Abstract
RNA G-quadruplex (G4) structures are thought to affect biological processes, including translation and pre-mRNA splicing, but it is not possible at present to demonstrate that they form naturally at specific sequences within long functional RNA molecules. We have developed a novel strategy, footprinting of long 7-deazaguanine substituted RNAs (FOLDeR), that allows the formation of G4s to be confirmed in long RNAs and in functional conditions.
G-rich sequences in DNA or RNA have the potential to form four-stranded structures known as G-quadruplexes (G4)1. DNA G4 formation has been demonstrated in vivo2 and implicated in a very wide range of gene expression processes, notably telomere maintenance, transcription and genome instability3. RNA G4s exist in vivo4 and have been implicated in various post-transcriptional processes5–10. They would be expected to form more readily in RNA than in DNA, but it is still difficult to predict whether they do form in any given RNA sequence.
G4s have been suggested previously to be of functional significance in pre-mRNA splicing. Most of these studies used similar approaches, using bioinformatic tools to identify well-characterized G4-forming sequences and biophysical methods to show that the predicted G4 can form in isolated short sections of RNA11–13. However, such sequences are naturally part of longer pre-mRNAs in which the propensity of a sequence to form a G4 is determined by competition with secondary structures and protein binding. Other methods for identifying G4s include the use of ligands or antibodies that bind selectively to G4s4,13,14, but a general hazard is that the binding reagent affects the equilibrium between different structures and thus might induce the formation of otherwise unstable non-functional G4s. Mutagenesis is often used for confirmation of G4 formation in vivo12,15, but mutants need to be designed so as not to affect the binding of regulatory proteins or perturb likely secondary structures.
We describe here a method in which we probed the secondary structure of an entire functional unit of pre-mRNA, comparing native RNA with 7-deazaguanine (7-deaza-G) substituted RNA, which can form the same secondary structures but not G4s16. Differences highlighted regions in which G4 might form. These differences were confirmed in functional splicing conditions by ribonuclease H digestion in nuclear extracts. This method allowed the identification of functionally relevant G4s.
The splicing reaction we have studied is that of the human B-cell lymphoma-extra large (Bcl-x) pre-mRNA, which has two alternative 5’ splice sites (5’SS) in its exon 2. The major isoform, Bcl-xL (XL), is an anti-apoptotic factor, while the alternative isoform, Bcl-xS (XS), is pro-apoptotic17. There are a number of guanine-rich (G-rich) sequences in the regions around the Bcl-x alternative splice sites that could form G4s.
We designed a functional splicing transcript, Bcl-x-681, by preserving the sequence between the alternative XS and XL 5’SS, shortening the intron and the exon 3 and adding an additional 5’SS at the 3’end of the construct (Fig. 1a). Splicing of this transcript substantially favored the Bcl-xL isoform, recapitulating the preference for Bcl-xL observed in HeLa cells18 (Fig. 1b, Supplementary Results, Supplementary Fig. 1).
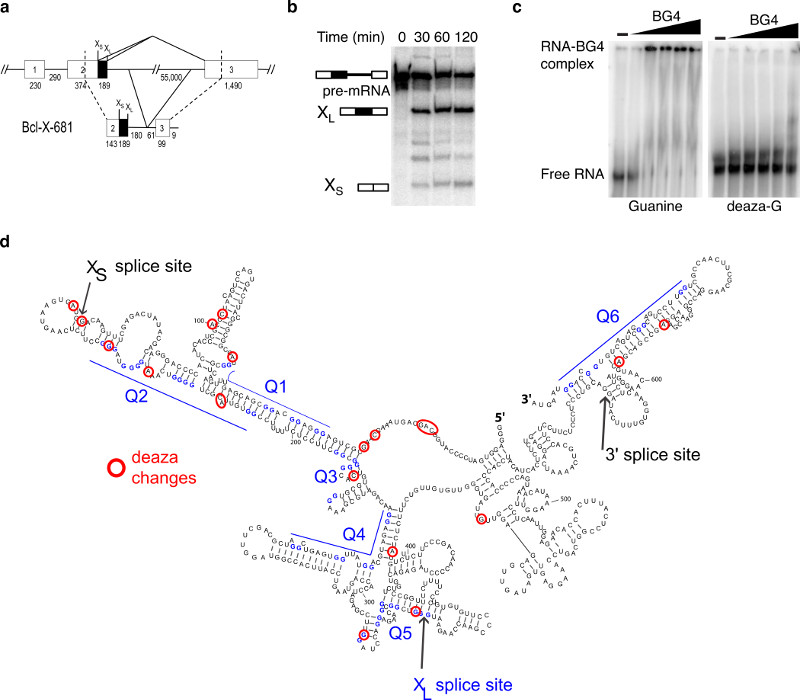
Figure 1. The functional Bcl-x-681 RNA contains G4s in vitro.
(a) Schematic representation of native Bcl-x (top) and Bcl-x-681 transcript (bottom) used in this study. The size of introns and exons are indicated in nucleotides below the diagrams. XL and XS 5’ splice sites are indicated above the diagrams. (b) In vitro splicing assay of Bcl-x-681. Bands corresponding to the unspliced transcript, the XL and the XS products are labelled. The full-length gel is displayed in Supplementary Fig. 11 (c) Electrophoretic-mobility shift assay of native (left) and 7-deaza-G substituted (right) Bcl-x-681 in the absence or presence of increasing amount of BG4 antibody (0, 150 pM, 1.5 nM, 15 nM, 150 nM and 1.5 μM). The full-length gel is displayed in Supplementary Fig. 11. (d) Mapping of RNA footprinting of Bcl-x-681. Nucleotides having a different footprinting pattern upon 7-deaza-GTP substitution are circled and are located mainly to the XS and XL domains. Putative G4s are labelled Q1 to Q6.
The presence of G4s in this functional RNA was assessed by an electrophoretic mobility shift assay (EMSA) using a previously described G4-specific antibody, BG42,4, that induced a shift in the migration of the Bcl-x-681 RNA (Fig. 1c, left). This transcript contains 6 G-rich sequences predicted to potentially form a G419, one upstream of the XS splice site (Q1), three between the XS and the XL splice sites (Q2, Q3 and Q4), and one downstream of the 3’ splice site (3’SS) (Q6). All of these short sequences displayed a circular dichroism spectrum that is typical for parallel G4s, with a positive signal at 265 nm and a negative signal at 240 nm (Supplementary Fig. 2). The stability of these G4s ranged from 37°C (Q4) to >70°C (Q2). However, these experiments did not provide information on whether these sequences do form G4 in the face of competing secondary structures in functional Bcl-x pre-mRNA.
Therefore, our strategy for identifying the location of G4 elements in this RNA was to (i) map structured regions using ribonucleases (RNases), and then (ii) repeat the assay with 7-deaza-G RNA, in which G4s would not form16. For (i), we performed RNA footprinting using RNases T1, T2 and V1, providing structural restraints at nucleotide resolution (Supplementary Figs. 3 and 4). In total, we defined 163 and 91 nucleotides as single-stranded and double-stranded, respectively (Supplementary Fig. 5, Supplementary Dataset 1). We used these restraints with a secondary structure prediction software (Mfold20) to derive the best fit model (Supplementary Fig. 6). This model predicted that the Bcl-x-681 RNA is highly structured and suggested the presence of four structurally independent domains, denoted as XS, XL, intron and 3’ss domains according to the splicing elements they contained (Supplementary Fig. 6). We transcribed each structural domain separately and RNA footprints of each domain showed a very similar pattern to that of the full-length RNA (Supplementary Fig. 7), supporting the structural model.
To identify potential G4s within this structure, we replaced all guanines by 7-deaza-G during in vitro transcription. The absence of the N7 in the 7-deaza-G was shown previously to abolish the possibility of Hoogsteen base pairing and thus of G4 formation, whilst still permitting Watson-Crick base-pairing and the formation of stem-loops16. We confirmed the absence of G4s in the 7-deaza-G Bcl-x-681 RNA by EMSA. As expected, the antibody BG42,4 did not induce a shift in the migration of the 7-deaza-G RNA (Fig. 1c, right).
We performed RNA footprinting on the 7-deaza-G RNA (Supplementary Fig. 4) and a comparison between the RNA footprinting of the 7-deaza-G-substituted transcripts and the native transcript was assessed by calculating an index factor for each nucleotide corresponding to (V1 7-deaza-G / V1 native) / (T2 7-deaza-G / T2 native) (Supplementary Dataset 2). The logarithms of the index were distributed as a Gaussian with a mean of -0.0416 and a standard deviation of 0.3051 (Supplementary Fig. 8). We considered the nucleotides whose index value had changed by more than two standard deviations from the mean (corresponding in this case to a difference of approximately three-fold or more in structural probing between the native and the 7-deaza-G RNA) to be disproportionately affected by 7-deaza-G substitution. Most of these were located in the XS and the XL domains, and many of these nucleotides coincided with two of the possible G4 regions (Q2, and Q5) (Fig. 1d). We therefore concluded that the Bcl-x-681 transcript contains G4s located near the XS and XL splice sites. Previous reports identified G4s by comparing structural probing of RNAs in the presence of either potassium (K+) or Lithium (Li+) ions8, although on much shorter sequences than Bcl-x-681. For comparison, we performed an RNA footprinting experiment of Bcl-x-681 in the presence of Li+ or K+ ions (Supplementary Dataset 3) and calculated an index as (V1 K+ / V1 Li+) / (T2 K+ / T2 Li+). By analysing the distribution of the logarithms of these scores as above, we found a mean of -0.02 and a standard deviation of 0.17. Only 19 nucleotides showed a change beyond two standard deviations, and these were not concentrated around potential G4s (Supplementary Fig. 9). These results showed that the use of 7-deaza-G substitution is more sensitive than the comparison of structural probing in K+ and Li+ containing buffers.
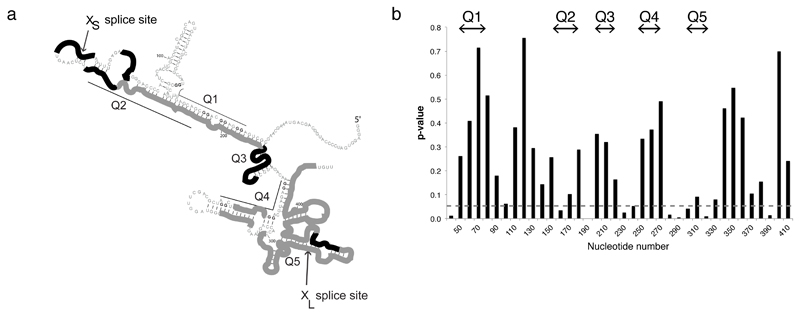
A major benefit of using 7-deaza-G RNA in structural probing is that, in contrast to the comparison between Li+ and K+ buffers, it can be done in functional conditions, such as in nuclear extracts. Therefore, we tested the accessibility of native and 7-deaza-G modified Bcl-x-681 RNAs using RNAse H cleavage directed by 10-mer DNA oligonucleotides in functional splicing conditions in nuclear extracts (Supplementary Fig. 10). We focused our analysis on the XS, and XL domains. We assigned protected or accessible regions on the basis of either < 40% or > 60% cleavage, respectively (Supplementary Fig. 10). The RNAse H cleavage patterns of native RNA in nuclear extract were in good agreement with the structure predicted in solution (Fig. 2a). Putative stems were protected in both the XS and XL domains, while several of the predicted loops were accessible. To compare the RNAse H patterns of the native and the 7-deaza-G RNA, each transcript was analysed three times and a Student’s t test was done for each oligonucleotide to identify whether there were significant differences between the native and deaza RNAs. The result (Fig. 2b) showed that most of the significant differences were found near the putative G4s, especially Q2 and Q5, consistent with the footprinting results. These data demonstrated that the use of 7-deaza-G RNA can identify G4s in functional splicing conditions.
Figure 2. The Bcl-x-681 RNA contains G4s in functional conditions.
(a) Schematic drawing of the RNAse H cleavage of native Bcl-x-681 on the secondary structure model of Bcl-x-681. Accessible regions (average >60% cleavage) are in black, and protected regions (average <40% cleavage) are in grey. RNAse cleavage experiments were done in triplicate and are presented in supplementary Figure 9A. (b) Significance of changes in RNAse H cleavage between 7-deaza-G and native RNA in nuclear extract. Three experiments were done on each transcript and, for each oligonucleotide, a Student’s t test was done to establish the probability that the two sets of results might come from the same population. Values of p below 0.05 indicate that the cleavage of the two transcripts was significantly affected by deaza substitution. Representative experiments on the two transcripts are shown in Supplementary Figure 10.
In conclusion, we have described FOLDeR, a new method that allowed us to demonstrate the presence of G4s within long RNAs by using 7-deaza-G RNA modifications in combination with secondary structure probing (Fig. 1). By coupling footprinting with ribonuclease H cleavage assays on the two different types of transcript, it was also possible to validate the findings in functional conditions, such as nuclear extracts (Fig. 2). Although we demonstrated the power of this method using nuclease digestions, its validity relies on the modification of the RNA and not on the structural probing method. Therefore, the use of 7-deaza-G RNAs to probe G4s could also be done in combination with other structural probing methods such as selective 2′-hydroxyl acylation analysed by primer extension (SHAPE) chemistry21 or dimethyl sulphate (DMS) footprinting22. It is now becoming clear that RNA G4s have a higher propensity to form than their DNA counterparts. Recently, structural data revealed the presence of RNA G4s that could not be predicted by bioinformatics tools13,23,24,25. The development of an experimental method for identifying and locating G4s is therefore a major step forward. The existence of G4s in a functional RNA can be tested using the BG4 antibody4 and their location established using 7-deaza-G-RNA as described here.
Online Methods
In Vitro Transcription of RNA
A transcription reaction containing 40 mM Tris (pH 7.5), 20 mM MgCl2, 10 mM NaCl, 2 mM spermidine HCl, 10 mM DTT, 4 mM NTPs, 1 μg DNA template (PCR product), 5% RNaseOut (Invitrogen), and 5% T7 polymerase (1:20 dilution) was incubated at 37 °C for 4 h. DNA fragments were removed using an S-300 column (GE Healthcare). The product was extracted by phenol-chloroform and precipitated by ethanol. The pellet was dissolved in the desired amount of TE buffer and stored at –20 °C. For radiolabelled RNA, A transcription reaction containing 40 mM Tris (pH 7.5), 6 mM MgCl2, 10 mM NaCl, 2 mM spermidine HCl, 0.5 mM ATP, 0.5 mM CTP, 0.5 mM UTP, 0.05 mM GTP, 50 ng DNA template (PCR product), 4% RNaseOut, 10% T7 (1:20 dilution), 1 mM Ribo m7G Cap analogue (Promega) and 0.33 μM [α-32P] GTP (10 mCi/ml, 3000 Ci/mmol) (Pelkin Elmer) was incubated at 37 °C for 1.5 h. Samples were then purified via a denaturing gel.
In vitro transcription of 7-deaza RNA
A transcription reaction containing 40 mM Tris (pH 7.5), 20 mM MgCl2, 10 mM NaCl, 2 mM spermidine HCl, 10 mM DTT, 4 mM NTPs (c7GTP instead of GTP), 1 μg DNA template (PCR product), 5% RNaseOut, 2 mM Ribo m7G Cap analogue (Promega) and 5% T7 (1:20 dilution) was incubated at 37 °C for 4 h. DNA fragments were removed using an S-300 column (GE Healthcare). The product was extracted by phenol-chloroform and precipitated by ethanol. The pellet was dissolved in the desired amount of TE buffer and stored at –20 °C. For radiolabelled deaza RNA, A transcription reaction containing 40 mM Tris (pH 7.5), 6 mM MgCl2, 10 mM NaCl, 2 mM spermidine HCl, 0.5 mM ATP, 0.5 mM CTP, 0.5 mM UTP, 0.05 mM GTP, 50 ng DNA template (PCR product), 4% RNaseOut, 10% T7 (1:20 dilution), 1 mM Ribo m7G Cap analogue (Promega) and 0.33 μM [α-32P] GTP (10 mCi/ml, 3000 Ci/mmol) (Pelkin Elmer) was incubated at 37 °C for 1.5 h. Samples were then purified via a denaturing gel.
In Vitro Splicing of RNA
Samples containing 1.53 mM ATP, 20 mM CrPi, 3.2 mM MgCl2, 20 mM Hepes (pH 7.5), 50 mM KGlu, 0.125% NP-40, 50% nuclear extract (Cilbiotech) and 10–20 cps of radiolabelled RNA transcript were set up in 10 μl reactions on ice then incubated at 30 °C for 2 h in a pre-heated waterbath. Aliquots of 2 μl were taken into a microtitre plate on dry ice at timepoints 0, 30, 60 and 120 min. Samples were then treated by addition of 50 µl of 0.4 mg/ml proteinase K and incubation for 10 mins at 37 °C. Samples were precipitated twice with 100% ethanol, and dissolved in 10 μl formamide dyes. To prepare for loading on an 8% denaturing polyacrylamide gel, samples were placed in boiling water for 30 sec and run until bromophenol blue has just run out of the gel. Gels were then fixed, dried and exposed to a phosphorimaging screen overnight. Quantification of pre-mRNA and both mRNA products was done using OptiQuant software, and intensities were normalised to account for the number of guanines in each RNA species.
RNase probing
Samples containing 400 ng RNA (either native or 7-deaza-substituted), 0.5 mg/ml yeast tRNA, and either 150 mM KCl, 1.5 mM MgCl2 and 20 mM Hepes pH 7.6 (K+ containing buffer) or 150 mM KCl, 1.5 mM MgCl2 and 20 mM Hepes pH 7.6 (Li+ containing buffer) in a total volume of 9 µl, were heated at 65 °C in a water bath for 10 min then slow cooled to 37 °C. 1 µl of either Tris buffer (0.5 mM Tris pH 7.6, 1 M KCl or 1M LiCl, and 25 mM MgCl2), RNase T1 (Roche Diagnostics), RNase T2 (Ambion) or RNase V1 (Mobitech) was added. Reactions were incubated at ambient temperature for 6 min then stopped with 101.8 µl of stop buffer (3.9 mM EDTA and 0.2 µg yeast tRNA) and 101.8 µl phenol-chloroform then spun at 13,500 rpm for 10 min. The top layer was removed and added to 700 µl ethanol (96%), 12 ng glycogen and 22 mM NaAc (pH 3.0). Samples were then ethanol-precipitated and dissolved in 4 µl potassium borate or sterile water.
Reverse transcription of probed RNA
A hybridization mix was made containing 0.25 µl hybridization buffer (reverse transcription buffer without MgCl2), 1 µl of 100 cps/µL radiolabelled primer and 0.25 µl water. 1 µl of probed RNA was incubated with 1.5 µl of hybridization mix at 65 °C for 10 min then cooled on ice. 2.5 µl of extension mix (0.1 µl 5 mM dNTPs, 0.25 µl reverse transcriptase buffer, 0.25 µl 1/10 diluted AMV reverse transcriptase (MP Biomedical) and 1.9 µl water) was added to each probed RNA mix and incubated at 42 °C for 30 min. 3 µl formamide dye was added and the sample was heated to 96 °C for 2 min, and then put on ice for 2 min. 2 µl was loaded onto 7% DNA sequencing gel and run for 2 h at 100 W. Gels were dried and exposed to a phosphorimaging screen overnight and quantified using SAFA. The data were analysed using a cleavage index to determine the change in accessibility of each nucleotide in the RNA, and the mean and standard deviation of this index were used as described in the Results to identify nucleotides that showed a significantly greater change. Footprinting of the native RNA was done twice and the structural model was confirmed by further footprinting experiments on the isolated structural domains.
RNase H assays
RNase H cleavage was done in splicing conditions by adding each of 44 DNA oligonucleotides at 10 µM after incubation for 30 min and continuing incubation for 5 min. Reactions were processed as for splicing reactions. Three independent sets of experiments were done for each RNA. The extent of cleavage was measured using a phosphorimager a Student’s t-test was performed to evaluate the significance of the changes between the 7-deaza-G and the native RNA triplicate experiments.
Electromobility Shift assays (EMSA)
The BG4 antibody was purchased from Absolute Antibody (Ab00174-1.1). Radiolabelled RNA at 10 count per seconds was incubated with various concentrations (0, 150 pM, 1.5 nM, 15 nM, 150 nM and 1.5 μM) of BG4 antibody in a microtitre plate. 3 µl of native gel dye was added, and 4 µl was run on a small 5% native poyacylamide gel at 4°C. The gel was run at 5 W for 1 h. Gels were dried and exposed to a phosphorimager screen overnight.
Circular Dichroism
RNA sequences corresponding to Q1, Q2, Q3, Q4, Q5, and Q6 were purchased from Dharmacon, GE Healthcare, deprotected according to the manufacturer instructions, lyophilised, and resuspended in 20mM KH2PO4, pH7, 100mM KCl at a final concentration of 20μM. Circular dichroism spectra were acquired on a Chirascan spectrometer at 20°C and data were collected between 350 and 220nm in triplicate and averaged. Buffer baseline was substracted for each spectrum. CD Melting curves were obtained by measuring the CD signal at 265nm from 20 to 80°C at 1°C heating rate.
Supplementary Material
Acknowledgments
This work was supported by a Medical Research Council Career Development Award (G1000526) to C.D. and a Sir Dudley Spurling Post Graduate Scholarship from the Bank of Butterfield Foundation in Bermuda to C.W. This work was also funded by CNRS and Lorraine University (UMR 7365 and previously 7214) and the European Alternative Splicing Network of Excellence (EURASNET, FP6 life sciences, genomics and biotechnology for health; LSHG-CT-2005-518238).
Footnotes
Author Contributions
C.W. performed all experiments under the guidance of I.C.E. and C.D. Footprinting experiments and structural model calculation were performed under the guidance of I.B.A. and C.B. G.A.B. and L.H.H. contributed to the development and the validation of the strategy. C.W., I.C.E., and C.D. interpreted the results and wrote the manuscript.
The authors declare no competing financial interests
References
- 1.Huppert JL. FEBS J. 2010;277:3452–3458. doi: 10.1111/j.1742-4658.2010.07758.x. [DOI] [PubMed] [Google Scholar]
- 2.Biffi G, Tannahill D, McCafferty J, Balasubramanian S. Nat Chem. 2013;5:182–186. doi: 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rhodes D, Lipps HJ. Nucleic Acids Res. 2015;43:8627–8637. doi: 10.1093/nar/gkv862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biffi G, Di Antonio M, Tannahill D, Balasubramanian S. Nat Chem. 2014;6:75–80. doi: 10.1038/nchem.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Agarwala P, Pandey S, Maiti S. Org Biomol Chem. 2015;13:5570–5585. doi: 10.1039/c4ob02681k. [DOI] [PubMed] [Google Scholar]
- 6.Millevoi S, Moine H, Vagner S. WIREs RNA. 2012;3:495–507. doi: 10.1002/wrna.1113. [DOI] [PubMed] [Google Scholar]
- 7.Kumari S, Bugaut A, Huppert JL, Balasubramanian S. Nat Chem Biol. 2007;3:218–221. doi: 10.1038/nchembio864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Darnell JC, et al. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- 9.Decorsiere A, Cayrel A, Vagner S, Millevoi S. Genes Dev. 2011;25:220–225. doi: 10.1101/gad.607011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wolfe AL, et al. Nature. 2014;513:65–70. doi: 10.1038/nature13485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marcel V, et al. Carcinogenesis. 2011;32:271–278. doi: 10.1093/carcin/bgq253. [DOI] [PubMed] [Google Scholar]
- 12.Ribeiro MM, et al. Hum Genet. 2015;134:37–44. doi: 10.1007/s00439-014-1485-6. [DOI] [PubMed] [Google Scholar]
- 13.Smith LD, et al. Cell Rep. 2014;9:193–205. doi: 10.1016/j.celrep.2014.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zizza P, et al. Nucleic Acids Res. 2015;44:1579–1590. doi: 10.1093/nar/gkv1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Munroe SH, Morales CH, Duyck TH, Waters PD. PLoS ONE. 2015;10:e0137893. doi: 10.1371/journal.pone.0137893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murchie AI, Lilley DM. Nucleic Acids Res. 1992;20:49–53. doi: 10.1093/nar/20.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boise LH, et al. Cell. 1993;74:597–608. doi: 10.1016/0092-8674(93)90508-n. [DOI] [PubMed] [Google Scholar]
- 18.Garneau D, Revil T, Fisette J-F, Chabot B. J Biol Chem. 2005;280:22641–22650. doi: 10.1074/jbc.M501070200. [DOI] [PubMed] [Google Scholar]
- 19.Kikin O, D'Antonio L, Bagga PS. Nucleic Acids Res. 2006;34:W676–W682. doi: 10.1093/nar/gkl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zuker M. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. J Am Chem Soc. 2005;127:4223–4231. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- 22.Kwok CK, Ding Y, Tang Y, Assmann SM, Bevilacqua PC. Nature Comms. 2013;4:1–12. doi: 10.1038/ncomms3971. [DOI] [PubMed] [Google Scholar]
- 23.Phan AT, et al. Nat Struct Mol Biol. 2011;18:796–804. doi: 10.1038/nsmb.2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang H, et al. Nat Chem Biol. 2014;10:686–691. doi: 10.1038/nchembio.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Warner KD, et al. Nat Struct Mol Biol. 2014;21:658–663. doi: 10.1038/nsmb.2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.