian systems by using haploid cells, chemical mutagenesis and next-generation sequencing, providing a new tool to explore mammalian genetic interactions.
Classical genetic screens with chemical mutagens assign functionality to genes in model organisms1,2. Since most mutagenic agents yield single-nucleotide variants (SNVs), mutation clustering provides information on the functionality of protein domains, and defines key amino acid residues within them3. RNA interference (RNAi) allows forward-genetic screening in human cell cultures3, and insertional mutagenesis in near-haploid human cancer cells4 and whole-genome CRISPR/Cas9 small-guide RNA (sgRNA) libraries have also been used for this purpose5,6. Although powerful, such loss-of-function (LOF) approaches miss phenotypes caused by separation-of-function or gain-of-function SNV mutations, are less informative on defining functional protein regions, and are not well suited to studying functions of essential genes7. Here, we describe the generation of chemically mutagenized mammalian haploid cell libraries, and establish their utility to identify recessive suppressor mutations by using resistance to 6-thioguanine (6-TG) as a proof-of-principle.
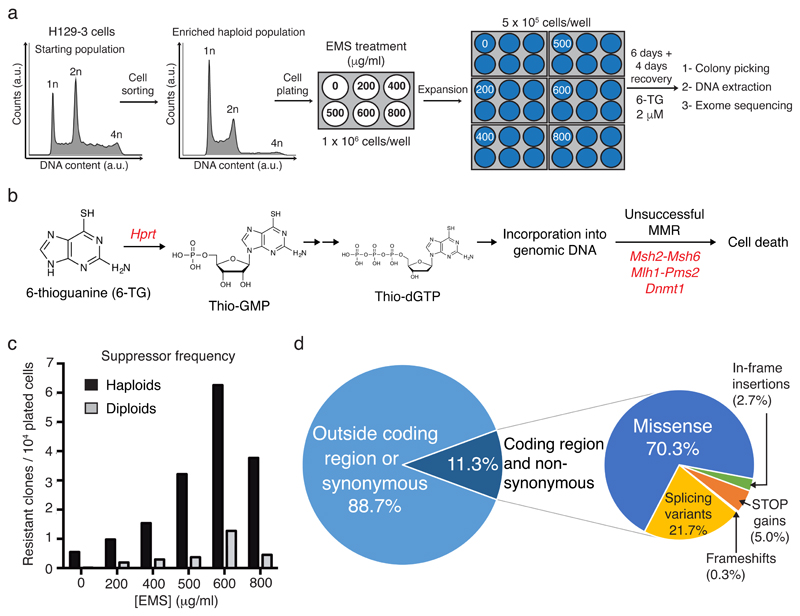
Comprehensive libraries of homozygous SNV-containing mutant clones are not feasible to obtain in cells with diploid genomes. To circumvent this, we used H129-3 haploid mouse embryonic stem cells (mESCs)8 that we had mock-treated or treated with varying doses of the DNA-alkylating agent ethylmethanesulfonate (EMS), a chemical inducer of SNVs9 (Fig. 1a; Supplementary Results, Supplementary Fig. 1a). For comparison, the same procedure was performed on diploid H129-3 mESCs (Supplementary Fig. 1b). Haploid and diploid mutant libraries were then screened for suppressors of cellular sensitivity to 6-TG (Fig. 1b). Ensuing analyses revealed EMS-dose dependent induction of 6-TG resistance, with more clones arising in haploid than in diploid cells (Fig. 1c), thus highlighting the advantage of identifying suppressor mutations in a haploid genetic background.
Figure 1. Generation of mutagenized libraries.
(a) Experimental workflow. (b) Schematic of 6-TG metabolism and genotoxicity. Inactivating mutations in genes highlighted in red have been shown to confer 6-TG resistance. (c) Suppressor frequencies to 6-TG treatment of different EMS-mutagenized libraries, represented as number of suppressor clones isolated per 10,000 plated cells. (d) Locations and consequences of identified mutations.
Next, we isolated 196 6-TG resistant clones from EMS-generated haploid cell libraries. To assess the feasibility of identifying causative suppressor mutations, we subjected DNA samples from seven resistant clones, and from control mESCs not treated with EMS, to whole-exome DNA sequencing. Ensuing analyses, comparing sequences from EMS-resistant clones with control mESCs and the 129S5 mouse genome (see Methods), identified homozygous base insertions/deletions (INDELs) and SNVs. Only 11.3% of these affected coding sequences and were non-synonymous (Fig. 1d). Thus, while each resistant clone had ~370 INDEL/SNV mutations (Supplementary Fig. 1c), on average only ~40 of these were in coding sequences and non-synonymous.
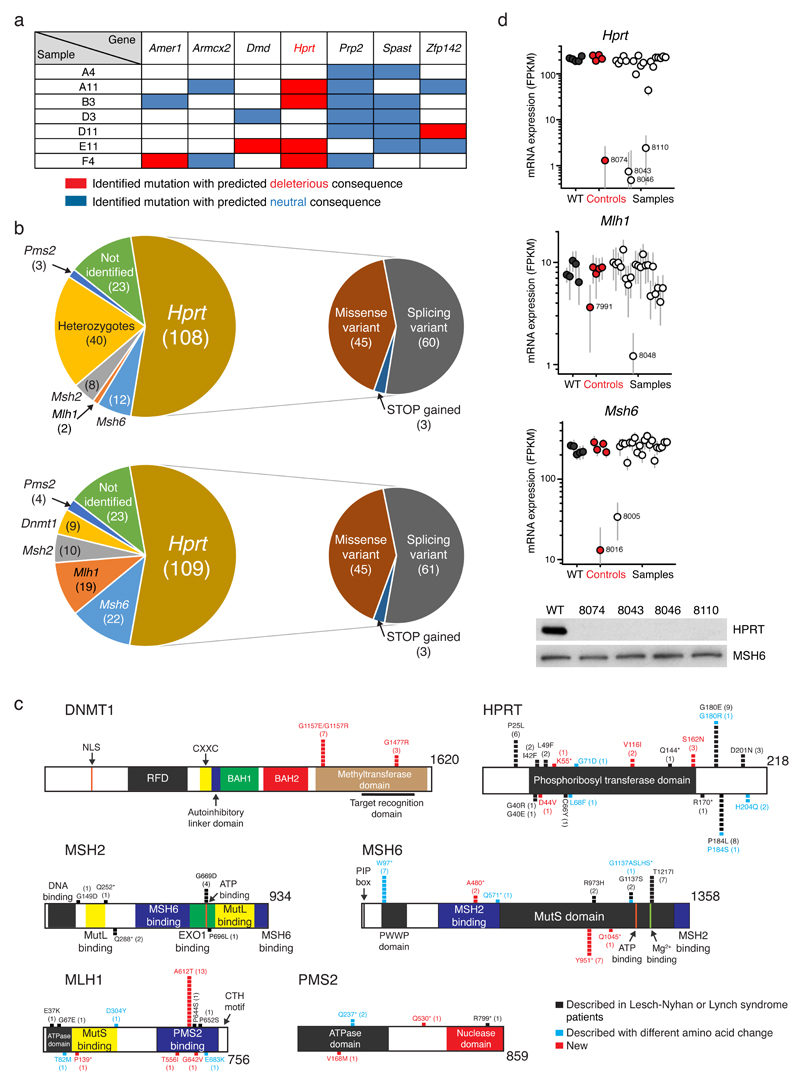
We then identified candidate suppressor genes by analyzing this set of non-synonymous mutations. We defined suppressor gene candidates as those being mutated in multiple independent clones and harboring multiple potential deleterious mutations as assigned by prediction software (see Methods and Supplementary Data Set 1). Hprt, the gene encoding the sole 6-TG target10 (Fig. 1b), was mutated in five of the seven sequenced clones (Supplementary Data Set 1). Moreover, it was the only gene mutated in multiple clones that carried likely deleterious mutations in all cases (Fig. 2a). Furthermore, these Hprt mutations affected different residues of the coding sequence (Supplementary Data Set 1). By contrast, only three non-synonymous mutations in other genes mutated in more than one clone were predicted to be deleterious, and no other gene contained a likely deleterious mutation in more than one clone (Fig. 2a, Supplementary Data Set 1). This analysis established that, without using any previous knowledge regarding the nature of suppressor loci, sequencing just a few clones identified Hprt as the top suppressor-gene candidate.
Figure 2. Identification of suppressor mutations.
(a) Genes identified through whole-exome sequencing of seven 6-TG resistant clones, harboring at least two independent mutations in different clones. Mutations were assigned as deleterious or neutral according to PROVEAN and SIFT software (see Methods). (b) Top panels. Distribution of mutations identified in suppressor gene candidates; numbers of independent clones are in brackets and types of Hprt mutations are shown in more detail on the pie-chart to the right. Bottom panels. Distribution of all suppressor gene candidate mutations identified, including heterozygous deleterious mutations. (c) Distribution of point mutations on DNMT1, HPRT and MMR proteins; each square represents an independent clone. Asterisks (*) denote STOP-codon gains. Except for HPRT, all proteins are shown at a proportional scale. (d) Hprt, Mlh1 and Msh6 mRNA expression levels (fragments per kilobase per million reads). Numbers next to dots are clone identifiers (see Supplementary Data Set 2). Black dots indicate wild-type (WT) samples, red dots represent clonal samples whose mutations were identified via targeted exon capture sequencing (controls; see Supplementary Data Set 2), and white dots represent samples for which no causative mutations were identified. Error bars represent uncertainties on expression estimates. Lower panel. Reduced Hprt mRNA levels correspond to reduced protein production as detected by western blot. Uncut gel images are available in Supplementary Fig. 7.
In addition to HPRT inactivation, mutations in genes for DNA mismatch repair (MMR) proteins confer 6-TG resistance11, as does inactivation of the DNA methyltransferase DNMT112. Notably, the two whole-exome sequenced clones that did not carry Hprt mutations contained nonsense mutations in MMR genes (Supplementary Data Set 1, Supplementary Fig. 1d). To further analyze coverage of our mutant libraries, we subjected the 189 additional suppressor clones we retrieved to targeted exon sequencing of the six known suppressor genes (Fig. 1b). With the exception of Dnmt1 (see below), we identified predicted deleterious mutations in all known suppressor genes in homozygosis in two or more resistant clones (Fig. 2b top panels, Supplementary Data Set 2). Importantly, introducing wild-type versions of Hprt or Mlh1 into resistant clones containing mutations in these genes restored 6-TG sensitivity (Supplementary Figure 2), confirming them as phenotypic drivers. Thus, if the non-targeted whole-exome sequence approach that we carried out in the initial analysis of seven clones had been applied to all 196 suppressor clones, Hprt, Msh2, Msh6, Mlh1 and Pms2 would have been identified as suppressor gene candidates, confirming the feasibility of the approach to identify most or all resistance loci.
Interestingly, ~20% (40) of clones presented two or more heterozygous deleterious mutations in the same suppressor gene (Supplementary Data Set 2). We note that haploid cell cultures cannot be maintained indefinitely and become diploid over time8,13. Accordingly, identified heterozygous mutations could have arisen after diploidization of the original EMS-treated haploid populations, or could have occurred in the small proportion of diploid H129-3 cells in the EMS-treated enriched haploid populations (Fig. 1a). Regardless of their origin, deleterious heterozygous mutations could only generate 6-TG resistance if each affected one allele of the gene, effectively inactivating both copies. Heterozygous mutations that we observed in Dnmt1 occurred in such close proximity that they could be analyzed from the same sequencing reads. As we observed no co-occurrence in the same reads (Supplementary Fig. 3a), we concluded that Dnmt1 mutants were compound heterozygotes, and confirmed this through Sanger sequencing (Supplementary Fig. 3b). Furthermore, as these mutations all scored as potentially deleterious for DNMT1 protein function (Supplementary Data Set 2), it is likely that they caused 6-TG resistance (see below). Dnmt1 would thus be included in the list of suppressor gene candidates when considering deleterious heterozygous mutations. Furthermore, this analysis increased the numbers of clones identified with mutations in other suppressor loci (Fig. 2b, lower panels).
Highlighting the applicability of our methodology to identify functionally important protein regions, we retrieved variants linked to Hprt mutations causative of Lesch-Nyhan syndrome14, as well as mutations in MMR genes linked to Lynch syndrome15 (Fig. 2c). Partially reflecting the mutational preferences of EMS (see below), we found mRNA splicing variant mutations potentially affecting total protein levels (Supplementary Data Set 2). These were particularly prevalent in Hprt (Fig. 2b), and a detailed analysis confirmed their impacts on reducing HPRT protein levels (Supplementary Figure 4). These results highlight how production of aberrant mRNA splicing and associated reduction of protein product is an important consequence of EMS mutagenesis.
We also identified mutations that had not been previously reported, the majority of which were predicted to have deleterious effects on protein function (Supplementary Fig. 5a, Supplementary Data Set 2). To verify their impacts, we introduced newly identified MLH1 (A612T) and DNMT1 (G1157E) mutations into wild-type mESCs by CRISPR/Cas9 gene editing (Supplementary Fig. 5b,c). H129-3 mESCs carrying these mutations were more resistant to 6-TG than their wild-type counterparts (Supplementary Fig. 5d), supporting these mutations being causative of the suppressor phenotype. mESCs carrying targeted mutations in Dnmt1 and Mlh1 also allowed examination of their effects on cell proliferation. As observed under non-selective conditions, mutations in Mlh1, and especially in Dnmt1, impaired cell proliferation (Supplementary Fig. 5e), potentially helping to explain the low proportion of Dnmt1 mutant suppressors arising from our screen. DNMT1-deficient cells exhibit 6-TG resistance, but the mechanism for this is not completely understood12,16. Our results point to an important role of Dnmt1 methyltransferase activity in mediating 6-TG sensitivity, as suppressor mutations identified in our screen localized to that domain (Fig. 2c). Collectively, these results further validated our pipeline to identify suppressor mutations.
Around 12% of resistant clones (23) did not present mutations in any of the known suppressor genes (Fig. 2b). We subjected these clones to whole-exome DNA and RNA sequencing. DNA sequencing of the unassigned clones and control samples allowed an unprecedented description of EMS mutagenic action, confirming its preference for producing SNVs and transition rather than transversion mutations (Supplementary Fig. 6). Although whole-exome sequencing retrieved causative mutations in all control 6-TG resistant samples, no other gene candidate could be identified from the remaining orphan suppressors (Supplementary Data Set 3). RNA sequencing, however, revealed reduced expression levels of Hprt, Mlh1 or Msh6 as likely causes of suppression in several such clones (Fig. 2d; Supplementary Data Set 4). Further studies will be required to define whether epigenetic alterations or mutations outside of exon regions, and hence not covered by exome-targeted DNA sequencing, could explain the nature of remaining orphan suppressor clones.
Collectively, our findings establish that classical genetic screening can be effectively performed in mammalian systems by combining use of haploid cells, chemical SNV induction, and next-generation sequencing. The use of haploid cells when creating SNV mutant libraries identifies recessive suppressor point-mutations, in contrast to diploid cell screening where only dominant mutations are retrieved17. Furthermore, EMS induction of SNVs generates complex mutant libraries, increasing the probability of identifying suppressor loci compared to isolation of rare, spontaneous suppressor events18. Through screening for cellular resistance to 6-TG, we identified point mutations in all described suppressor genes. This highlights the power of our approach to comprehensively identify suppressor loci with low error rates, as no false positive suppressor candidate genes were found. Moreover, as we have established for 6-TG suppressor loci, our methodology has value in delineating key amino-acid residues required for protein function, thus helping to explain molecular mechanisms of suppression. We note that SNV-based mutagenesis will be useful to identify separation-of-function and gain-of-function mutations, including those in essential genes. Also, through studies performed in cells bearing mutations in another gene, our approach has the potential to investigate gene-gene interactions in a comprehensive manner. In addition, we envisage the applicability of this approach in human haploid cells19,20. Chemical mutagenesis of haploid cells, either alone or in combination with LOF screens, has the potential to bring functional genomics in mammalian systems to a hitherto unachieved comprehensive level.
Online Methods
Cell lines and culture conditions
H129-3 haploid mouse embryonic stem cells (mESCs)8 were used for the experiments described in this paper. When pure haploid content was required, cells were grown in chemically defined 2i medium plus LIF as described previously8. In all other cases, cells were grown in DMEM high glucose (Sigma) supplemented with glutamine, streptomycin, penicillin, non-essential amino acids, sodium pyruvate, β-mercaptoethanol and LIF. All plates and flasks were gelatinized prior to cell seeding. All cells used in this study were mycoplasma free.
Cell sorting
Cell sorting for DNA content was performed after staining with 15 μg ml−1 Hoechst 33342 (Invitrogen) on a MoFlo flow sorter (Beckman Coulter). The haploid 1n peak was purified. Analytic flow profiles of DNA content were recorded after fixation of the cells in ethanol, RNase digestion and staining with propidium iodide (PI) on a Fortessa analyzer (BD Biosciences). Cell cycle profiles were produced using FlowJo software (Tree Star).
Ethylmethanesulfonate (EMS) treatment
Mutagenesis with EMS and measurement of killing and suppression frequency was performed as described previously9, with the following modifications. After cell sorting, haploid cells were grown in 2i medium plus LIF and changed to DMEM plus LIF for the overnight EMS treatment. After EMS treatment, cells were cultured for 5 passages in DMEM plus LIF and plated into 6-well plates at a density of 5 x 105 cells per well. Cells were treated with 2 μM 6-thioguanine (6-TG; Sigma) for 6 days, supplying new media with drug daily. Cells were then grown in medium without 6-TG until mESC colonies could be picked.
DNA isolation and exome sequencing
mESC clones were grown into 12-well plates. Genomic DNA was extracted from confluent wells using QIAamp DNA Blood Mini Kit (QIAGEN) and cleaned performing a proteinase K (QIAGEN) digestion step. Genomic DNA (approximately 1 μg) was fragmented to an average size of 150 bp and subjected to DNA library creation using established Illumina paired-end protocols. Adapter-ligated libraries were amplified and indexed via PCR. A portion of each library was used to create an equimolar pool comprising 8 indexed libraries. For whole-exome sequencing, each pool was hybridized to SureSelect RNA baits (Mouse_all_exon; Agilent Technologies). Whole-exome sequencing was performed with 8 DNA samples per sequencing lane (first 7 suppressors plus control) or 15 DNA samples per sequencing lane (subsequent 66 suppressors analysed). For the exon-capture experiment, samples were hybridized with a specific array of RNA baits (Agilent) covering the exonic sequences of Dnmt1, Hprt, Mlh1, Mlh3, Msh2, Msh3, Msh4, Msh5, Msh6, Pms1, Pms2 and Setd2 genes. Sequence targets were captured and amplified in accordance with manufacturer’s recommendations. Enriched libraries were subjected to 75 base paired-end sequencing (HiSeq 2500; Illumina) following manufacturer’s instructions. A single sequencing library was created for each sample, and the sequencing coverage per targeted base per sample is given in Supplementary Data Set 5. All raw sequencing data is available from ENA under accession numbers ERP003577 and ERP005179.
DNA sequence analysis
Sequencing reads were aligned to the Mus musculus GRCm38 (mm10) assembly (Ensembl version release 68) using BWA (v0.5.10-tpx). All lanes from the same library were merged into a single BAM file with Picard tools (http://broadinstitute.github.io/picard), and PCR duplicates were marked by using ‘MarkDuplicates’21. SNVs and INDELs were called using SAMtools (v1.3) mpileup followed by BCFtools (v1.3)22. The following parameters were used for Samtools mpileup: -g -t DP,AD -C50 -pm3 -F0.2 -d10000. BCFtools call parameters were: -vm -f GQ. The variants were annotated using the Ensembl Variant Effect Predictor23. Variants were filtered to remove any variants detected outside the bait regions and any heterozygous variants where appropriate. Additionally, variants were filtered using VCFtools (v0.1.12b) vcf-annotate with options -H -f +/q=25/SnpGap=7/d=5 and custom filters were written to exclude variants with a GQ score of less than 10 24. INDELs were left aligned using BCFtools norm. VCFtools vcf-isec was used to remove variants present in the control sample from all other samples as well as variants present in sequencing of a mouse strain from the 129S5 background25. INDELs called from whole exome sequencing data were further verified using the microassembly based caller Scalpel26 and discarded from the data if not identified by both callers. All remaining variants were used to generate a visualization of mutational patterns. All SNVs were assigned to one of 96 possible triplet channels using the GRCm38 assembly to identify flanking bases.
Antibodies
Rabbit anti-HPRT (Abcam ab10479, 1: 10 000 dilution), mouse anti-MSH6 (BD Biosciences 610919, 1: 2 000), mouse anti-PMS2 (BD Biosciences 556415, 1: 1 000), rabbit anti-MRE11 (Abcam ab33125, 1: 10 000) and mouse anti-MLH1 (BD Biosciences 554073, 1: 1 000) were used for western blot analysis.
Complementation assays
Human MLH1 was amplified from pEGFP-MLH127 and cloned into pPB-CMV-HA-pA-IN28 using EcoRI and MluI sites to generate pPB-Tet-MLH1. Cells from the SC_6TG5758127 Mlh1 mutant clone (see Supplementary Data Set 2) were transfected with a combination of pCMV-HyPBase29, pPB-CAG-rtTAM2-IP (a derivative of pPBCAG-rtTAIRESNeo28 where the neomycin resistance cassette was replaced by a puromycin resistance one, gift from J. Hackett) and pPB-CMV-HA-pA-IN or pPB-Tet-MLH1, in a 1:1:10 ratio using TransIT-LT1 transfection reagent (Mirus) and following manufacturer’s instructions. 48 h after transfection, selection was applied with 3 μg/ml puromycin for 6 days. Resistant cell populations were plated into 6-well plates (125 000 cells per well) and MLH1 expression was induced by the addition of 1 μg/ml doxycycline. 24 h after doxycycline induction, cells were left untreated or treated with 2 μg/ml 6-TG for 6 days. Surviving cells were stained using crystal violet.
Cells from SC_6TG5758069 and SC_6TG5758117 Hprt mutant clones (see Supplementary Data Set 2) were transfected with pEGFP-C1 (Clontech) or pCMV6-AC-Hprt-GFP (OriGene MG202453) using TransIT-LT1 transfection reagent (Mirus) and following manufacturer’s instructions. 48 h after transfection, selection was applied with 175 μg/ml G418 for several days, until GFP-positive colonies were picked. Cells were left untreated or treated with 2 μg/ml 6-TG for 6 days. Surviving cells were stained using crystal violet. Microscopy images were obtained from an Olympus IX71 microscope using CellF imaging software (Olympus).
Prediction of mutation consequences on protein function
Amino acid mutations were analysed using PROVEAN30 and SIFT31 software. Scores below -2.5 for PROVEAN and 0.05 for SIFT indicate likely deleterious effects.
Sanger sequencing
PCR amplifications from genomic DNA were performed using the following oligonucleotides: Dnmt1-1157F 5'- CGAGATGCCTGGTAGACACA -3', Dnmt1 1157R 5'- GAGTAGGCCTGAGGAGAGCA -3', Dnmt1 1477F 5'- GCTACAAAACCCCAGGAAGC -3', Dnmt1 1477R 5'- CAGGATCAGATTGGCGTGAC -3'. PCR products from SC_6TG5758159 and SC_6TG5758161 Dnmt1 mutant suppressors (see Supplementary Data Set 2) were cloned using Zero Blunt TOPO PCR cloning kit (Thermo Fisher Scientific) and following manufacturer’s instructions.
Gene editing
Sequences for DNA templates for small guide RNAs were generated using CRISPR Design (http://crispr.mit.edu) and cloned into pAiO-Cas9 D10A32. Sequences of the guides were the following: Dnmt1-1 5'- TCGGAAGGATTCCACCAAGC -3', Dnmt1-2 5'-ACATCCAGGGTCCGGAGCTT -3', Mlh1-1 5'- AGGACGACGGCCCGAAGGAA -3'; Mlh1-2 5'- GCCACTTTCAGGACTGTCTA -3'. H129-3 cells were transfected with Dnmt1 or Mlh1 targeting plasmids and single-stranded DNA oligonucleotides (200 nt, IDT Technologies) containing the desired mutations using TransIT-LT1 transfection reagent (Mirus) and following manufacturer’s instructions. 48 h after transfection GFP-positive cells were sorted on a MoFlo flow sorter (Beckman Coulter) and seeded into gelatinized plates. Colonies forming after 5-6 days were picked into 96-well plates, DNA was isolated using QuickExtract DNA extraction solution (Epicentre Biotechnologies) and PCR amplifications of the edited regions were performed. Sequences of the oligonucleotides used were as follows: Dnmt1-F 5'-CGAGATGCCTGGTAGACACA -3', Dnmt1-R 5'- GAGTAGGCCTGAGGAGAGCA -3', Mlh1-F 5'- TGTCCCAACCTAGGGACTTG -3', Mlh1-R 5'- TGCTGGCCTTAGACAGTCCT -3'. PCR products (358 bp for Dnmt1, 287 bp for Mlh1) were digested with EcoRI restriction enzyme and run on a 3% agarose 1xTAE gel for 1.5 h at 150 V. Positive clones (those producing two DNA fragments after EcoRI digestion of approx. 180 bp (Dnmt1) or 200 and 80 bp (Mlh1)) were confirmed by Sanger sequencing of the PCR products, and tested for resistance to 6-TG as described for the screen.
Population doublings
Each cell line was seeded in duplicate into 2 rows of a 24-well plate at a density of 25 000 cells/well. Cells were collected daily and cell counts were measured using a Countess II Automated Cell Counter (ThermoFisher Scientific) using Trypan Blue staining to discard dead cells.
RNA isolation and sequencing
mESC clones were grown in 24-well plates. Total RNA was extracted from confluent wells using RNeasy Mini Kit (QIAGEN). Libraries for RNA-seq were prepared from 500 ng total RNA using the QuantSeq 3' mRNA-Seq kit (Lexogen) according to manufacturer's instructions. An exception to the instruction was the application of 13 instead of the recommended 12 PCR cycles for library amplification. Libraries were pooled in equal concentrations. Prior to sequencing, a T-fill reaction was performed on a cBot as described previously33, providing the T-fill solution in a primer tube strip. Finally, sequencing was carried out using an Illumina Hiseq-2500 using 50 bp single read v3 chemistry. Raw sequencing data is available from ENA under accession number ERP014134.
RNA sequence analysis
Reads were trimmed of adapter sequences using Cutadapt (v.1.2.1). High-quality reads were extracted using TriageTools34 (v0.2.2, long reads –length 35, high-quality bases –quality 9, and complex sequences –lzw 0.33). Alignments onto the mm10 genome were carried out using GSNAP35 (v2014-02-28) with Gencode gene splice junctions. Expression levels were obtained using Exp3p (github.com/tkonopka/Exp3p v0.1) and then processed with custom R scripts (Supplementary Data Set 6).
Statistical analyses
All groups analysed showed comparable variances.
Supplementary Material
Acknowledgements
We thank all S.P.J. lab members for discussions, especially A. Blackford, F. Puddu, C. Schmidt and P. Marco-Casanova for critical reading of the manuscript, and C. Le Sage and T.-W. Chiang for advice with CRISPR/Cas9 gene editing. We thank M. Leeb for H129-3 cells and advice on haploid ES cell culture conditions, and J. Hackett for advice in generating stable ES cell lines. We thank C.D. Robles-Espinoza for helping designing the array of baits for the exon-capture experiment, and J. Hewinson for technical support. Research in the S.P.J. laboratory is funded by Cancer Research UK (CRUK; programme grant C6/A11224), the European Research Council and the European Community Seventh Framework Programme (grant agreement no. HEALTH-F2-2010-259893; DDResponse). Core funding is provided by Cancer Research UK (C6946/A14492) and the Wellcome Trust (WT092096). S.P.J. receives salary from the University of Cambridge, supplemented by CRUK. J.V.F. was funded by Cancer Research UK programme grant C6/A11224 and the Ataxia Telangiectasia Society. J.C. was funded by Cancer Research UK programme grant C6/A11224. D.J.A. is supported by CRUK. Research leading to these results has received funding from the European Research Council under the European Union's Seventh Framework Programme (FP7/2007-2013)/ERC grant agreement no. [311166]. B.V.G. is supported by a Boehringer Ingelheim Fonds PhD fellowship.
Footnotes
Author contributions
J.V.F. and S.P.J. designed the project. J.V.F mutagenized haploid cells, performed 6-TG selection and isolated suppressor clones. J.V.F. and J.C. expanded suppressor clones, isolated gDNA and prepared samples for sequencing. M.H. analyzed DNA sequencing data, supervised by T.M.K. and D.J.A. J.V.F. and J.C. produced stable cell lines and CRISPR/Cas9 knock-ins. J.V.F. and J.C. isolated RNA from suppressor clones and prepared samples for sequencing. B.V.G. produced RNA sequencing libraries and T.K. analyzed RNA sequencing data, supervised by S.M.N. J.V.F and S.P.J wrote the manuscript, with input from all authors.
Accession codes
DNA sequencing data is available from ENA under accession numbers ERP003577 and ERP005179. RNA sequencing data is available from ENA under accession number ERP014134.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Forsburg SL. The art and design of genetic screens: yeast. Nat Rev Genet. 2001;2:659–668. doi: 10.1038/35088500. [DOI] [PubMed] [Google Scholar]
- 2.Johnston D. The art and design of genetic screens: Drosophila melanogaster. Nat Rev Genet. 2002;3:176–188. doi: 10.1038/nrg751. [DOI] [PubMed] [Google Scholar]
- 3.Boutros M, Ahringer J. The art and design of genetic screens: RNA interference. Nat Rev Genet. 2008;9:554–566. doi: 10.1038/nrg2364. [DOI] [PubMed] [Google Scholar]
- 4.Carette JE, et al. Haploid genetic screens in human cells identify host factors used by pathogens. Science. 2009;326:1231–1235. doi: 10.1126/science.1178955. [DOI] [PubMed] [Google Scholar]
- 5.Koike-Yusa H, Li Y, Tan E-P, Velasco-Herrera MDC, Yusa K. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat Biotechnol. 2014;32:267–273. doi: 10.1038/nbt.2800. [DOI] [PubMed] [Google Scholar]
- 6.Shalem O, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rolef Ben-Shahar T, et al. Eco1-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science. 2008;321:563–566. doi: 10.1126/science.1157774. [DOI] [PubMed] [Google Scholar]
- 8.Leeb M, Wutz A. Derivation of haploid embryonic stem cells from mouse embryos. Nature. 2011;479:131–134. doi: 10.1038/nature10448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Munroe RR, Schimenti JJ. Mutagenesis of mouse embryonic stem cells with ethylmethanesulfonate. Methods Mol Biol. 2009;530:131–138. doi: 10.1007/978-1-59745-471-1_7. [DOI] [PubMed] [Google Scholar]
- 10.LePage GA, Jones M. Purinethiols as feedback inhibitors of purine synthesis in ascites tumor cells. Cancer Res. 1961;21:642–649. [PubMed] [Google Scholar]
- 11.Swann PF, et al. Role of postreplicative DNA mismatch repair in the cytotoxic action of thioguanine. Science. 1996;273:1109–1111. doi: 10.1126/science.273.5278.1109. [DOI] [PubMed] [Google Scholar]
- 12.Guo G, Wang W, Bradley A. Mismatch repair genes identified using genetic screens in Blm-deficient embryonic stem cells. Nature. 2004;429:891–895. doi: 10.1038/nature02653. [DOI] [PubMed] [Google Scholar]
- 13.Elling U, et al. Forward and reverse genetics through derivation of haploid mouse embryonic stem cells. Cell Stem Cell. 2011;9:563–574. doi: 10.1016/j.stem.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jinnah HA, De Gregorio L, Harris JC, Nyhan WL, O'Neill JP. The spectrum of inherited mutations causing HPRT deficiency: 75 new cases and a review of 196 previously reported cases. Mutat Res. 2000;463:309–326. doi: 10.1016/s1383-5742(00)00052-1. [DOI] [PubMed] [Google Scholar]
- 15.Jiricny J. Postreplicative mismatch repair. Cold Spring Harb Perspect Biol. 2013;5:a012633. doi: 10.1101/cshperspect.a012633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loughery JEP, et al. DNMT1 deficiency triggers mismatch repair defects in human cells through depletion of repair protein levels in a process involving the DNA damage response. Hum Mol Genet. 2011;20:3241–3255. doi: 10.1093/hmg/ddr236. [DOI] [PubMed] [Google Scholar]
- 17.Kasap C, Elemento O, Kapoor TM. DrugTargetSeqR: a genomics- and CRISPR-Cas9-based method to analyze drug targets. Nat Chem Biol. 2014;10:626–628. doi: 10.1038/nchembio.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smurnyy Y, et al. DNA sequencing and CRISPR-Cas9 gene editing for target validation in mammalian cells. Nat Chem Biol. 2014;10:623–625. doi: 10.1038/nchembio.1550. [DOI] [PubMed] [Google Scholar]
- 19.Blomen VA, et al. Gene essentiality and synthetic lethality in haploid human cells. Science. 2015;350:1092–1096. doi: 10.1126/science.aac7557. [DOI] [PubMed] [Google Scholar]
- 20.Sagi I, et al. Derivation and differentiation of haploid human embryonic stem cells. Nature. 2016;532:107–111. doi: 10.1038/nature17408. [DOI] [PubMed] [Google Scholar]
- 21.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McLaren W, et al. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26:2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Danecek P, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keane TM, et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477:289–294. doi: 10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Narzisi G, et al. Accurate de novo and transmitted indel detection in exome-capture data using microassembly. Nat Methods. 2014;11:1033–1036. doi: 10.1038/nmeth.3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hong Z, et al. Recruitment of mismatch repair proteins to the site of DNA damage in human cells. J Cell Sci. 2008;121:3146–3154. doi: 10.1242/jcs.026393. [DOI] [PubMed] [Google Scholar]
- 28.Murakami K, et al. NANOG alone induces germ cells in primed epiblast in vitro by activation of enhancers. Nature. 2016;529:403–407. doi: 10.1038/nature16480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yusa K, Zhou L, Li MA, Bradley A, Craig NL. A hyperactive piggyBac transposase for mammalian applications. Proc Natl Acad Sci USA. 2011;108:1531–1536. doi: 10.1073/pnas.1008322108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choi Y, Chan AP. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics. 2015;31:2745–2747. doi: 10.1093/bioinformatics/btv195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 32.Chiang T-WW, le Sage C, Larrieu D, Demir M, Jackson SP. CRISPR-Cas9(D10A) nickase-based genotypic and phenotypic screening to enhance genome editing. Sci Rep. 2016;6:24356. doi: 10.1038/srep24356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilkening S, et al. An efficient method for genome-wide polyadenylation site mapping and RNA quantification. Nucleic Acids Res. 2013;41:e65–e65. doi: 10.1093/nar/gks1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fimereli D, Detours V, Konopka T. TriageTools: tools for partitioning and prioritizing analysis of high-throughput sequencing data. Nucleic Acids Res. 2013;41:e86–e86. doi: 10.1093/nar/gkt094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010;26:873–881. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.