Abstract
The cellular sources of interleukin-6 (IL-6) that are relevant for the differentiation of TH17 cells remain unclear. Here, we used a novel strategy of IL-6 conditional deletion of distinct IL-6-producing cell types to show that Sirpα+ dendritic cells (DC) were essential for the generation of pathogenic TH17 cells. During the process of cognate interaction, Sirpα+ DCs trans-presented IL-6 to T cells using their own IL-6Rα. While ambient IL-6 was sufficient to suppress the induction of the transcription factor Foxp3 in T cells, IL-6 trans-presentation by DC-bound IL-6Rα (here defined as IL-6 cluster signaling) was required to prevent premature induction of IFN-γ in T cells and to generate pathogenic TH17 cells in vivo. These findings will guide therapeutic approaches for TH17-mediated autoimmune diseases.
During antigen-specific priming, CD4+ T helper (TH) cells differentiate into distinct subsets characterized by specific master transcription factors and signature cytokines. The differentiation process is controlled by various cytokines present in the micro-environment, in which CD4+ T cells cognately interact with antigen presenting cells. Because T cell receptor (TCR) stimulation in the presence of the ubiquitously expressed cytokine TGF-β results in induction of the transcription factor Foxp3, productive effector T cell responses require efficient ways to suppress Foxp3 induction in T cells during priming. Although interleukin 27 (IL-27) has additional regulatory functions 1, IL-27 and IL-4 are strong inhibitors of Foxp3 induction during the development of TH1 cells and TH2 cells, respectively 2–4. During TH17 cell development, IL-6 prevents the transcription of Foxp3 and at the same time induces IL-17 5,6. However, it is unclear whether these functions of IL-6 are connected with each other or are independent events.
TH17 cells are categorized into pathogenic vs non-pathogenic depending on whether or not they have sensed IL-23 7,8. However, single cell analysis of TH17 cells isolated from the inflamed CNS reveals that individual TH17 cells can exibit a non-pathogenic gene signature including transcription factors (Eomes, Irf8, c-Maf), cytokines (Il24, Il9) and surface receptors (Cxcr6 and Cd96) although they express Il23r 9. Thus, the decision of whether a TH17 cell will become pathogenic can be taken independently of IL-23 and might be installed early during priming.
Because IL-6 is the dominant factor to initiate the transcriptional program of TH17 cells, we speculated that intrinsic properties of IL-6 might be a major determinant of priming pathogenic T cells. In order to signal into target cells, IL-6 first binds to the IL-6Rα subunit. This complex then associates with gp130, the signaling subunit of the IL-6 receptor, resulting in a heterohexameric signaling complex (IL-6, IL-6Rα, gp130 in a 2:2:2 stoichiometry) that triggers productive IL-6 signaling into the target cell 10,11. In addition to the membrane form, IL-6Rα can be shed and bind IL-6 as a soluble receptor. The IL-6-sIL-6Rα soluble complex associates with gp130 and initiates IL-6 signaling in gp130+ cells. This type of IL-6 signaling is termed IL-6 trans-signaling 12,13. In contrast to classic IL-6 signaling, IL-6 trans-signaling can be blocked by soluble gp130 which acts as a decoy receptor for the soluble IL-6-IL-6Ra complex 14.
Here, in order to investigate whether the cellular source of IL-6 was a determinant of TH17 cell fate, we developed a novel IL-6 reporter strategy that allowed for the IL-6 conditional deletion of distinct cell types. We show that a subset of CD11b+ DCs that are Sirpα+ are indispensable for priming of myelin peptide specific encephalitogenic T cells. Our data indicate that CD11b+Sirpα+ DCs are able to trans-present IL-6 through a complex containing DC-expressed IL-6Rα bound to IL-6 that can interact with gp130 expressed on T cells. We define this mode of IL-6 signaling as ‘cluster signaling’ and propose that imprinting of encephalitogenic properties in effector T cells is dependent on IL-6 cluster signaling, while classic IL-6 signaling through its membrane bound receptor complex is sufficient to suppress the TGF-β-induced expression of Foxp3, but fails to prime pathogenic TH17 cells.
Results
DCs are the relevant source of IL-6 for priming pathogenic TH17 cells
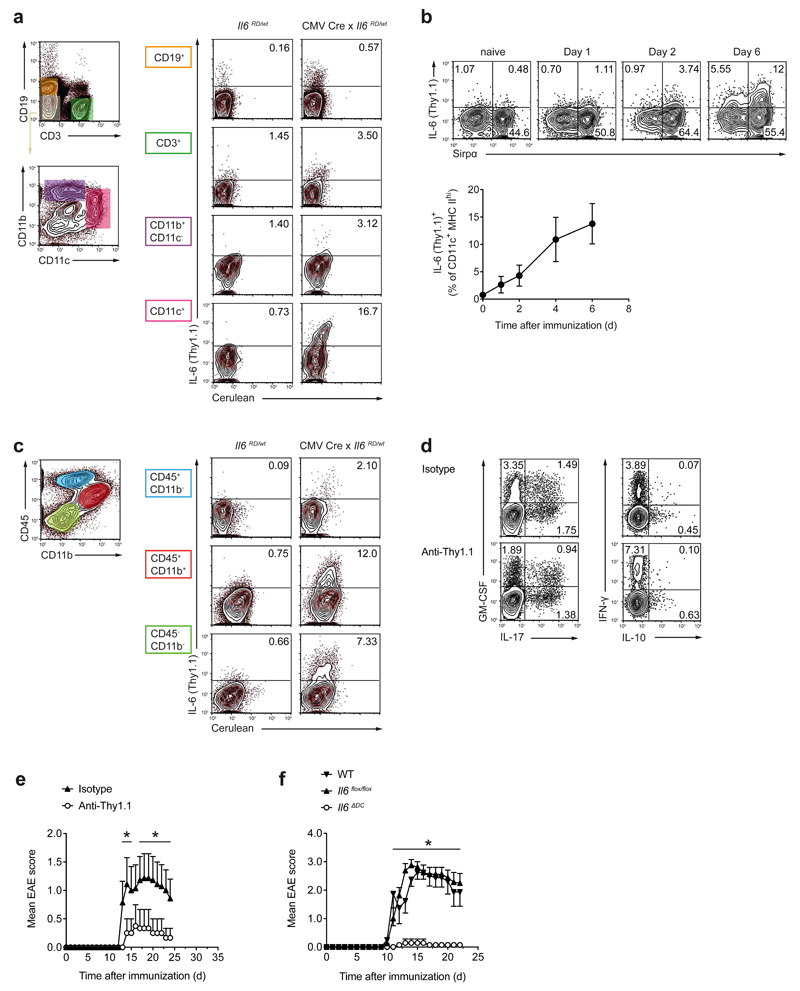
Because IL-6 is produced by different hematopoietic and non-hematopoietic cells, we sought to define the relevant cellular source of IL-6 for the differentiation of pathogenic TH17 cells. We generated an IL-6 reporter knock-in allele (Il6RD), in which IL-6 expression is reported by cerulean and Thy1.1 expression (Supplementary Fig. 1). In addition, the reporter contains a floxed stop cassette that allows a cell type-specific expression of the reporter cassette depending on the Cre driver used in various mouse strains. First, we crossed Il6RD/wt mice with a CMV-Cre deleter strain to allow for unrestricted expression of the IL-6 reporter cassette and immunized these mice with MOG35-55 in CFA. On day 7 after immunization, Thy1.1 (IL-6) was exclusively produced by CD45+ hematopoietic cells in draining lymph node cells and spleen of CMV-Cre x Il6RD/wt mice. CD11c+ cells contained the largest frequencies of Thy1.1 (IL-6)+ cells (Fig. 1a). Subgroup analysis revealed that Thy1.1 (IL-6) expression was restricted to CD11b+Sirpα+CD103-SiglecH- DCs (Supplementary Fig. 2). In draining lymph nodes, some DCs were Thy1.1+ already on the first day after immunization with MOG35-55 in CFA. The subset of Thy1.1+ DCs was maintained at least through day 6 after immunization (Fig. 1b). At the peak of EAE (day 16 post immunization), Thy1.1+ cells in the CNS were mainly CD45+CD11b+ myeloid cells (Fig. 1c). Nevertheless, and in contrast to the peripheral immune compartment, a substantial fraction of IL-6 in the CNS appeared to be produced by non-hematopoietic cells. Importantly, specific ablation of IL-6-producing DCs in CD11c-Cre x Il6RD/wt mice using an anti-Thy1.1 antibody (Supplementary Fig. 2) resulted in the priming of MOG35-55-specific T cells with reduced IL-17 and increased IFN-γ production (Fig. 1d, Supplementary Fig. 3) and abrogated the development of EAE (Fig. 1e). These data suggested that either IL-6 production by DCs or the physical presence of IL-6-producing DCs were required for the induction of EAE. In order to discriminate between these possibilities, we conditionally deleted Il6 in DCs using CD11c-Cre and Il6flox alleles. Loss of Il6 in dendritic cells in CD11c-Cre x Il6flox/flox mice, herein called Il6ΔDC, resulted in complete resistance to EAE, despite the continued presence of CD11b+Sirpα+ DCs in these mice (Fig. 1f). Indeed, Il6ΔDC mice phenocopied Il6-/- mice in their resistance to EAE. Apart from DCs, some Thy1.1 (IL-6) was expressed by T cells, B cells and macrophages (Fig. 1a). Conditional deletion of Il6 in these cells modulated disease severity, but did not abrogate EAE development (Supplementary Fig. 4). Thus, DC-derived IL-6 is essential for priming pathogenic T cell responses in EAE.
Figure 1. IL-6-producing cells during MOG35-55 induced EAE.
Using a novel reporter mouse, IL-6 producing cells were identified by Thy1.1 and cerulean. (a) Control animals or CMV Cre x Il6RD/wt mice were immunized and splenocytes were analyzed on day 7 for IL-6 (Thy1.1) expression in the indicated cell populations (of CD45+ cells) after 4 h ex vivo PMA/ionomycin stimulation. Representative cytograms out of two experiments. (b) Kinetics of IL-6 (Thy1.1) expression in draining lymph node (dLN) DCs of DC conditional IL-6 reporter mice (CD11c Cre x Il6RD/wt x R26 Stopflox/flox YFP) on different days after immunization. DCs were defined as YFP+CD11c+MHC class IIhigh and analyzed for IL-6 (Thy1.1) and Sirpα (CD172a) after 4 h ex vivo stimulation with PMA/ionomycin. Mean ± SD, n=4 (c) IL-6 (Thy1.1)-expressing cells in the CNS at the peak of EAE (day 16) after ex vivo PMA/ionomycin stimulation. Representative cytograms out of two experiments. (d) DC conditional IL-6 reporter mice were immunized followed by treatment with isotype (mouse IgG2a) or anti-Thy1.1 (19E12) to deplete IL-6 (Thy1.1)+ DCs. On day 7, CD4+ T cells from dLN were assessed for cytokine production after ex vivo re-stimulation with PMA/ionomycin. Representative cytograms out of five mice analyzed per group. (e) EAE in control treated or anti-Thy1.1 treated DC conditional IL-6 reporter mice. Representative of two experiments. Mean EAE scores + SEM, n=6. (f) EAE in mice with DC conditional deletion of Il6 (CD11c Cre x Il6flox/flox, Il6ΔDC). Representative of four experiments. Mean EAE scores + SEM, n=7. *P<0.05, ANOVA plus Fisher's LSD test for individual days.
DC derived IL-6 is sensed by interacting T cells in a specific manner
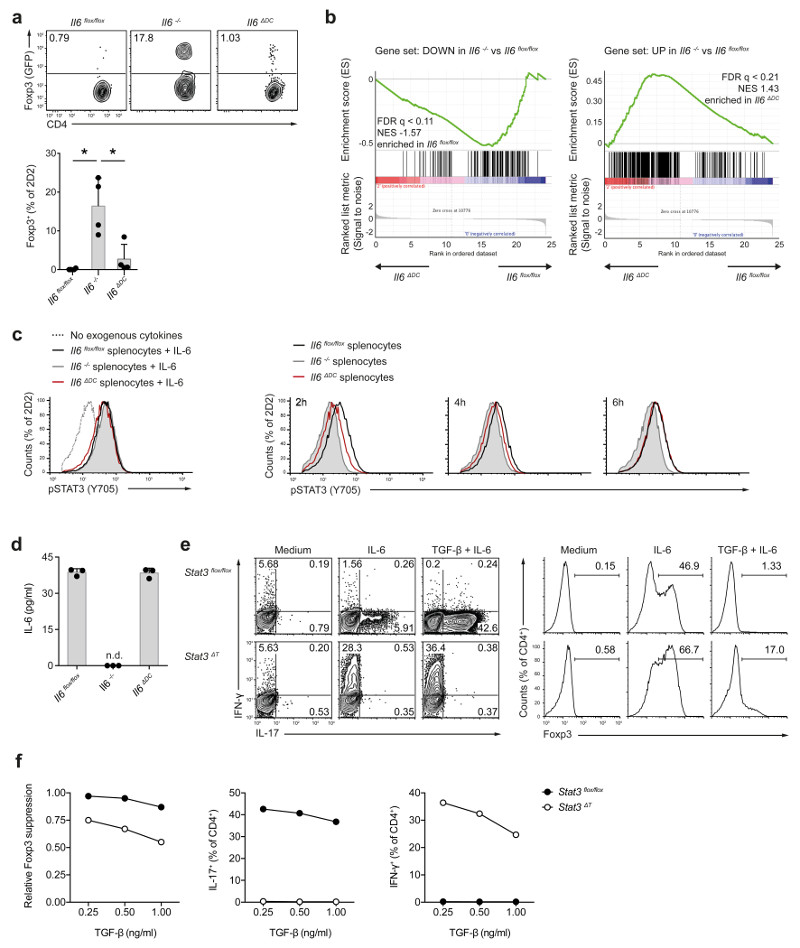
Our data would be consistent with the idea that DC-derived IL-6 acted back on DCs in an autocrine manner to boost their ability to prime pathogenic TH17 cells. However, no major differences in the induction of Il1b, Il12 or Il23 were observed between wild-type and IL-6Rα-deficient BMDCs, which cannot respond to soluble IL-6, upon exposure to exogenous IL-6 (Supplementary Fig. 5). Thus, we explored alternative modes of action of DC-derived IL-6 during cognate interaction with T cells. Naive (Foxp3-) CD4+ T cells from 2D2 x Foxp3gfp.KI mice , which express a MOG35-55-specific transgenic TCR and in which Foxp3 expression is reported by GFP, were transferred into Il6flox/flox control, Il6-/- or Il6ΔDC mice followed by subcutaneous immunization with MOG35-55 in CFA. As previously reported 15, priming of transgenic T cells in an IL-6-deficient environment in the Il6-/- mice resulted in the conversion of about 20% 2D2 T cells into GFP (Foxp3)+ Treg cells (Fig. 2a). In contrast, we did not observe conversion of GFP- 2D2 T cells into GFP+ 2D2 T cells during priming of naive 2D2 T cells in the Il6ΔDC mice, indicating that IL-6 from sources other than DCs was sufficient to suppress the conversion of conventional T cells into Foxp3+ Treg cells during antigen specific priming. Indeed, the systemic amount of IL-6 measured in the serum after subcutaneous immunization with MOG35-55 peptide in CFA was similar in Il6flox/flox and Il6ΔDC animals (Supplementary Fig. 6).
Figure 2. Conditional ablation of Il6 in DCs results in delayed Stat3 activation in antigen specific T cells but does not impair the suppression of Foxp3 induction.
(a) Naive MOG35-55 specific T cells (2D2) were transferred into control hosts (Il6flox/flox), Il6-/- mice, or DC conditional IL-6 deficient mice (Il6ΔDC) followed by immunization with MOG35-55. After 3 weeks, donor cells were re-isolated from dLN and analyzed for Foxp3 expression. Representative of two experiments. Mean + SD, n=4, *P<0.005, ANOVA plus Tukey´s test. (b) RNA seq was performed in re-isolated Foxp3- 2D2 T cells, and IL-6 hallmark genes were analyzed in cells derived from control vs Il6ΔDC host mice. (c) Splenocytes from Il6flox/flox control mice, Il6-/- mice, or Il6ΔDC mice were co-cultured with naive 2D2 T cells in the presence of antigen and Marimastat. In control cultures exogenous recombinant IL-6 was added instead of LPS (left histogram). At several time points after addition of LPS, Stat3 phosphorylation was analyzed in 2D2 T cells. p-Stat3 was delayed in cultures with Il6ΔDC splenocytes compared to control splenocytes (2h) although the amount of soluble IL-6 was similar at this time point (d). Representative of two experiments. Mean + SD, n=3. (e, f) Naive control (Stat3flox/flox) or Stat3 deficient (CD4 Cre x Stat3flox/flox, Stat3ΔT) T cells were activated with anti-CD3/anti-CD28 in the presence of TGF-β plus IL-6. Intracellular cytokine staining after PMA/ionomycin stimulation (e, left) and nuclear staining for Foxp3 (e, right) on day 3 of culture. (f) Relative suppression of Foxp3 and frequencies of cytokine positive cells in control or Stat3ΔT T cells in response to IL-6 (50 ng/ml) and graded amounts of TGF-β. Representative of three experiments.
Although the emergence of Foxp3+ Treg cells was equally suppressed in Il6ΔDC as in IL-6-sufficient (Il6flox/flox) mice, Il6flox/flox mice developed EAE following immunization with MOG35-55 in CFA, while Il6ΔDC mice did not (Fig. 1f). Thus, to search for effector T cell-intrinsic features that would explain the inability of Il6ΔDC primed T cells to induce EAE, we performed RNA sequencing analysis of GFP (Foxp3)- 2D2 T cells that were re-isolated from the draining lymph nodes of Il6flox/flox, Il6-/- or Il6ΔDC mice on day 6 after immunization with MOG35-55 in CFA. The set of genes differentially expressed in 2D2 T cells isolated from Il6-/- vs Il6flox/flox control mice was defined as IL-6 target genes (Supplementary Table 1). Within this set, we identified those genes enriched in 2D2 T cells primed in Il6ΔDC vs Il6flox/flox mice (Fig. 2b, Supplementary Tables 2 and 3) and analyzed upstream pathways compatible with this enrichment by ingenuity pathway analysis (Supplementary Fig. 6). We found that differential activation of Stat3 was the major predictor of the distinct gene enrichment profiles of 2D2 T cells primed in Il6ΔDC vs Il6flox/flox (Supplementary Fig. 6). Moreover, when we directly tested a published Stat3 dependent gene set 16, Stat3 target genes were enriched in 2D2 T cells primed in Il6flox/flox over Il6ΔDC mice (Supplementary Fig. 6 and Supplementary Table 4). Thus, we tested the activation of Stat3 in T cells in Il6flox/flox, Il6-/- or Il6ΔDC mice. Naive 2D2 T cells were co-cultured with Il6flox/flox, Il6-/- or Il6ΔDC splenocytes in the presence of cognate antigen (MOG35-55). After LPS stimulation, Stat3 activation was negligible in T cells cultured with Il6-/- APCs. Stat3 activation was delayed in T cells primed with Il6ΔDC APCs compared to wild-type APC-primed T cells (Fig. 2c) despite similar amounts of soluble IL-6 in the supernatant of wild-type APC-T cell and Il6ΔDC APC-T cell co-cultures (Fig. 2d). Thus, IL-6 induced an early and robust activation of Stat3 in antigen-specific 2D2 T cells only when provided by DCs.
In addition, because the in vivo priming of 2D2 T cells in Il6ΔDC mice resulted in a highly efficient suppression of Foxp3 induction (Fig. 2a), we investigated whether activation of Stat3 had differential effects on the IL-6-mediated suppression of Foxp3 expression and the induction of effector properties in conventional T cells. Naive CD44-CD25-CD4+ T cells from Stat3flox/flox control mice or CD4-Cre x Stat3flox/flox mice (Stat3ΔT) were activated in vitro with anti-CD3 and anti-CD28 in the presence of TGF-β without and with IL-6 followed by assessment of Foxp3 induction and effector cytokines. As expected, Stat3 was indispensable for the induction of IL-17 (Fig. 2e). However, Stat3 deficient T cells produced high amounts of IFN-γ when differentiated under TH17 conditions (Fig. 2e, f). The TGF-β-induced expression of Foxp3 was (at least in part) suppressed by IL-6 in Stat3 deficient T cells (Fig. 2e, f), indicating that lack of Stat3 in T cells resulted in a TH1-like phenotype under TH17 differentiation conditions. In summary, these data suggest that T cells can distinguish between DC-derived IL-6 and IL-6 from other sources due to different Stat3 activation kinetics translating into different T cell phenotypes.
IL-6 cluster signaling is an efficient signaling mode
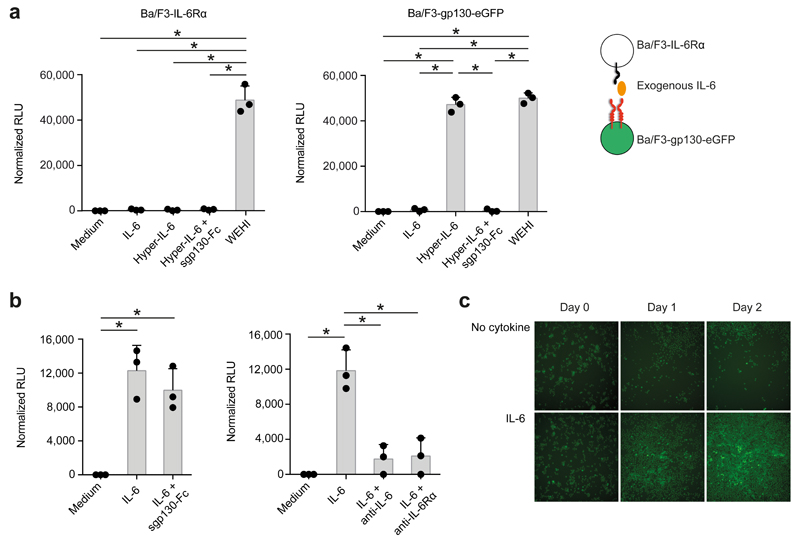
Besides classic IL-6 signaling through soluble IL-6 (sIL-6) binding to its membrane-bound receptor, soluble IL-6Rα-IL-6 complexes engage gp130 on target cells that lack IL-6Rα, a process called IL-6 trans-signaling 13. We tested whether during a cognate DC-T cell encounter, DC-bound IL-6Rα trans-presents DC-derived IL-6 to T cells through a distinct IL-6 trans-presentation process, which requires the clustering of donating and receiving cell - hence named here IL-6 cluster signaling. First, we established the functionality of IL-6 cluster signaling by retrovirally expressing either IL-6Rα (without eGFP) or gp130-eGFP in Ba/F3 cells, an IL-3-dependent murine pro-B-cell line that lacks both endogenous IL-6Rα and gp130 expression 17,18, followed by testing their proliferative response to IL-6 signaling (Fig. 3a). While in separate cultures, Ba/F3-IL-6Rα cells, which lacked gp130, did not proliferate in response to either sIL-6 or a sIL-6-soluble IL-6Rα complex (hereafter hyper-IL-6 19) (Fig. 3a), Ba/F3-gp130-eGFP cells, which lacked IL-6Rα, proliferated in response to hyper-IL-6, but not sIL-6 (Fig. 3a), suggesting that hyper-IL-6 formed a functional signaling complex with membrane bound gp130-eGFP in Ba/F3-gp130-eGFP cells. Notably, when Ba/F3-gp130-eGFP cells were co-cultured with Ba/F3-IL-6Rα cells, sIL-6 alone induced the proliferation of Ba/F3-gp130-eGFP cells (Fig. 3b, c), suggesting that Ba/F3-IL-6Rα cells trans-presented IL-6 to Ba/F3-gp130-eGFP cells, inducing the proliferation of the latter cells. This IL-6 cluster signaling requires the clustering of Ba/F3-IL-6Rα cells and Ba/F3-gp130-eGFP cells in the same co-culture. Exogenous IL-6Rα Abs, but not soluble gp130 (sgp130-Fc), a strong inhibitor of IL-6 trans-signaling by hyper-IL-6, neutralized IL-6 cluster signaling (Fig. 3b), indicating that IL-6 trans-signaling and IL-6 cluster signaling are functionally different. Together, these results suggest that IL-6 bound to IL-6Rα on the surface of IL-6Rα-expressing cells signals through membrane-bound gp130 in cells that do not express IL-6Rα when both cell types are in physical proximity.
Figure 3. IL-6 presentation in trans is a functional IL-6 signaling modality in engineered murine pre-B-cells (Ba/F3) and is not blocked by sgp130-Fc.
Ba/F3 cells stably transduced with the human IL-6Rα (termed Ba/F3-IL-6Rα) show no GFP fluorescence whereas Ba/F3-gp130-eGFP cells stably transduced with gp130-eGFP (termed Ba/F3-gp130-eGFP) appear bright green. (a) Equal amounts of either Ba/F3-IL-6Rα (left) or Ba/F3-gp130-eGFP cells (right) were cultured for two days with either 10 ng/ml human IL-6, 10 ng/ml hyper-IL-6, 10 ng/ml hyper-IL-6 plus 1 μg/ml sgp130-Fc, 1% conditioned WEHI-supernatant (containing IL-3), or left untreated (Medium). Mean RLU ± SD (n=3, representative of three experiments). (b) Ba/F3-IL-6R and Ba/F3-gp130-eGFP cells were co-cultured in a 1:10 ratio for two days. Cells were stimulated with 10 ng/ml IL-6, 10 ng/ml IL-6 plus 1 μg/ml sgp130-Fc, 10 ng/ml IL-6 plus 1 µg/ml of the anti-IL-6 antibody mAb#8, 10 ng/ml IL-6 plus anti-IL-6R antibody tocilizumab, or were left untreated. Mean RLU ± SD (n=3, representativeof three experiments). (c) Ba/F3-IL-6R and Ba/F3-gp130-eGFP cells were co-cultured in a 1:10 ratio for two days either without or with 10 ng/ml IL-6. Microscopic pictures were taken on days as indicated. Pictures are representative of three independent experiments. *P<0.05, ANOVA plus Tukey's multiple comparisons test.
DCs present IL-6 in trans and induce IL-6 cluster signaling
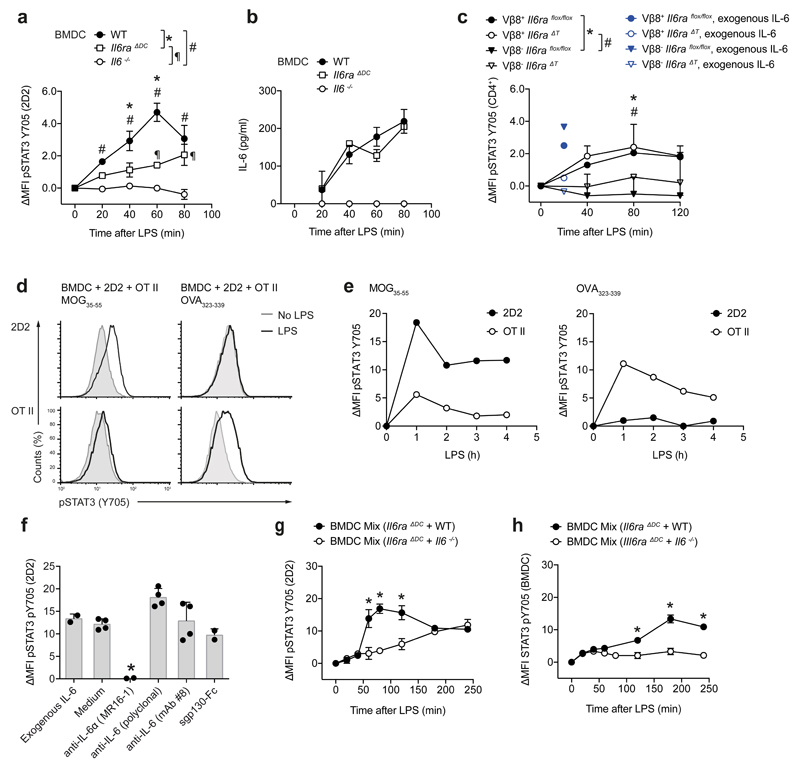
Next, we tested the ability of DCs to present IL-6 in trans during a cognate interaction with T cells (Supplementary Fig. 7). Naive (CD4+CD44-GFP (Foxp3)-) 2D2 T cells were co-cultured with BMDCs in the presence of cognate antigen (MOG35-55) and LPS to induce IL-6 production in BMDCs. GFP (Foxp3)- effector 2D2 T cells showed robust Stat3 activation assessed by flow cytometry when they were co-cultured with wild-type BMDCs, but not after culture with Il6ra-/- BMDCs, although the amount of soluble IL-6 produced by wild-type and Il6ra-/- BMDCs was similar (Fig. 4a, b). To test if IL-6 cluster signaling occurred during cognate DC-T cell interactions, we co-cultured wild-type BMDCs with naive T cells isolated either from wild-type or CD4-Cre x Il6raflox/flox (Il6raΔT) mice, and used Staphylococcal enterotoxin B (SEB), a superantigen that activates TCR-Vβ8+ T cells in an MHC class II dependent manner 20. TCR-Vβ8+ T cells exhibited an increased p-Stat3 signal compared to TCR-Vβ8- T cells present in the same culture (Fig. 4c). In addition, Stat3 activation occurred in both wild-type and Il6raΔT TCR-Vβ8+ T cells. In contrast, exogenous soluble IL-6 induced p-Stat3 in wild-type, but not by Il6raΔT T cells (Fig. 4c). In addition, when MOG35-55-specific transgenic 2D2 T cells and OVA323-339-specific transgenic OT II T cells where co-cultured with wild-type BMDCs in a triple culture, early Stat3 phosphorylation was exclusively observed in T cells whose cognate antigen was present in the culture but not in co-cultured T cells with irrelevant specificity (Fig. 4d, e), suggesting that IL-6 cluster signaling did not result in bystander activation of T cells that did not interact with DCs in a cognate manner.
Figure 4. DCs perform IL-6 cluster signaling during antigen specific priming of T cells.
(a) 2D2 T cells were cultured in the presence of MOG35-55 with WT, Il6-/-, or Il6ra-/- BMDCs. Subsequent to LPS stimulation, Stat3 phosphorylation was assessed in 2D2 T cells and plotted as ΔMFI. Representative of four experiments. Mean + SD, n=2, P<0.05, ANOVA plus Tukey's multiple comparisons test. (b) IL-6 was measured by ELISA in the supernatants of (a). (c) WT BMDCs were co-cultured with either control CD4+ T cells or IL-6Rα deficient T cells in the presence of SEB followed by LPS stimulation. Stat3 activation was measured in TCR-Vβ8- vs TCR-Vβ8+ T cells (SEB responders). As control, p-Stat3 triggered by exogenous IL-6 was assessed (blue). Representative of two experiments. Mean + SD, n=2, P<0.05, ANOVA plus Tukey's multiple comparisons test. (d, e) Triple cultures of WT BMDCs with 2D2 T cells and OT II T cells were stimulated with either MOG35-55 or OVA323-339. At different time points after addition of LPS p-Stat3 was measured in 2D2 or OT II T cells. Representative histograms at 2 h (d) and time course (e). Representative of two experiments. (f) WT BMDCs were cultured with 2D2 T cells and MOG35-55 in the presence of exogenous IL-6 or with LPS and 1µg/ml blocking agents (anti-IL-6R, anti-IL-6, or sgp130-Fc). After 2h, p-Stat3 was analyzed. Representative of four experiments. Mean + SD, n=4. *P<0.04, ANOVA plus Tukey post test. (g, h). 2D2 T cells were cultured with mixtures of BMDCs followed by assessment of p-Stat3 in 2D2 T cells (g) and BMDCs (h). Representative of three experiments, mean + SD, n=2. *P<0.05, ANOVA plus Tukey's multiple comparisons test.
In order to further characterize IL-6 cluster signaling during a DC-T cell interaction, we used a variety of IL-6 blocking reagents at the time of LPS activation of BMDCs co-cultured with 2D2 T cells in the presence of MOG35-55. The monoclonal antibody MR16-1, which binds IL-6Rα, completely abolished the p-Stat3 signal in 2D2 T cells (Fig. 4f). In contrast, sgp130-Fc did not inhibit p-Stat3 in 2D2 T cells (Fig. 4f), indicating that sgp130-Fc cannot engage the membrane bound IL-6-IL-6Rα complex interacting with gp130 in trans. In addition, antibodies to IL-6, and in particular mAb #8, a monoclonal IL-6 antibody that binds the interaction site between IL-6 and IL-6Rα 10, failed to block Stat3 activation in T cells in response to LPS activated co-cultured BMDCs (Fig. 4f). Because IL-6 bound in a complex with IL-6Rα would be less easily neutralized by IL-6 antibodies, we tested whether IL-6 was loaded onto the IL-6Rα in the intracellular compartment and transported to the cell membrane of DCs as a complex. We incubated equal mixtures of Il6ra-/- and wild-type BMDCs or Il6ra-/- and Il6-/- BMDCs with naive (CD44-GFP (Foxp3)-) 2D2 T cells in the presence of MOG35-55 and stimulated the cultures with LPS. Within 120 min after LPS stimulation, Il6ra-/- + wild-type BMDCs elicited Stat3 activation in GFP (Foxp3)- effector T cells comparable to wild-type BMDCs alone, while Il6ra-/- + Il6-/- BMDCs failed to induce p-STAT3, similar to Il6ra-/- BMDCs alone (Fig. 4g), indicating that Il6-/- BMDCs, which express IL-6Rα, do not pick up ambient IL-6 supplied by Il6ra-/- BMDCs and do not trans-present ambient IL-6 to T cells. Notably, there was no clustered IL-6 feedback Stat3 signal into any of the BMDCs present in the co-cultures but only a smoldering Stat3 activation in BMDCs beyond 120 min of LPS stimulation (Fig. 4h). The late p-Stat3 activation in BMDCs was higher in the Il6ra-/- + wild-type BMDC co-cultures than in the Il6ra-/- + Il6-/- BMDC co-cultures (Fig. 4h), suggesting that higher amounts of soluble IL-6 in the Il6ra-/- + wild-type BMDC co-cultures (data not shown) were responsible for classic IL-6 signaling into BMDCs. Taken together, DCs load IL-6 onto IL-6Rα in their intracellular compartment and use the IL-6-IL-6Rα complex to perform IL-6 cluster signaling when cognately interacting with T cells. IL-6 cluster signaling then leads to the targeted activation of Stat3 in antigen specific T cells but not in bystander T cells.
IL-6 and IL-6Rα co-localize in DCs
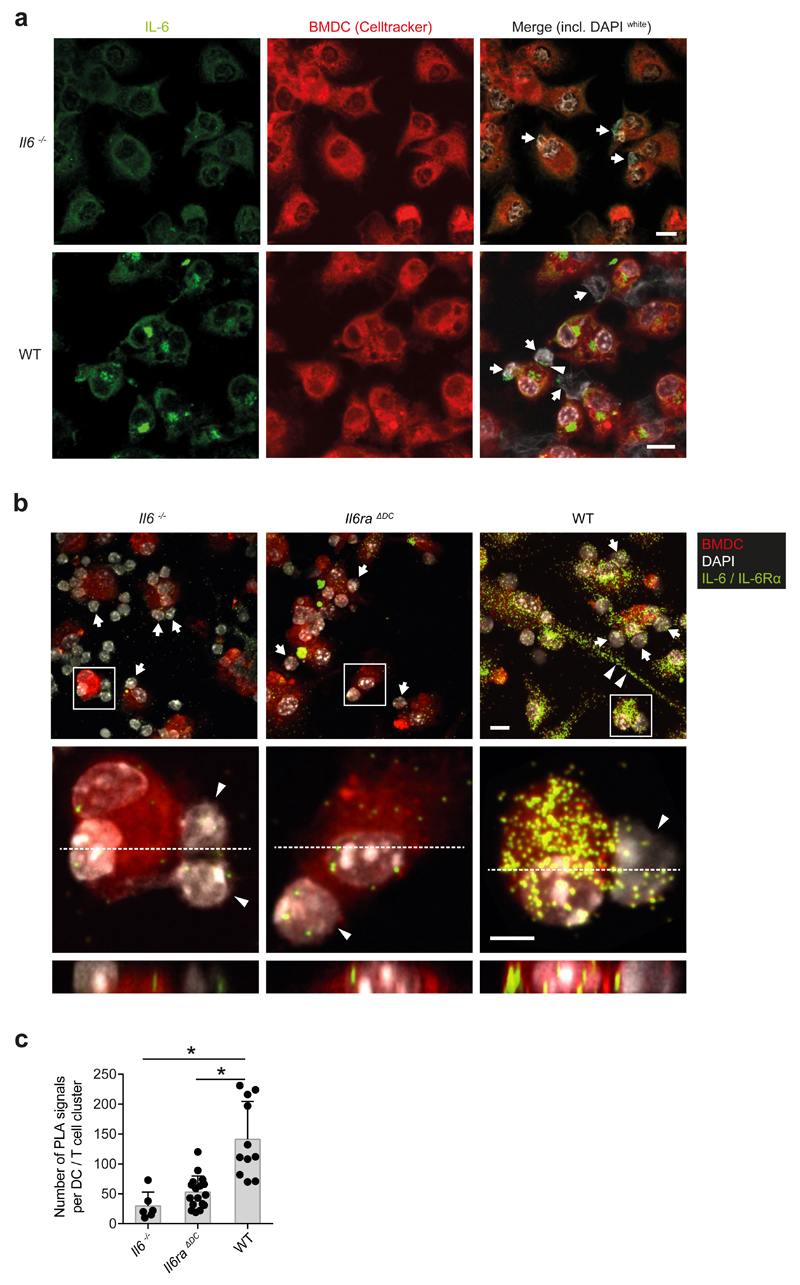
Next, we used confocal microscopy to visualize IL-6-IL-6Rα complexes in BMDCs that interacted with 2D2 T cells in a cognate manner. BMDCs were co-cultured with 2D2 T cells in the presence of MOG35-55 and stimulated with LPS in the absence of exogenous IL-6. We detected "clustered" IL-6 on the surface of LPS-stimulated BMDCs interacting with 2D2 T cells, with some IL-6 locating at the BMDC-T cell interaction interface (Fig. 5a). To test whether IL-6 and IL-6Rα interact on the surface of BMDCs, we performed a proximity ligation assay (PLA) 21. IL-6 deficient, IL-6Rα deficient, or wild-type BMDCs were co-cultured with 2D2 T cells in the presence of MOG35-55 and stimulated with LPS. IL-6 and IL-6Rα were simultaneously labeled by antibodies followed by a proximity ligation assay to visualize IL-6-IL-6Rα complexes. Positive PLA signals indicating colocalisation of IL-6 and IL-6Rα at a distance of less than 40 nm were detected in wild-type BMDC-T cell co-cultures (Fig. 5b) while we observed only few PLA signals in Il6-/- or Il6ra-/- BMDC-T cell co-cultures (Fig. 5b, c), suggesting that the background by unspecific PLA amplification reactions was low. Co-localization of IL-6 and IL-6Rα occurred in the cytoplasm and at the membrane surface of wild-type BMDCs, with some signal localized at the DC-T cell interaction zone (see xz-projections in Fig. 5b). Thus, DC-associated IL-6-IL-6Rα complexes can be visualized in DCs.
Figure 5. The IL-6-IL-6Rα complex is formed in intracellular compartments of DCs and presented in trans to cognately interacting T cells.
2D2 T cells were cultured in the presence of MOG35-55 with WT, Il6-/-, or Il6ra-/- BMDCs that were pre-stained with CellTracker. Subsequent to LPS stimulation for 80 min, cells were fixed and permeabilized. (a) Immunofluorescence staining of IL-6 (green) in co-cultures of T cells (arrows) with Il6-/- BMDCs (upper row) or WT BMDCs (lower row). Nuclei of both cell types were stained with DAPI (white). The arrow head highlights an IL-6 signal at the DC-T cell interface. Scale bar = 10 μm. Representative of four experiments. (b) Proximity ligation assay analyses for IL-6-IL-6Rα interactions (green) in BMDCs interacting with 2D2 T cells. Co-cultures of 2D2 T cells with Il6-/- BMDCs, Il6ra-/- BMDCs, or WT BMDCs were stimulated with LPS. After 80 min cells were fixed and subjected to PLA. Nuclei (DAPI), BMDCs (CellTracker), and IL-6-IL-6Rα complexes (green). Overview (upper row): Arrows indicate T cells, arrow heads highlight a cytoplasm bridge of a WT DCs. Scale bar = 10 μm. Confocal images of individual DC-T cell (arrow heads) interactions. Middle row (xy plain) and lower row (xz plain): Scale bar = 5 μm. Representative of two experiments. (c) Quantification of IL-6-IL-6Rα complexes (PLA signals) per DC. Mean + SD, *P<0.0001, ANOVA plus Tukey post test.
IL-6 cluster signaling during antigen-specific T cell priming in vivo
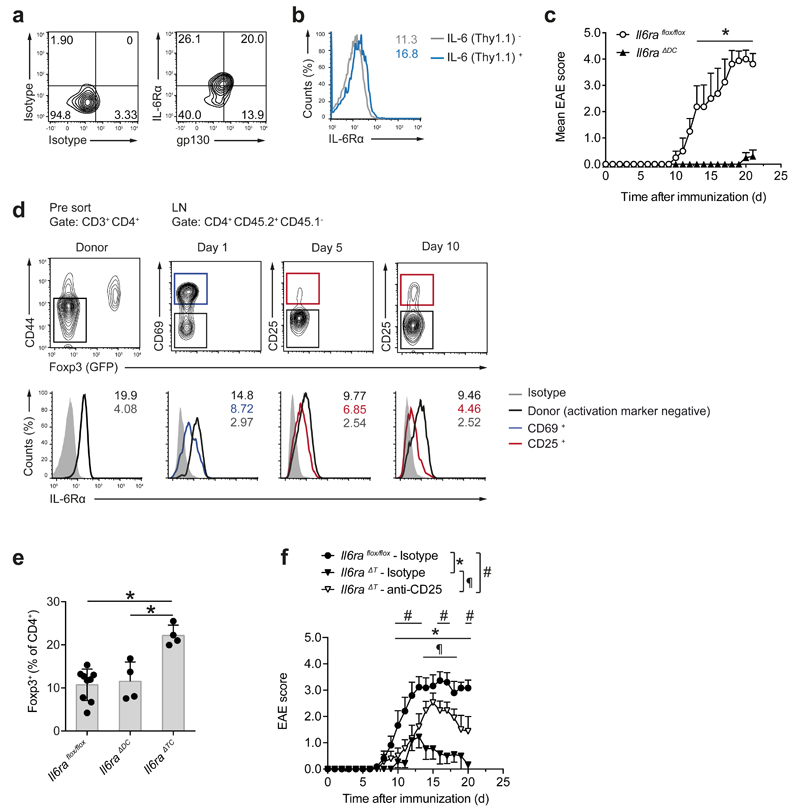
Next, we analyzed whether lack of DC-mediated IL-6 cluster signaling explained the lack of EAE development in Il6ΔDC mice. CD11b+ DCs isolated from the draining lymph nodes of MOG35-55-CFA-immunized wild-type mice showed surface and intracellular expression of IL-6Rα (Fig. 6a and Supplementary Fig. 7). In MOG35-55-CFA-immunized CD11c-Cre x Il6RD/wt mice, the surface expression of IL-6Rα in Thy1.1+ DCs isolated from the draining lymph nodes was higher than in Thy1.1- DCs (Fig. 6b), indicating that DCs that expressed IL-6Rα and IL-6 simultaneously can be detected in the draining lymph nodes of immunized mice. In order to investigate the requirement for DC mediated IL-6 cluster signaling in the priming of pathogenic T cells in vivo, we immunized CD11c-Cre x Il6raflox/flox mice (termed Il6raΔDC), which lack IL-6Rα specifically in DCs, and are thus unable to trans-present IL-6 during antigen specific priming, with MOG35-55 in CFA. Compared to Il6raflox/flox control mice, Il6raΔDC mice were resistant to EAE (Fig. 6c). Thus, IL-6 cluster signaling is required to promote the differentiation of pathogenic T cells during antigen-specific priming in a DC proximal manner.
Figure 6. IL-6 cluster signaling is operational in vivo.
(a) Ex vivo flow cytometric assessment of surface expression of the IL-6R subunits IL-6Rα and gp130 in CD11c+MHC class IIhigh CD11b+DCs isolated from the dLN of MOG35-55 immunized WT mice on day 10. Representative cytograms out of two experiments. (b) CD11c Cre x Il6RD/wt x R26 Stopflox/flox YFP mice were treated with Flt3L cells as described in Supplementary Fig. 2. Two days after LPS injection, LN cells were prepared and stained ex vivo. Surface IL-6Rα expression is depicted in IL-6 (Thy1.1)- vs IL-6 (Thy1.1)+ CD11b+ DCs (Numbers in histogram plots depict MFI). (c) Mean clinical EAE scores of Il6raflox/flox control and Il6raΔDC mice + SEM, n≥4. *P<0.05, ANOVA plus Sidak post test. Representative of four experiments. (d) Naive T cells (CD4+CD44-GFP (Foxp3)-; left column) from 2D2 x Foxp3gfp.KI mice were transferred into WT congenic (CD45.1) mice followed by MOG35-55 immunization. At the indicated time points, 2D2 T cells were re-isolated from dLN and surface IL-6Rα expression was measured in activated (CD69+or CD25+) effector (GFP (Foxp3)-) 2D2 T cells vs non-activated 2D2 T cells (black). (e) Frequencies of Foxp3+ Tregs in dLN CD4+ T cells of control mice (Il6raflox/flox) vs Il6raΔDC, and Il6raΔT animals. Mean + SD, n≥4, *P<0.003 (ANOVA plus Tukey's multiple comparisons test). (f) Course of EAE in Il6raflox/flox control mice vs isotype treated and Treg-depleted Il6raΔT mice. To deplete Tregs, 0.5 mg of anti-CD25 (PC61) were injected i.p. on days -5 and -3 prior to immunization. Mean EAE scores + SEM, n≥9. *P<0.05, ANOVA plus Sidak post test.
In vivo, sorted naive (CD44-GFP (Foxp3)-) 2D2 T cells, which had been transferred into congenic CD45.1 host mice followed by immunization with MOG35-55 in CFA, showed a down-regulation of the surface expression of IL-6Rα in the draining lymph nodes as of day 1 after immunization through at least day 5 when engaged in an antigen specific encounter as defined by upregulation of CD69 or CD25, respectively (Fig. 6d). In addition, sgp130-Fc, which blocks IL-6 trans-signaling22, did not ameliorate EAE in sgp130-Fc transgenic mice as compared to wild-type control animals (Supplementary Fig. 8), indicating that soluble IL-6-IL-6Rα complexes are irrelevant in EAE. We next tested whether IL-6Rα-deficient T cells could respond to IL-6 cluster signaling by DCs and become pathogenic effector T cells by using T cell conditional IL-6Rα-deficient mice (Il6raΔT). We found higher frequencies of Foxp3+ Treg cells in the draining lymph node CD4+ T cell compartment of Il6raΔT mice immunized with MOG35-55 in CFA compared to Il6raflox/flox control animals while MOG35-55 immunized Il6raΔDC mice harbored similar Foxp3+ Treg cell frequencies as Il6raflox/flox control mice (Fig. 6e), consistent with the observation that ambient IL-6, and thus classic IL-6 signaling, significantly contributes to suppressing the induction of Foxp3+ Treg cells from conventional T cells in a polyclonal repertoire. Depletion of Treg cells with an anti-CD25 antibody prior to MOG35-55-CFA immunization in Il6raΔT mice lead to an increase in the severity of EAE compared to non-Treg cell depleted Il6raΔT mice (Fig. 6f and Supplementary Fig. 9), indicating that Il6raΔT TH17 cells could be primed to become pathogenic.
Pathogenic TH17 cells can be differentiated with IL-21 in vivo, in the absence of IL-6 15,23. To evaluate the contribution of IL-21 in the pathogenic priming of T cells in Il6raΔT mice, we adoptively transferred naive polyclonal CD4+ T cells from Il21r-/- x Il6raΔT mice, in which T cells are deficient in the expression of both IL-6Rα and IL-21R, into Rag1-/- host mice, followed by subcutaneous immunization with MOG35-55 in CFA. Rag1-/- mice transferred with Il21r-/- x Il6raΔT T cells mounted a TH17 response in response to MOG35-55 immunization and developed EAE (Supplementary Fig. 10). Thus, T cells lacking IL-21 responsiveness and IL-6Rα expression that mediates classic IL-6 signaling can still become pathogenic TH17 cells.
Pathogenic TH17 cells are dependent on IL-6 cluster signaling
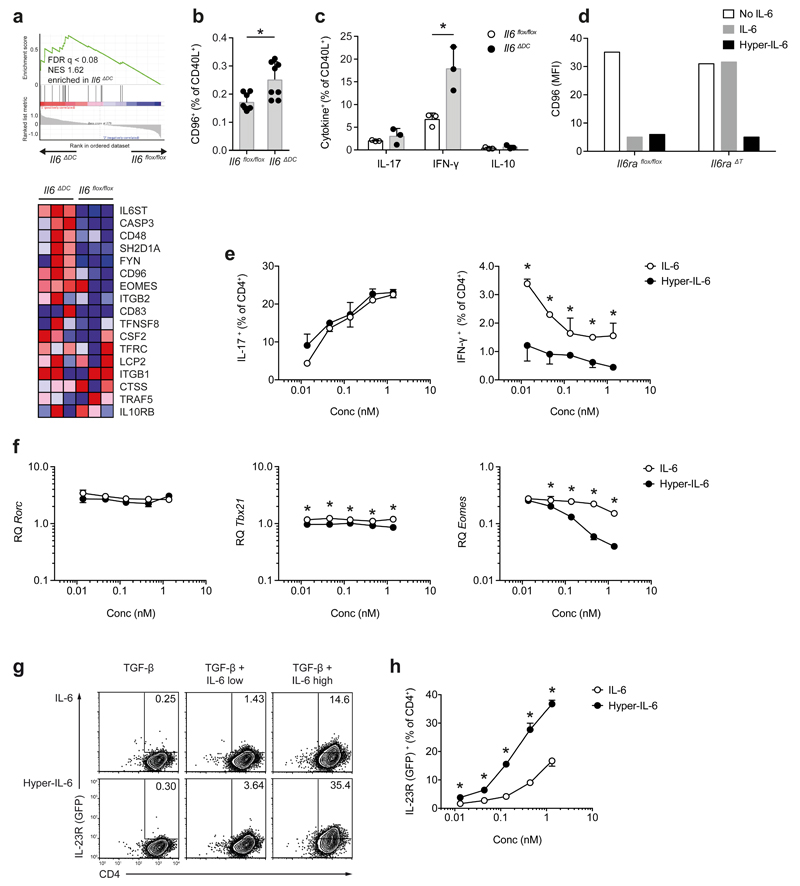
Il6-/- mice develop an exaggerated Treg cell response upon antigen specific priming with adjuvant 15. Because Il6ΔDC mice show normal Foxp3+ Treg responses, but do not develop EAE we tested whether effector T cell response in Il6ΔDC mice were intrinsically inefficient. Il6flox/flox control mice, Il6-/- and Il6ΔDC mice were sensitized with MOG35-55 in CFA and CD4+Foxp3- T cells were isolated from draining lymph nodes for transcriptome analysis. Because EAE is essentially a TH17 cell mediated model of CNS autoimmunity, we further tested a transcriptional module previously associated with “non-pathogenic” TH17 cells 8 in Foxp3- T cells primed in the Il6ΔDC mice. Gene set enrichment analysis indicated that conventional Foxp3- T cells primed in Il6ΔDC mice had a significantly higher expression of Il6st (gp130), Cd96, and Eomes as compared to effector T cells primed in a Il6flox/flox control environment (Fig. 7a, b). Notably, CD40L+Foxp3- T cells primed in the Il6ΔDC mice and re-stimulated with MOG35-55 in vitro exhibited significantly higher fractions of IFN-γ single producers compared to CD40L+ effector T cells primed in Il6flox/flox control mice (Fig. 7c), indicating that naïve T cells primed by IL-6 deficient DCs differentiated preferentially into CD96+IFN-γ+ T cells, which were incapable of inducing EAE.
Figure 7. T cell priming is aberrant in an Il6ΔDC environment and results in non-pathogenic T cells.
CD4+Foxp3- effector T cells were purified from dLN of MOG35-55 immunized control (Il6flox/flox), Il6-/-, or Il6ΔDC mice and analyzed by nanostring. (a) GSEA for a non-pathogenic TH17 gene set 8 was performed. Il6st (encoding gp130), Eomes, and Cd96 were highly enriched in effector T cells primed in Il6ΔDC mice. (b, c) Control mice or Il6ΔDC mice were immunized with MOG35-55 and after 7 days, the expression of CD96 (b) and intracellular cytokines (c) were assessed on CD40L+ T cells upon restimulation with MOG35-55. Representative of two experiments, mean + SD, n=4, (b) *P=0.0066, unpaired t-test; (c) *P <0.005, ANOVA plus Sidak's multiple comparisons test. (d) Naïve WT or IL-6Rα deficient T cells were stimulated in the presence of TGF-β (0.1 ng/ml) and either IL-6 or hyper-IL-6 followed by assessment of CD96 expression. Representative of two experiments. (e, f) Naïve WT T cells were activated in the presence of TGF-β and equimolar concentrations of either soluble IL-6 or hyper-IL-6. Subsequent to PMA/ionomycin restimulation, cells were stained for IL-17 and IFN-γ (e). Representative of three experiments, mean + SD, n=2, *P<0.04, ANOVA plus Holm-Sidak post test. Rorc, Tbx21 and Eomes mRNA expression was analyzed (f). Mean + SD of technical replicates. Representative of of two experiments. *P<0.04, ANOVA plus Holm-Sidak post test. (g, h) Naïve T cells from GFP (IL-23R) reporter mice were stimulated in the presence of TGF-β plus equimolar concentrations of IL-6 or hyper-IL-6. After 2 days, GFP (IL-23R) expression was measured (g). Mean + SD, n=3. *P<0.03, ANOVA plus Holm-Sidak post test. Representative of two experiments (h).
In vitro, hyper-IL-6 suppressed the upregulation of CD96 in IL-6Rα-deficient T cells during TH17 differentiation (Fig. 7d), indicating that IL-6 presented in trans was efficient in preventing the TGF-β mediated induction of CD96. We used hyper-IL-6 as a surrogate for IL-6 presented in trans and compared its effect to soluble IL-6 (and thus classic IL-6 signaling) during APC free differentiation of naive sorted (CD4+CD44-CD25-) T cells into TH17 cells. When using equimolar amounts of soluble IL-6 or hyper-IL-6, the induction of IL-17 in T cells was similar with both reagents (Fig. 7e). However, the suppression of IFN-γ expression during TH17 differentiation was significantly more efficient with hyper-IL-6 than with soluble IL-6 (Fig. 7e). Because under certain conditions, Eomes is an inducer of IFN-γ in CD4+ T cells 24,25, we tested whether the increased fraction of IFN-γ+ T cells induced by TGF-β plus soluble IL-6 was due to high Eomes expression. During TH17 differentiation, Tbx21 and in particular Eomes mRNA were less efficiently suppressed by IL-6 than by hyper-IL-6, with no modulation of Rorc (Fig. 7f). Conversely, hyper-IL-6 was significantly more potent than soluble IL-6 in inducing robust GFP (IL-23R) expression during in vitro TH17 cell differentiation of naive CD4+ T cells isolated from Il23rgfp/+ mice 26 (Fig. 7g, h), indicating that the higher IFN-γ expression in T cells primed in Il6ΔDC mice was linked to lack of IL-6 trans-presentation in these mice.
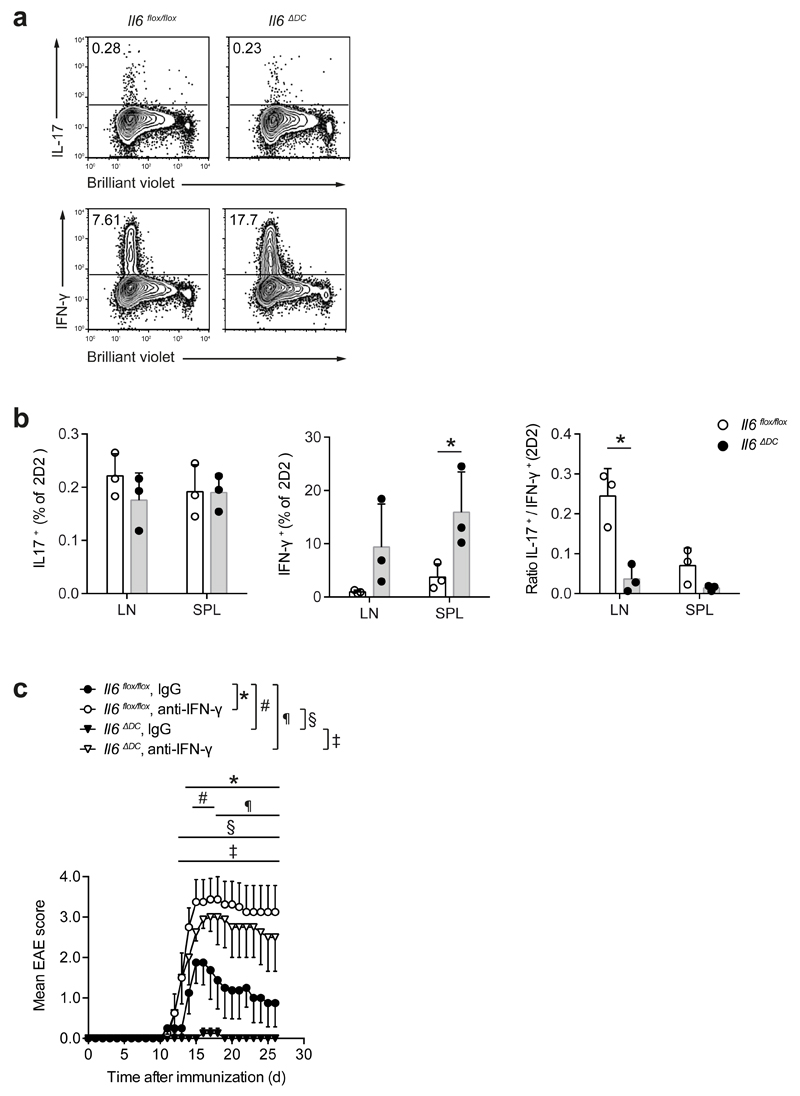
Because high IFN-γ expression in myelin-specific-T cells is associated with protection from EAE 27, we tested whether the high IFN-γ expression observed in Foxp3- effector T cells in the Il6ΔDC mice might determine the resistance of Il6ΔDC mice to EAE. As measured by dye dilution, 2D2 T cells primed in Il6ΔDC mice proliferated as well as 2D2 T cells primed in Il6flox/flox control mice but showed higher production of IFN-γ (Fig. 8a, b). Neutralization of IFN-γ in Il6ΔDC mice with a monoclonal antibody to IFN-γ fully restored EAE severity in Il6ΔDC mice compared to Il6ΔDC mice treated with a control antibody (Fig. 8c). In summary, these data imply that priming of TH17 cells in Il6ΔDC mice in the absence of DC-mediated IL-6 cluster signaling results in the exaggerated expression of IFN-γ in CD4+ T cells, most likely due to aberrant Stat3 activation in these cells.
Figure 8. Aberrant priming of TH17 cells in the absence of DC mediated IL-6 cluster signaling is reversed by neutralization of IFN-γ in vivo.
(a, b) Naive 2D2 T cells were labeled with a proliferation dye and transferred into either Il6flox/flox control mice or Il6ΔDC animals followed by immunization with MOG35-55. After re-isolation on day 7 after immunization, proliferation and intracellular cytokines were assessed in 2D2 T cells. Representative of two experiments, mean + SD, n=4. *P<0.04, t-test. (c) The disease phenotype of Il6ΔDC mice is rescued upon neutralization of IFN-γ. Control mice (Il6flox/flox) or Il6ΔDC mice were immunized with MOG35-55 and either control treated (rat IgG1) or injected i.p. with 200 μg anti-IFN-γ every other day until day 11 after immunization. Representative of two experiments. Mean EAE scores + SEM, n=4 per group, *P<0.05, ANOVA plus Tukey post test.
Discussion
Here, we show that T cells respond to IL-6 in the absence of IL-6Rα expression through a process that we call IL-6 cluster signaling, in which DC-membrane bound IL-6Rα complexed with IL-6 is presented in trans and sensed by gp130 molecules expressed on T cells. IL-6 cluster signaling not only substitutes for classic IL-6 signaling, but leads to qualitatively different T cell responses. The prototypic cytokine to be trans-presented by auxiliary cells via its high affinity receptor is IL-15 28. It is likely that similar to IL-15, IL-6 is loaded onto IL-6Rα in endosomal compartments. Trans-presentation has also been proposed for another IL-6 family member, cardiotrophin-like cytokine, through binding to the CNTF receptor 29. Although IL-6 forms a stable complex with soluble IL-6Rα, the affinity of IL-6 for IL-6Rα is only about 0.5 to 1 nM 13, and thus two orders of magnitude lower than the affinity of IL-15 for IL-15Rα. However, membrane bound cytokine trans-presentation was also reported for IL-2, whose affinity for IL-2Rα is in the same range as the affinity of IL-6 for IL-6Rα 13,30.
When interacting with T cells, DC mediated IL-6 cluster signaling can restrict the IL-6 signal to cognate T cells, with high temporal synchronization with the TCR signal. Such coordination of signals leads to the imprinting of pathologic properties in recipient T cells. Notably, Foxp3 suppression is induced in T cells with reduced or even absent Stat3 signaling, and can be fully supported by classic IL-6 signaling. Therefore, the IL-6 that prevents the upregulation of Foxp3 in T cells does not have to be derived from the priming DC, and can be derived in soluble form from the micro-milieu. Hence, it is possible that the suppression of Foxp3 induction in T cells occurs as a "bystander" effect of ambient IL-6. However, the IL-6-dependent induction of inflammatory properties in Foxp3- T cells is only efficient in the context of antigen-presentation, and likely does not occur as a bystander effect of ambient IL-6. This suggests that IL-6-mediated pathogenic T cell differentiation passes through two checkpoints: the suppression of Foxp3 induction via classic IL-6 signaling and the initiation of a pathogenic effector T cell transcriptional program via IL-6 cluster signaling. Therefore, the ability to induce pathogenic TH17 cells is dependent on antigen presenting cells that can co-present IL-6 in trans.
Here, we show that IL-6+IL-6Rα+ DCs that are able to perform cluster signaling are CD103- and belong to the subset of CD11b+Sirpα+ DCs that were recently classified as cDC2 31 and were previously associated with pathogenic TH17 responses 32. Because no mechanistic underpinning of the unique instructive capacity of this DC subset to instruct TH17 responses has been identified, IL-6 cluster signaling in the context of synchronized antigen presentation might be an intriguing hypothesis. In contrast to classic IL-6 signaling, IL-6 cluster signaling leads to an earlier and more robust expression of IL-23R in antigen-activated T cells. Therefore, facilitated sensing of IL-23 further supports their pathogenic phenotype 33. Vice versa, in the absence of IL-6 cluster signaling, IL-23 is unable to compensate for the impaired initiation of a pathogenic transcriptional program in antigen specific T cells, suggesting that the synchronization of antigen-specific priming and IL-6-mediated Stat3 signaling is fundamental for the pro-inflammatory phenotype of T cells. Because T cell blasts extensively downregulate cell surface expression of IL-6Rα, it is plausible that IL-6 cluster signaling is a means to combine a “pathogenic” IL-6 signal with cognate re-activation. The APC type and the anatomical compartment of this process have not been exactly defined.
We provide evidence that lack of IL-6 cluster signaling during TH17 priming deviates the cytokine phenotype into IFN-γ production, which is in part dependent on Eomes expression. Although there has been some controversy 34, IL-6 has been shown to suppress the development of Th1 cells via direct and indirect mechanisms 35. Here we demonstrate that IL-6 cluster signaling in particular is an efficient means to prevent the induction of IFN-γ during TH cell differentiation. In our model, in the absence of IL-6 cluster signaling, insufficient Stat 3 activation, and thus a Stat1-Stat3 imbalance, allows for the expression of Eomes and IFN-γ in TH17 priming conditions and results in impaired encephalitogenicity.
Finally, the possibility of IL-6 cluster signaling has implications for therapeutic interventions. Anti-IL-6 antibodies fail to inhibit IL-6 cluster signaling. The IL-6-IL-6Rα complex formation occurs within DCs, most likely followed by targeted shuttling to plasma membrane regions involved in the cognate interaction with T cells, with immediate accessibility to gp130 molecules on the T cell side. Therefore, site I of the IL-6 molecule 10, which mediates IL-6 binding to IL-6Rα and is targeted by most anti-IL-6 antibodies, is already buried in the IL-6-IL-6Rα complex when it appears at the plasma membrane. Although sgp130-Fc suppresses IL-6 trans-signaling 14, IL-6 cluster signaling is not inhibited by sgp130-Fc, and we hypothesize that sgp130-Fc cannot access the IL-6-IL-6Rα complex at the DC-T cell interaction zone. However, similar to classic IL-6 signaling and IL-6 trans-signaling, IL-6 cluster signaling remains amenable to neutralization via anti-IL-6Rα antibodies. Thus, IL-6 targeting strategies have to be carefully re-evaluated for the differential signaling modalities when designing therapeutic strategies for autoimmunity, chronic inflammation, and cancer.
Online methods
Generation of Il6RD mice
To generate a new conditional IL-6 reporter/depleter strain [Il6RD; Il6tm3307(Cerulean-P2A-CD90.1)Arte] allowing for detection and specific depletion of IL-6 producing cells, we targeted Exon 2 of the Il6 locus on chromosome 5 by insertion of a loxP-flanked transcription termination cassette (STOP) followed by a reporter cassette. The reporter expression cassette comprised (i) fluorescent reporter Cerulean, PCR-amplified from the cloning vector pDH51-GW-CFP; (ii) 2A peptide from Porcine teschovirus-1 (P2A), the aa sequence GSGATNFSLLKQAGDVEENPGP 36 was synthetically attached to the adjacent sequences using oligonucleotides; (iii) Thy1.1 (CD90.1) surface protein, that is in C57BL/6 mice (endogenously expressing allele Thy1.2 [CD90.2]) exclusively expressed on Il6 reporter positive cells. The sequence was provided by M. Schmidt-Supprian and is identical to the GenBank # AAR17087.1; (iv) a polyadenylation signal BGH pA, derived from the plasmid pcDNA3.1(+). A targeting vector MW21214 containing the floxed STOP cassette and the reporter expression cassette was generated. To target this construct to the Il6 locus, homologous arms were amplified from BAC clone C57BL/6J RPCIB-731. The SHA (short homologous arm) encompassed ~4kb upstream of Il6, Exon1, Intron 1 and 31 nucleotides of Exon 2. The natural Exon 1 contains the 5´UTR and the translation initiation codon (START) which was mutated in the targeting vector (noSTART) by site directed mutagenesis (from ATG into AAG) in order to prevent transcription of the remaining mouse Il6 gene. Downstream of the expression cassette, a positive selection marker (CAGGS promoter driven puromycin resistance) was inserted, that was flanked by FRT sites allowing for Flp-mediated deletion of the selection marker. The downstream LHA (long homologous arm) consisting of 5.7 kb of the Il6 gene encompassed the remaining 153 nucleotides of exon 2, the complete intron 2, exon 3, intron 3, exon 4, and a partial intron 4. With the exception of the mutated start codon in exon 1, the genomic sequence was left intact, in order to preserve all potential regulatory elements of Il6. For negative selection, a PGK promoter driven HSV TK was inserted. The targeting vector MW21214 was approved by complete sequencing, linearized by NotI and transfected into the C57BL/6N ES cell line Art B6 3.6 by electroporation to generate heterozygous targeted ES cells. Homologous recombinant clones were isolated using positive (PuroR) and negative (TK) selections. Clones were screened by PCR, resulting in 22% targeting frequency. The majority of the clones analyzed carried the inserted point mutation of the start codon. Four clones were expanded and validated by Southern Blot and PCR analyses. Targeted ES cells were injected into blastocysts (isolated at dpc 3.5 from superovulated, mated BALB/c females). Injected blastocysts were transferred into pseudopregnant NMRI females (2.5 days post coitum). Chimerism was evaluated by coat color and highly chimeric mice were bred to Flp-Deleter mice (C57BL/6-Tg(CAG-Flpe)2Arte), allowing for (i) identification of germline transmission and (ii) Flp-mediated deletion of the positive selection marker. Il6RD mice were crossed with CMV-Cre [Tg(CMV-cre)1Cgn] for ubiquitous expression or with CD11c-Cre [Tg(Itgax-cre)1-1Reiz] mice for DC-specific, conditional expression of the reporter cassette after Cre-mediated removal of the STOP cassette. Under control of the endogenous Il6 promoter, a chimeric transcript harboring the Cerulean ORF fused to the P2A sequence and the CD90.1 ORF is expressed. The P2A provides effective co-translational cleavage in vitro and in vivo, resulting in strict stoichiometric co-expression of individual Cerulean and CD90.1 proteins with correct subcellular localization (cytoplasm or cell surface, respectively).
Mice
Il6flox/flox [Il6tm1.1Jho] 8,37, Foxp3.gpf KI [Foxp3tm1Kuch] 5,36,38, Il23r.gfp KI [Il23rtm1Kuch] 26 and MOG35-55 specific TCR transgenic 2D2 mice [Tg(Tcra2D2,Tcrb2D2)1Kuch] 39 were described previously. Il6-/- [Il6tm1Kopf] were obtained from Jackson Laboratory. Il6raflox/flox mice were kindly provided by Thomas Wunderlich 40. OVA323-339 specific TCR transgenic OT II mice [Tg(TcraTcrb)425Cbn] with the congenic marker CD45.1 were kindly provided by D. Busch (Institute for Medical Microbiology, Immunology and Hygiene, TU Munich). Stat3flox/flox mice [Stat3tm2Aki] were received from F. Greten (Institute for Tumor Biology and Experimental Therapy, Georg-Speyer Haus, Frankfurt). sgp130-Fc transgenic mice were previously described 41. For the EAE experiments reported in supplementary Fig. 10, opt_sgp130-Fc mice were used. To generate mice with cell-type specific excision of floxed cassettes, floxed mice were bred with CD11c Cre [Tg(Itgax-cre)1-1Reiz], CD4 Cre [Tg(Cd4-cre)1Cwi], CD19 Cre [Cd19tm1(cre)Cgn] or LysM Cre [Lyz2tm1(cre)Ifo], respectively (all from Jackson Laboratory). To visualize Cre-expressing cells, for some experiments a Cre reporter allele Gt(ROSA)26Sortm1(EYFP)Cos was crossed in. All mouse strains were on pure C57BL/6 background. Animals were kept in a specific pathogen-free facility at the TU Munich or Johannes Gutenberg University Mainz. All experimental protocols were approved by the standing committee for experimentation with laboratory animals of the Bavarian or Rhine Palatinate state authorities and carried out in accordance with the corresponding guidelines (AZ 55.2-1-54-2532-29-13, AZ 55.2-1-54-2532-95-14 and 23 177-07/G 13-1-099).
Induction of EAE
To induce EAE, mice were immunized s.c. (base of tail) with 200 µl of an emulsion containing 200 µg MOG35-55 (MEVGWYRSPFSRVVHLYRNGK; Auspep, Tullamarine, Australia) and 500 µg M. tuberculosis H37Ra (BD Difco) in CFA and received 200 ng PTx (Sigma) i.v. on the same day and 2 d after the immunization. Clinical signs of disease were monitored as described previously 42. To ablate Il6 reporter (Thy1.1) expressing cells, mice were treated with i.p. injections of anti-Thy1.1 antibodies (19E12) or isotype control antibodies (mouse IgG2a; C1.18.4). For in vivo neutralization of IFN-γ, mice received anti-IFN-γ antibodies (R4-6A2) or isotype control antibodies (rat IgG1; HRPN). Antibodies were obtained from BioXCell and were injected at a dose of 200 µg starting one day after immunization followed by injections every other day until the development of first signs of disease. For adoptive transfer experiments, mice of the indicated genotypes received 2.5 x 106 naïve GFP (Foxp3)- 2D2 cells i.v. one day prior to immunization.
Preparation of CNS mononuclear cells
At the peak of disease, CNS-infiltrating cells were isolated after perfusion through the left cardiac ventricle with PBS. Brain and spinal cord were dissected and digested with collagenase D (2.5 mg/ml) and DNase I (1 mg/ml) at 37°C for 45 min. After passing the tissue through a 70 µm cell strainer, cells were separated by discontinuous Percoll gradient (70%/37%) centrifugation. Mononuclear cells were isolated from the interphase.
Preparation of naïve T cells and differentiation of T helper cell subsets
Naïve T helper cells were isolated from peripheral lymph nodes and spleens. Naïve T cells were isolated using magnetic beads (Naïve CD4+ T Cell Isolation Kit, Miltenyi Biotec) or subsequent to enrichment of T helper cells (CD4+ T Cell Isolation Kit, untouched, Miltenyi Biotec) by FACS (CD4+CD44lowGFP (Foxp3)-). For T helper cell differentiation, naïve T cells were stimulated for 3 days with plate-bound antibody to CD3 (145-2C11, 4 μg/ml) and soluble antibody to CD28 (PV-1, 2 μg/ml). Recombinant cytokines were added to the differentiation cultures as indicated: human TGF-β1 (0.25 to 2 ng/ml) and/or mouse IL-6 (50 ng/ml), all R&D Systems. Cells were cultured in Dulbecco’s Modified Eagle medium (DMEM) supplemented with 10% FCS, 50 µM β-mercaptoethanol, nonessential amino acids (MEM-NEAA), MEM Vitamins, Folic Acid, 50 U/ml penicillin, 50 µg/ml streptomycin and 0.1 mg/ml Gentamicin. In some experiments, we compared the capacity of soluble IL-6 vs hyper-IL-6 19. Here, we differentiated naïve T cells of the indicated genotypes in the presence of TGF-β (0.1 to 0.5 ng/ml) and equimolar amounts of hyper-IL-6 and human IL-6.
Isolation of DCs
Peripheral lymph nodes and spleens were injected with digestion mix (1mg/ml Collagenase D and 20 μg/ml DNase I [Roche], 2% FCS in HEPES-buffered RPMI-1640). Organs were fragmented and incubated at 37°C for 45 min. To disrupt cell complexes, 10 mM EDTA was added for additional 5 min. Cell suspensions were passed through 100 µm cell strainers.
Generation and culture of BMDCs
Bone marrow derived DCs were generated by flushing out femora and tibiae. Viable cells were cultured in clone medium supplemented with GM-CSF and IL-4 (both 20 ng/ml, R&D Systems) for 6 days with splitting every other day. DCs were removed by vigorous pipetting and cultured in the presence of TLR ligands CpG [ODN 1826] from InVivoGen or LPS [E.coli 0111:B4 Lipopolysaccharide], Sigma-Aldrich.
BMDC / T cell co-culture
Naïve 2D2 T cells were pre-cultured overnight (at a ratio of 1:4) with splenocytes or BMDCs from Il6flox/flox mice, Il6-/- mice, or Il6ΔDC mice in the presence of 50 µg/ml MOG35-55. For some experiments naïve 2D2 and OT II T cells were pre-cultured with BMDCs of the indicated genotypes in the presence of either MOG35-55 or OVA323-339 (ISQAVHAAHAEINEAGR; Auspep, Tullamarine, Australia). LPS (0.5 µg/ml) was added and cells were harvested at the indicated time points for analysis. In some experiments, Marimastat (3 μM, Sigma-Aldrich) was added to block IL-6Rα shedding. To analyze different IL-6 signaling modes, blocking agents were added in equimolar concentrations: anti-IL6 (Polyclonal Goat IgG, R&D or monoclonal mAb#8, Ref. 43), anti-IL6R (MR16-1), and sgp130-Fc 14.
Flow cytometry and intracellular cytokine staining
Cells were stained with live/dead fixable dyes (Aqua [405 nm exc] or Near-IR [633 nm exc], Invitrogen) and antibodies to surface markers: CD3 (clone 145-2C11), CD4 (GK1.5 or RM4-5), CD11b (M1/70), CD11c (HL3), CD19 (6D5), CD25 (PC61), CD44 (IM7), CD45.1 (A20), CD45.2 (104), CD69 (H1.2F3), Thy1.1 (CD90.1, Ox-7), CD96 (3.3), CD103 (2E7), CD126 (IL-6Rα, D7715A7), CD130 (gp130, KGP130), CD172 (SIRPα, P84), 2D2 TCR Vα3.2 (RR3-16) Vβ11 (RR3-15), Ly6G (1A8 ), MHC II (M5/114.15.2), NK1.1 (PK136), SiglecH (eBio440c); all BD Biosciences, eBioscience or BioLegend. For intracellular cytokine staining, cells were re-stimulated with 50 ng/ml PMA (Sigma-Aldrich), 1 μg/ml ionomycin (Sigma-Aldrich) and monensin (1 μl/ml BD GolgiStop) at 37 °C for 2.5 h. Subsequent to live/dead and surface staining, cells were fixed and permeabilized (Cytofix/Cytoperm and Perm/Wash Buffer; BD Biosciences), and stained for cytokines IL-17A (TC11-18H10.1; BioLegend), IFN-γ (XMG1.2, eBioscience), IL-10 (JES5-16E3, BD Biosciences), and GM-CSF (MP1-22E9, BD Biosciences). For intracellular staining of IL-6 in BMDCs, cells were stimulated with TLR ligands for 3 h before adding 5 μg/ml Brefeldin A (Sigma-Aldrich) for further 2 h. Subsequent to live/dead and surface staining, fixation and permeabilization, cells were stained for IL-6 (MP5-20F3, BD). For intranuclear staining of Foxp3 (FJK-16s, eBioscience), cells were stained for live/dead discrimination and surface markers. Fixation, permeabilization, and intranuclear staining were performed according to the manufacturer’s instructions (Transcription Factor Staining Buffer Set, eBioscience). To analyze Stat3 activation, cells were fixed with Phosflow Lyse/Fix Buffer (BD) for 10 min at 37 °C, permeabilized with PhosFlow Perm Buffer III (BD) for 30 min on ice and stained simultaneously for surface markers and Tyrosine 705 phosphorylated Stat3 (4/P-STAT3, BD) according to the manufacturer’s instructions. Cells were analyzed using a CyAn ADP 9 flow cytometer (Beckmann/Coulter) and FlowJo software (Tree Star).
Detection of antigen specific T cells ex vivo
MOG specific cytokine producing T helper cells were analyzed around day 7 to 8 after immunization. Single cell suspensions from draining lymph nodes or spleens were restimulated with 30 μg/ml MOG35-55 for 6 h in the presence of 5 μg/ml Brefeldin A during the last 3 h of incubation followed by surface and intracellular cytokine staining as described above. MOG specific T cells were detected via staining of fixed and permeabilized cells with biotinylated anti-CD40L (CD154, clone MR1, eBioscience) and fluorochrome-coupled Streptavidin.
ELISA
IL-6 amounts in cell culture supernatant and in the serum of immunized mice were quantified using standard sandwich ELISA (R&D Systems, DuoSet). Standard curves and sample concentrations were calculated based on the mean of triplicates for each dilution or sample.
Quantitative PCR analysis
For quantitative PCR, RNA was extracted using RNeasy columns (Qiagen). cDNA was transcribed using TaqMan Reverse Transcription Reagents as recommended (Applied Biosystems) and used as template for quantitative PCR. Primer plus FAM-labelled probe mixtures were obtained from Applied Biosystems (Il1b: Mm00434228_m1; Il6: Mm99999064_m1; Il12a (p35): Mm00434165_m1; Il12b (p40): Mm01288989_m1; Il23a (p19): Mm00518984_m1; Eomes: Mm01351985_m1; Rorc: Mm00441144_g1; Tbx21: Mm00450960_m1). The gene expression was normalized to the expression of β-actin (ACTB, VIC-labelled probe).
RNA Seq
Total RNA was isolated from FACS sorted 2D2 GFP (Foxp3)- T cells re-isolated from draining lymph nodes after priming in Il6flox/flox host mice, Il6-/- host mice, or Il6ΔDC host mice. Three biological replicates were prepared for each condition. Total RNA was further purified with AmpureXP beads to remove impurities detected by UVvis spectrometry. This RNA was used to construct sequencing libraries using the Encore Complete RNA-Seq library system (NuGen Inc. Santa Clara, USA) according to the instructions of the manufacturer. In brief 125 ng of total RNA were treated with heat-labile ds DNAse (Thermo Scientific Inc., MA, USA) to remove residual DNA and used for mixed random-/polyA-primed first-strand cDNA synthesis. After second strand synthesis the double-stranded cDNA was fragmented by sonication (Covaris M220; power 50 W, duty factor 20, 200 cycles p. burst, 115 s treatment time). The fragmented cDNA was end repaired and ligated with sample-specific barcoded adaptors. After strand selection, the library was amplified (4 cycles, 94 °C 30 sec, 55 °C 30 sec, 72 °C 60 sec and 12 cycles 94 °C 30 sec, 63 °C 30 sec, 72 °C 60 sec) and purified and size-selected with AMPure XP beads (Beckman Coulter, CA, USA). Resulting libraries were quantified on a Bioanalyzer 2100 (Agilent, Santa Clara, USA). The barcoded libraries were pooled at 10 nM concentration for multiplexed sequencing. All libraries were sequenced on a HiSeq 1500 as 100 b single reads to a depth of 25-30 Mio raw reads per sample. After demultiplexing of sequence data, adaptor sequences and polyA tails were removed and each library was mapped to the murine reference genome mm10 with the spliced-read aligner STAR 44. Based on the mapping results, sequence reads for annotated genes were counted with HTSeq 45. Comparative analyses of gene expression were performed with the DEseq2 package 46 as pairwise comparison of two groups for each combination of interest. The false discovery rate for detection of differentially abundant transcripts was set to 5 %.
For gene set enrichment analysis (GSEA) a ranked list of the fold change of RNA Seq read values or nanostring signals between wild primed and Il6ΔDC primed effector T cells isolated from draining lymph nodes was calculated. The java GSEA Desktop Application v2.2.1, was used in conjunction with the Molecular Signatures Database v45.1 to run the analysis. Data analysis of differentially enriched genes in wild type-primed and Il6ΔDC primed Foxp3- 2D2 T cells was performed through the use of QIAGEN’s Ingenuity® Pathway Analysis (IPA®, QIAGEN Redwood City,www.qiagen.com/ingenuity). The Molecule Activity Predictor (MAP) tool was used to investigate the effect of IL-6 signaling pathways on identified IL-6 target genes.
Immunofluorescence, proximity ligation assay (PLA), and confocal microscopy
Naïve 2D2 T cells were cultured at a ratio of approximately 4:1 with CellTracker Deep Red (Molecular Probes) labeled BMDCs in chamber slides in the presence of MOG35-55 to form immunological synapses. Subsequent to LPS stimulation for 80 min, cells were fixed with ice-cold methanol, blocked and permeabilized in PBS containing 10% normal goat serum, 1% BSA and 0.2% Triton X-100. Incubation with primary antibody specific for IL-6 (polyclonal goat IgG, R&D) diluted in PBS supplemented with 1% BSA and 0.1% Tween 20 was carried out overnight at room temperature. Secondary Antibody (chicken anti-goat IgG_AF 488 conjugate; Invitrogen) was diluted in PBS supplemented with 1% BSA and 0.1% Tween 20 and incubated for one hour at room temperature. Samples were mounted with ProLong Gold Antifade Reagent with DAPI (Molecular Probes) for nuclear counterstaining.
To visualize IL-6-IL-6Rα complexes, a proximity ligation assay (PLA) was performed according to the manufacturer’s instructions (Duolink In Situ Red Kit Goat/Rabbit, Sigma-Aldrich) using primary antibodies specific for IL-6 (polyclonal goat IgG, R&D) and IL-6Rα (polyclonal rabbit IgG M-20, Santa Cruz).
Confocal image stacks were recorded on a confocal microscope (Olympus FV1000) equipped with x60/1.42 N.A. oil-immersion objective. All images were acquired sequentially. Maximum intensity projections and xz representations were generated using the open-source image analysis software Fiji 47. Gamma was adjusted nonlinearly to enhance visibility of morphological details.
Statistical analysis
Statistical evaluations of cell frequency measurements, cell numbers, mRNA amounts, and protein levels were performed with the unpaired Student’s t test when two populations were compared. Two-tailed p values < 0.05 were considered significant. Multiple comparisons were performed with one-way ANOVA followed by post-hoc multiple comparisons tests as indicated in the legends to the figures. EAE scores between groups were analyzed as disease burden per individual day with one-way-ANOVA and post-hoc test as indicated.
Data availability statement
For materials, protocols, and supporting data please place requests to the corresponding author TK (thomas.korn@tum.de).
Supplementary Material
Acknowledgments
The authors thank all members of the Korn laboratory for their various inputs. We also thank B. Kunze, B. Lunk and colleagues for expert mouse care. We are grateful to B. Stockinger (MRC National Institute for Medical Research, London) for kindly providing the 19E12 hybridoma and to M. Kopf (ETH, Zürich) for providing Il21r-/- mice. This work was supported by the Deutsche Forschungsgemeinschaft (TRR128 to AW and TK, TRR156 to AW, WA1600/8-1 to AW, SFB1054-B07 to TK, SyNergy to TK, SFB877-A01 to SJR, SFB877-A10 to Christoph Garbers (CG), and the cluster of excellence “Inflammation at interfaces” to SRJ and CG) and by the ERC (CoG 647215 to TK). JH is supported by SAF2011-23272 and SAF2014-56546-R.
Footnotes
Author contributions
S.H. designed, performed and analyzed most experiments and drafted the manuscript. N.Y. performed and analyzed key in vivo experiments, C. Garbers, M.H., L.A., V.H., A.L.C., K.M.-H., and T.R. performed or contributed to specific experiments. C. Gasperi performed bioinformatic analysis. H.S.B. and K.S. performed and analyzed nanostring experiments. S.K. and H.B. performed and analyzed RNAseq experiments. T.W., B.H., T.M., J.H., M.O., S.R-J., M.S.-S. provided reagents, advice, design, and supervision to experiments. A.W. supervised experiments, analyzed data, and wrote the manuscript. T.K. conceptualized the study, designed and supervised the experiments, analyzed data, and wrote the manuscript.
The authors declare no competing financial interest.
References
- 1.Hall AO, et al. The Cytokines Interleukin 27 and Interferon-γ Promote Distinct Treg Cell Populations Required to Limit Infection-Induced Pathology. Immunity. 2012;37:511–523. doi: 10.1016/j.immuni.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neufert C, et al. IL-27 controls the development of inducible regulatory T cells and Th17 cells via differential effects on STAT1. Eur J Immunol. 2007;37:1809–1816. doi: 10.1002/eji.200636896. [DOI] [PubMed] [Google Scholar]
- 3.Huber M, et al. IL-27 inhibits the development of regulatory T cells via STAT3. Int Immunol. 2008;20:223–234. doi: 10.1093/intimm/dxm139. [DOI] [PubMed] [Google Scholar]
- 4.Dardalhon V, et al. IL-4 inhibits TGF-beta-induced Foxp3+ T cells and, together with TGF-beta, generates IL-9+ IL-10+ Foxp3(-) effector T cells. Nat Immunol. 2008;9:1347–1355. doi: 10.1038/ni.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bettelli E, et al. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441:235–238. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- 6.Serada S, et al. IL-6 blockade inhibits the induction of myelin antigen-specific Th17 cells and Th1 cells in experimental autoimmune encephalomyelitis. Proc Natl Acad Sci USA. 2008;105:9041–9046. doi: 10.1073/pnas.0802218105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghoreschi K, et al. Generation of pathogenic T(H)17 cells in the absence of TGF-β signalling. Nature. 2010;467:967–971. doi: 10.1038/nature09447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee Y, et al. Induction and molecular signature of pathogenic TH17 cells. Nat Immunol. 2012;13:991–999. doi: 10.1038/ni.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaublomme JT, et al. Single-Cell Genomics Unveils Critical Regulators of Th17 Cell Pathogenicity. Cell. 2015;163:1400–1412. doi: 10.1016/j.cell.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boulanger MJ, Chow D-C, Brevnova EE, Garcia KC. Hexameric structure and assembly of the interleukin-6/IL-6 alpha-receptor/gp130 complex. Science (New York NY) 2003;300:2101–2104. doi: 10.1126/science.1083901. [DOI] [PubMed] [Google Scholar]
- 11.Babon JJ, Varghese LN, Nicola NA. Inhibition of IL-6 family cytokines by SOCS3. Semin Immunol. 2014;26:13–19. doi: 10.1016/j.smim.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tamura T, et al. Soluble interleukin-6 receptor triggers osteoclast formation by interleukin 6. Proc Natl Acad Sci USA. 1993;90:11924–11928. doi: 10.1073/pnas.90.24.11924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rose-John S, Heinrich PC. Soluble receptors for cytokines and growth factors: generation and biological function. Biochem J. 1994;300(Pt 2):281–290. doi: 10.1042/bj3000281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jostock T, et al. Soluble gp130 is the natural inhibitor of soluble interleukin-6 receptor transsignaling responses. Eur J Biochem. 2001;268:160–167. doi: 10.1046/j.1432-1327.2001.01867.x. [DOI] [PubMed] [Google Scholar]
- 15.Korn T, et al. IL-21 initiates an alternative pathway to induce proinflammatory T(H)17 cells. Nature. 2007;448:484–487. doi: 10.1038/nature05970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Durant L, et al. Diverse targets of the transcription factor STAT3 contribute to T cell pathogenicity and homeostasis. Immunity. 2010;32:605–615. doi: 10.1016/j.immuni.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baran P, Nitz R, Grötzinger J, Scheller J, Garbers C. Minimal interleukin 6 (IL-6) receptor stalk composition for IL-6 receptor shedding and IL-6 classic signaling. Journal of Biological Chemistry. 2013;288:14756–14768. doi: 10.1074/jbc.M113.466169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palacios R, Steinmetz M. Il-3-dependent mouse clones that express B-220 surface antigen, contain Ig genes in germ-line configuration, and generate B lymphocytes in vivo. Cell. 1985;41:727–734. doi: 10.1016/s0092-8674(85)80053-2. [DOI] [PubMed] [Google Scholar]
- 19.Fischer M, et al. I. A bioactive designer cytokine for human hematopoietic progenitor cell expansion. Nat Biotechnol. 1997;15:142–145. doi: 10.1038/nbt0297-142. [DOI] [PubMed] [Google Scholar]
- 20.White J, et al. The V beta-specific superantigen staphylococcal enterotoxin B: stimulation of mature T cells and clonal deletion in neonatal mice. Cell. 1989;56:27–35. doi: 10.1016/0092-8674(89)90980-x. [DOI] [PubMed] [Google Scholar]
- 21.Söderberg O, et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods. 2006;3:995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- 22.Linker RA, et al. IL-6 transsignalling modulates the early effector phase of EAE and targets the blood-brain barrier. J Neuroimmunol. 2008;205:64–72. doi: 10.1016/j.jneuroim.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 23.Nurieva R, et al. Essential autocrine regulation by IL-21 in the generation of inflammatory T cells. Nature. 2007;448:480–483. doi: 10.1038/nature05969. [DOI] [PubMed] [Google Scholar]
- 24.Suto A, Wurster AL, Reiner SL, Grusby MJ. IL-21 inhibits IFN-gamma production in developing Th1 cells through the repression of Eomesodermin expression. J Immunol. 2006;177:3721–3727. doi: 10.4049/jimmunol.177.6.3721. [DOI] [PubMed] [Google Scholar]
- 25.Yagi R, et al. The transcription factor GATA3 actively represses RUNX3 protein-regulated production of interferon-gamma. Immunity. 2010;32:507–517. doi: 10.1016/j.immuni.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Awasthi A, et al. Cutting edge: IL-23 receptor gfp reporter mice reveal distinct populations of IL-17-producing cells. J Immunol. 2009;182:5904–5908. doi: 10.4049/jimmunol.0900732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chu CQ, Wittmer S, Dalton DK. Failure to suppress the expansion of the activated CD4 T cell population in interferon gamma-deficient mice leads to exacerbation of experimental autoimmune encephalomyelitis. J Exp Med. 2000;192:123–128. doi: 10.1084/jem.192.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dubois S, Mariner J, Waldmann TA, Tagaya Y. IL-15Ralpha recycles and presents IL-15 In trans to neighboring cells. Immunity. 2002;17:537–547. doi: 10.1016/s1074-7613(02)00429-6. [DOI] [PubMed] [Google Scholar]
- 29.Plun-Favreau H, et al. The ciliary neurotrophic factor receptor alpha component induces the secretion of and is required for functional responses to cardiotrophin-like cytokine. EMBO J. 2001;20:1692–1703. doi: 10.1093/emboj/20.7.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wuest SC, et al. A role for interleukin-2 trans-presentation in dendritic cell-mediated T cell activation in humans, as revealed by daclizumab therapy. Nat Med. 2011;17:604–609. doi: 10.1038/nm.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tussiwand R, Gautier EL. Transcriptional Regulation of Mononuclear Phagocyte Development. Front Immun. 2015;6:533. doi: 10.3389/fimmu.2015.00533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lewis KL, et al. Notch2 receptor signaling controls functional differentiation of dendritic cells in the spleen and intestine. Immunity. 2011;35:780–791. doi: 10.1016/j.immuni.2011.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Croxford AL, Mair F, Becher B. IL-23: One cytokine in control of autoimmunity. Eur J Immunol. 2012;42:2263–2273. doi: 10.1002/eji.201242598. [DOI] [PubMed] [Google Scholar]
- 34.Tanaka T, et al. Enhancement of T helper2 response in the absence of interleukin (IL-)6; an inhibition of IL-4-mediated T helper2 cell differentiation by IL-6. Cytokine. 2001;13:193–201. doi: 10.1006/cyto.2000.0828. [DOI] [PubMed] [Google Scholar]
- 35.Diehl S, Rincón M. The two faces of IL-6 on Th1/Th2 differentiation. Mol Immunol. 2002;39:531–536. doi: 10.1016/s0161-5890(02)00210-9. [DOI] [PubMed] [Google Scholar]
- 36.Kim JH, et al. High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS ONE. 2011;6:e18556. doi: 10.1371/journal.pone.0018556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quintana A, et al. Astrocyte-specific deficiency of interleukin-6 and its receptor reveal specific roles in survival, body weight and behavior. Brain Behav Immun. 2013;27:162–173. doi: 10.1016/j.bbi.2012.10.011. [DOI] [PubMed] [Google Scholar]
- 38.Korn T, et al. Myelin-specific regulatory T cells accumulate in the CNS but fail to control autoimmune inflammation. Nat Med. 2007;13:423–431. doi: 10.1038/nm1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bettelli E, et al. Myelin oligodendrocyte glycoprotein-specific T cell receptor transgenic mice develop spontaneous autoimmune optic neuritis. J Exp Med. 2003;197:1073–1081. doi: 10.1084/jem.20021603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wunderlich FT, et al. Interleukin-6 signaling in liver-parenchymal cells suppresses hepatic inflammation and improves systemic insulin action. Cell Metab. 2010;12:237–249. doi: 10.1016/j.cmet.2010.06.011. [DOI] [PubMed] [Google Scholar]
- 41.Rabe B, et al. Transgenic blockade of interleukin 6 transsignaling abrogates inflammation. Blood. 2008;111:1021–1028. doi: 10.1182/blood-2007-07-102137. [DOI] [PubMed] [Google Scholar]
- 42.Korn T, et al. IL-6 controls Th17 immunity in vivo by inhibiting the conversion of conventional T cells into Foxp3+ regulatory T cells. Proc Natl Acad Sci USA. 2008;105:18460–18465. doi: 10.1073/pnas.0809850105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brakenhoff JP, Hart M, De Groot ER, Di Padova F, Aarden LA. Structure-function analysis of human IL-6. Epitope mapping of neutralizing monoclonal antibodies with amino- and carboxyl-terminal deletion mutants. J Immunol. 1990;145:561–568. [PubMed] [Google Scholar]
- 44.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
For materials, protocols, and supporting data please place requests to the corresponding author TK (thomas.korn@tum.de).