Abstract
Background & Aims
The inflammatory bowel diseases (IBD) ulcerative colitis (UC) and Crohn’s disease (CD) cause significant morbidity and are increasing in prevalence among all populations, including African Americans. More than 200 susceptibility loci have been identified in populations of predominantly European ancestry, but few loci have been associated with IBD in other ethnicities.
Methods
We performed 2 high-density, genome-wide scans comprising 2345 cases of African Americans with IBD (1646 with CD, 583 with UC, and 116 inflammatory bowel disease unclassified [IBD-U]) and 5002 individuals without IBD (controls, identified from the Health Retirement Study and Kaiser Permanente database). Single-nucleotide polymorphisms (SNPs) associated at P<5.0×10−8 in meta-analysis with a nominal evidence (P<.05) in each scan were considered to have genome-wide significance.
Results
We detected SNPs at HLA-DRB1, and African-specific SNPs at ZNF649 and LSAMP, with associations of genome-wide significance for UC. We detected SNPs at USP25 with associations of genome-wide significance associations for IBD. No associations of genome-wide significance were detected for CD. In addition, 9 genes previously associated with IBD contained SNPs with significant evidence for replication (P<1.6×10−6): ADCY3, CXCR6, HLA-DRB1 to HLA-DQA1 (genome-wide significance on conditioning), IL12B, PTGER4, and TNC for IBD; IL23R, PTGER4, and SNX20 (in strong linkage disequilibrium with NOD2) for CD; and KCNQ2 (near TNFRSF6B) for UC. Several of these genes, such as TNC (near TNFSF15), CXCR6, and genes associated with IBD at the HLA locus, contained SNPs with unique association patterns with African-specific alleles.
Conclusions
We performed a genome-wide association study of African Americans with IBD and identified loci associated with CD and UC in only this population; we also replicated loci identified in European populations. The detection of variants associated with IBD risk in only people of African descent demonstrates the importance of studying the genetics of IBD and other complex diseases in populations beyond those of European ancestry.
Keywords: SNP, genetic analysis, Risk factor, Trans-Ethnic
Inflammatory bowel disease (IBD), a chronic intestinal inflammatory disorder, affects over 1.4 million people in the US alone and is a significant burden on resources with healthcare costs estimated at greater than $6 billion/year1. Historically a disease of the developed world and European ancestry populations, recent years have seen a rising prevalence in non-European populations including African-Americans (AAs). Association studies have previously identified 200 genome-wide significant (GWS) IBD susceptibility loci in European ancestry populations2, 3. At least thirty-five loci have been identified in Asians and a handful appear Asian specific3–7.
We recently performed the first large-scale evaluation of established IBD genetic loci in the understudied African American (AA) population, using the Immunochip genotyping platform (Illumina San Diego, CA), in 1,511 cases and 1,797 controls4. We found significant replication in AAs for maximal established SNPs at 5 European loci for IBD and CD (FCGR2A and PTGER4 for IBD; IL23R, NOD2, and IKZF3 for CD) and for UC at HLA rs9271366, congruent with the maximal East Asian HLA association. We also observed strong association signals at PTGER4, IL12B, and STAT3A/STAT5 for SNPs independent of established European association signals. African-specific IBD risk SNPs (i.e. SNPs present only in African populations or in populations with African ancestry, and monomorphic or unknown in other 1000 genome populations) were detected for STAT3A/STAT5. No loci, however, had SNPs with evidence of GWS association nor were any African-specific loci established, although we found suggestive evidence (p<4×10−5 with consideration for the relatively small number of SNPs evaluated) for 3 potential novel loci (i.e. C2orf43, HDAC11, and LINC00994)4.
It is hoped that genetic advances will enable more personalized approaches to managing IBD. Given that significant differences in pathological and molecular mechanisms may exist in AAs, who have a higher risk for developing disease complications in IBD and worse disease outcome, it is imperative to use more comprehensive genotyping platforms in more highly powered samples to detect population specific IBD loci and associations. 8–11. We hypothesized that high-density GWAS of IBD in AAs could identify population specific variants, further define IBD genetic architecture, and expose novel disease mechanisms.
MATERIALS AND METHODS
Study design
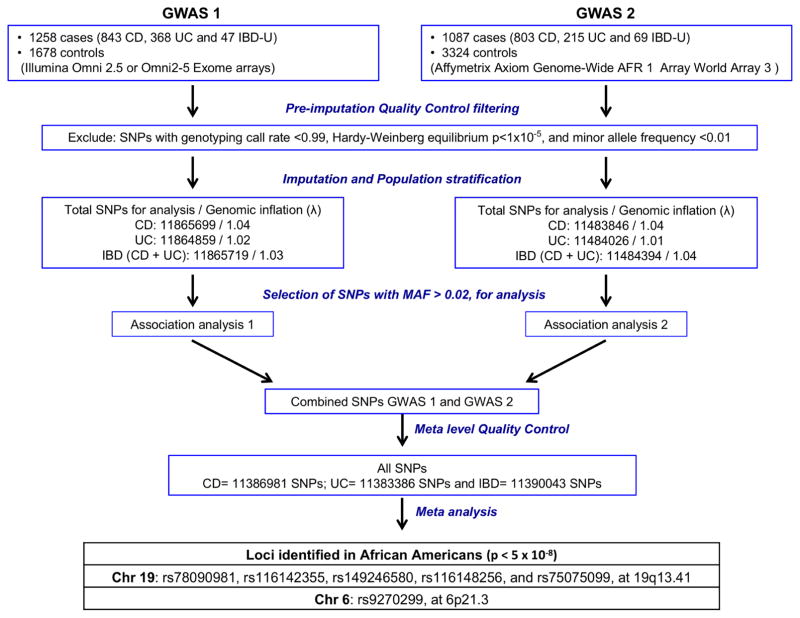
We conducted two GWAS using independent case-control datasets, totaling 2345 AA IBD cases (1646 CD, 583 UC and 116 IBD-U) and 5002 controls population (Figure 1) from unrelated, self-identified AAs individuals. Samples with IBD from GWAS1 (n=1258 IBD cases [843 CD, 368 UC, 47 IBD-U]) were recruited by Johns Hopkins Multicenter African American IBD Study (MAAIS) (coordinated by Johns Hopkins IBD Genetics Research Center [GRC] of the NIDDK IBD Genetics Consortium [IBDGC] with recruitment from 13 collaborating IBD centers and 4 other IBDGC GRCs12) and at Cedars-Sinai Medical Center IBD Center. Control AA subjects of GWAS1 (n=1678) were derived from the dbGaP Health and Retirement Study (HRS), a longitudinal panel study sponsored by the National Institute on Aging. Samples with IBD from GWAS2 (n=1087 IBD cases [803 CD, 215 UC, 69 IBD-U]) were obtained by Emory University from the GENESIS study (an ancillary study of the NIDDK IBDGC, coordinated by Emory University with recruitment of IBD cases and matched controls from 12 of their collaborating IBD centers and the RISK study, a large pediatric CD inception cohort with recruitment of IBD cases from 29 IBD centers13). The GWAS2 AA control subjects (n=3324) were obtained from the Kaiser RPGEH study (a research program at Kaiser Permanente in California with the goal of discovering which genes and environmental factors linked to specific diseases).
Figure 1.
Experiment design flowchart. Two independent GWAS were performed and included 1258 cases/1678 controls (GWAS 1 genotyped on Illumina Omni2.5) and 1087 cases/3324 controls (GWAS 2 genotyped on Affymetrix Axiom Genome-Wide AFR 1 Array). After quality control and imputation based association analysis of CD, UC and IBD for each GWAS, a combined meta-analysis of observed and imputed SNPs identified 6 SNPs above genome threshold that are associated with UC in African Americans.
Genotyping, Quality Control (QC) and Population Ancestry
All DNA samples for GWAS1 were genotyped on the Illumina Omni 2.5 (~2.3 million SNPs) or Omni 2.5 Exome (~2.6 million SNPs) arrays according to the manufacturer’s protocol. The two channel raw data files (.idat) for all samples were transferred to a central location and assembled into a single project for joint genotype calling. Samples for GWAS2 were genotyped on the Affymetrix Axiom Genome-Wide AFR 1 World Array 3 (African array) (Affymetrix, Inc., Santa Clara, California) according to the manufacturer’s protocol. The World Array 3 contains ~894,000 SNPs optimized for individuals of African ancestry.
For each GWAS, samples with low call rates (<97.5%) were excluded. SNPs that failed the HWE test (p<0.00001) in the controls were removed. We tested for agreement between X/Y genotypes and sex, and for unexpected relatedness between individuals by applying RELPAIR and GRR14 to 10,000 – 20,000 markers in linkage equilibrium (Pairwise r2<0.1) evenly distributed across the genome. Samples with discordant gender were excluded from analyses. For pairs of individuals that appeared to be genetically related, one of the pair was removed from subsequent analyses. One member from all first and second degree relative pairs (r >= 0.25) was dropped. To investigate population structure and identify population group outliers, we used principal components analysis (PCA). Beginning with all SNPs that passed QC, we first filtered down to a small (~20,000) subset of SNPs with moderate minor allele frequency and no linkage disequilibrium (LD) (r2<0.1). We inferred principal components (PCs) for all samples of each phase cohort using the method proposed by Price et al15 as implemented in the software package EIGENSTRAT. Samples were plotted in PC space, and outliers that exceeded thresholds of <−0.05 or >0.05 for both PC1 or PC2 were rejected (Supplementary figure 1). In all, we excluded 121 samples in GWAS1 and 126 samples in GWAS2. The genomic control values for CD, UC and IBD were 1.04, 1.02 and 1.03 in the GWAS1 analysis, and 1.04, 1.01 and 1.04 in the GWAS2 analysis, respectively, indicating little evidence of population stratification after controlling for global ancestries and suggesting that there was no inflation of false positives from confounding by ancestry. PCs were included as covariates in our downstream analyses.
Imputation-based association analysis, meta-analysis and conditional analysis
Before imputation, an initial quality control was performed separately on each set of case-control datasets using Plink16. The initial step included filtering out SNPs with genotyping call rate <0.99, Hardy-Weinberg equilibrium p<1×10−5, and minor allele frequency <0.01. Additionally, a filter that excluded all A/T or G/C SNPs was applied. To improve the coverage of genetic variants, the datasets were separately imputed to the 1000 Genomes Project Phase 3 integrated autosomal reference panel, using the software IMPUTE217. GWAS1 included 2.47 million genotyped SNPs imputed to 11.9 million SNPs and GWAS2 included 893,815 SNPs imputed to 11.5 million SNPs. After excluding SNPs with low imputation quality, indels and copy number variants (CNVs), imputed genotypes were combined with the observed sample genotype data set for association analysis. Genotype imputation clouds (i.e. the full genotype probability values, not single point estimates) from IMPUTE2 were directly used to assess association with SNPTEST (version 2.5.2 or a later)18 under an additive model (-frequentist 1, -method score parameter options). Ten PCs were included in all SNPTEST analyses. For each GWAS, association analyses were performed separately for UC, CD and IBD (CD and UC combined). Association P-values of <5×10−8 (corresponding to a genome-wide significance level of 0.05 after a Bonferroni correction for multiple testing of 1M SNPs) were considered statistically significant. A total of ~11.9 million and ~11.5 million markers were analyzed from the Illumina Omni 2.5 and Axiom arrays, respectively.
To combine data between the cohorts of GWAS1 and GWAS2, all results were meta-analyzed with the program METAL (versions 1.7 or later)19, using an inverse-variance, fixed-effects model, after controlling for residual test statistic inflation via genomic control. SNPs with minor allele frequency less than 0.01 or imputation quality scores less than 0.5 were excluded. Following exclusions, 11.4 milion SNPs were available for meta-analysis. The meta-analysis was well powered to identify common variants with OR ≥ 1.3. Owing to numerous single SNP associations within the extended linkage disequilibrium (LD) of HLA region (Supplementary figure 2A), we performed an exploratory analysis conditioned on SNP rs9270299 (a non-synonymous coding variant [c.179A>G, p.Ala29Thr]) to narrow associations to those with the best evidence for strength and independence. The conditional association analysis was performed using SNPTEST 2.5.218. SNPs were annotated using annovar20.
Validation of ZNF649 genotypes
The 6 SNPs most highly associated within the ZNF649 locus were in perfect LD with each other (r2~1). SNP rs75075099 and rs75577191 were genotyped by Taqman assay in 96 randomly selected samples. Genomic DNA of the 96 randomly selected AAs with UC was quantitated via UV absorbance using Nanodrop 1000 (Thermo Scientific, Wilmington, DE, USA) and 10 nanogram of DNA was used for allelic discrimination using TaqMan SNP genotyping assay (assay IDs C__27836655_10 and C__25965275_10, Applied Biosystems, Foster City, CA, USA). Genotypes were determined automatically using the ABI Prism 7900HT SDS software suite (SDS version 2.4, Applied Biosystems, Foster City, CA, USA). We compared the genotype probability from the imputation to the genotyped observed by the Taqman assay. Genotypes for 85 and for 9 DNAs from the 96 randomly selected AAs were high confidence homozygote reference and heterozygotes, respectively, and these were all confirmed by Taqman genotyping for both SNPs. The remaining 2 samples were only 20–30% confidence heterozygotes. One was confirmed by Taqman genotyping.
Significance
Significant genome-wide association was defined using the accepted standard of p≤5×10−8, a Bonferroni corrected association of p≤0.05, corrected for 1 million independent tests with the additional requirement of nominal association (p≤0.05) in each genotyping array. Given that we had two independent GWAS and genotyped datasets, we set an additional criterion of GWS association with internal replication to define a more rigorous meta-analysis association evidence than that observed in a single GWAS. This was developed as follows: the meta-analysis of the two GWASs incorporated 11.4 million observed and imputed markers present across both studies. With ~11 million SNPs analyzed in each GWAS, for a most conservative criterion (by not taking into account LD among SNPs), we would expect approximately 715 unassociated SNPs to achieve p<6.5×10−5 by chance. In a replication study of 715 SNPs, Bonferroni multitest corrected significance would correspond to p<7×10−5. Thus, setting a threshold of p<6.5×10−5 in both GWAS is comparable to taking forward SNPs with p<6.5×10−5 from a discovery study, and then demanding Bonferroni corrected significance for the carried forth SNPs in a replication study. Therefore, we defined GWS association with internal replication as SNPs achieving GWS of p≤5×10−8 in the meta-analysis and p≤6.5×10−5 in each individual GWAS.
Significant evidence for locus replication within 250 kb of known loci was defined as p≤1.5×10−6 as follows: Given 200 established IBD loci, there will be 100,000kb of genome incorporated within 250 kb of each association peak (500kb combined on each side). Given the estimated size of the autosomal genome at 2.9 million kb, 3.4% of the genome would be encompassed within 250 kb of all 200 loci. Taking 1 million independent SNPs as that being the standard for multiple test correction for (autosomal) GWS of 0.05, approximately 34,000 independent SNPs would be present amongst these 200 loci by chance, and association at a p-value of 0.05 would hence be corrected to 1.5×10−6 for any SNPs that are detected in these regions for IBD. Association corrected for all 11.4 million imputed SNPs at 0.05 significance would be 1.3×10−7, but this may be considered overly conservative given that the majority of SNPs are not independent (for example the 6 ZNF649 GWS SNPs as noted, were in perfect LD).
RESULTS
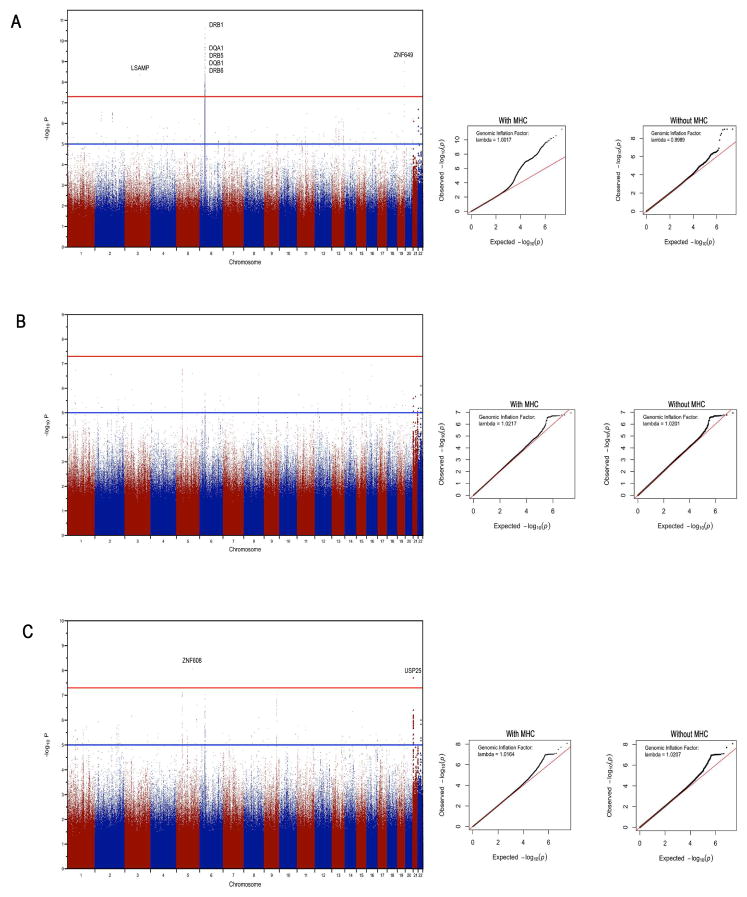
The top associations per locus for UC, CD and IBD are listed in Table 1. From the meta-analysis of UC, 5 SNPs on chromosome 19 and 41 SNPs in the HLA region achieved GWS with internal replication (Figure 2A and Supplementary Table 1). The chromosome 19 SNPs map to the transcriptional repressor ZNF649 (Supplementary figure 3) and overlap ZNF649 antisense RNA1 (ZNF649-AS1). A second novel GWS association was detected at African-specific variant rs72947885 in the axonal neuronal adhesion molecule gene LSAMP (Supplementary Figure 2B) in UC (meta p=4.5×10−9) but did not meet criteria for internal replication. In fact, SNP rs72947885 was genotyped on the Axiom array (see cluster plot in Supplementary figure 4) but not on the Omni array, its imputation quality score suggests high confidence (0.976 in the Omni dataset and 0.975 in the Axiom dataset) and the concordance rate between the genotyped and imputed data is 0.984. Both the ZNF649 and LSAMP SNPs are specific to Africans (i.e. present only in African populations or in populations with African ancestry, and monomorphic or unknown in other populations) as noted by the 1000 Genomes Project Phase3 population allele frequencies (Table 1). A side-by-side comparison of Q-Q plots with and without the HLA variants demonstrated that results follow the null expectation. The early upward departure of the observed values was being driven by the HLA signal in UC (Figure 2A and Supplementary Figure 2B). The top associations per locus for UC, CD and IBD are listed in Table 1.
Table 1.
Genome Wide Significant Associations and Maximal Established Loci Replications
Genome Wide Significant Associations and Established Loci Replications for UC, CD and IBD. The 1000Genomes Project phase 3 allele frequencies for various populations is included.
| Phenotyp e |
Chr | Position | A1 | A2 | SNP Name | GWAS1 (Omni2.5) | GWAS2 (Axiom) | Meta-analysis | Nearest Gene(s) | Annotatio n |
Significance Level |
MAF (Minor Allele) |
African Sub-populations with highest MAF |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||||||||||||||||||
| P-Value | Case Freq |
Control Freq |
OR | 95% CI | P-Value | Case Freq |
Control Freq |
OR | 95% CI | OR | P-value | YRI | EAS | EUR | SAS | ||||||||||
| UC | 6 | 32559063 | A | C | rs9270484 | 1.45E-07 | 0.448 | 0.356 | 0.68 | 0.58 – 0.8 | 4.41E-07 | 0.426 | 0.319 | 0.63 | 0.52 – 0.76 | 0.58 | 3.22E-12 | HLA-DRB1 | intergenic | GWS | 0.11 (C) | 0.31 (C) | 0.31 (C) | 0.3 (C) | |
| 6 | 32557435 | A | C | rs9270299 | 5.98E-08 | 0.284 | 0.207 | 0.66 | 0.55 – 0.79 | 1.06E-05 | 0.281 | 0.209 | 0.68 | 0.55 – 0.83 | 0.52 | 2.65E-11 | HLA-DRB1 | exonic | GWS | 0.15 (A) | 0.16 (A) | 0.12 (A) | 0.24 (A) | ||
| 6 | 32552617 | G | A | rs28724138 | 4.62E-07 | 0.412 | 0.487 | 0.74 | 0.62 – 0.87 | 2.75E-06 | 0.572 | 0.482 | 0.70 | 0.58 – 0.84 | 0.57 | 4.70E-11 | HLA-DRB1 | intronic | GWS | 50 (A) | 47 (A) | 47 (G) | 49 (A) | ||
| 19 | 52400451 | G | A | rs78090981 * | 9.65E-06 | 0.054 | 0.026 | 2.16 | 1.46 – 3.21 | 1.54E-05 | 0.051 | 0.022 | 2.42 | 1.55 – 3.78 | 3.64 | 9.93E-10 | ZNF649 | intronic | GWS | 0.06 (A) | 0 (A) | 0 (A) | 0 (A) | YRI - 0.06 (A) | |
| 3 | 115528296 | C | T | rs72947885 * | 5.19E-08 | 0.099 | 0.054 | 1.91 | 1.43 – 2.56 | 8.07E-03 | 0.080 | 0.051 | 1.62 | 1.14 – 2.3 | 2.42 | 4.48E-09 | LSAMP | UTR3 | GWS | 0.07 (T) | 0 (T) | 0 (T) | 0 (T) | LWK - 0.12 (T) | |
| 20 | 62100266 | T | G | rs914467 | 1.72E-02 | 0.101 | 0.071 | 1.46 | 1.11 – 1.93 | 1.23E-06 | 0.140 | 0.093 | 1.59 | 1.21 – 2.09 | 1.97 | 5.91E-07 | KCNQ2 | intronic | Locus Replication | 0.02 (G) | 0.15 (G) | 0.24 (G) | 0.14 (G) | ||
| 9 | 117811778 | C | T | rs114032850 * | 4.04E-05 | 0.078 | 0.042 | 1.91 | 1.38 – 2.64 | 1.78E-02 | 0.065 | 0.041 | 1.61 | 1.1 – 2.38 | 2.06 | 3.68E-06 | TNC | intronic | 0.06 (T) | 0 (T) | 0 (T) | 0 (T) | MAG - 0.09 (T) | ||
| 3 | 101641101 | A | T | rs141365838 | 4.72E-02 | 0.111 | 0.082 | 1.39 | 1.06 – 1.81 | 2.55E-06 | 0.166 | 0.106 | 1.68 | 1.3 – 2.17 | 1.67 | 3.90E-06 | NFKBIZ | intergenic | 0.1 (T) | 0.1 (T) | 0.22 (T) | 0.2 (T) | |||
| 20 | 62308570 | A | G | rs2738782 | 6.15E-06 | 0.152 | 0.099 | 0.61 | 0.48 – 0.78 | 7.20E-01 | 0.162 | 0.167 | 1.03 | 0.8 – 1.33 | 0.45 | 8.21E-06 | RTEL1:TNFRSF6B | intronic | 0.05 (A) | 0.42 (G) | 0.41 (A) | 0.37 (A) | |||
|
| |||||||||||||||||||||||||
| CD | 16 | 50709723 | G | A | rs6596 | 3.55E-05 | 0.058 | 0.032 | 1.91 | 1.42 – 2.56 | 3.14E-04 | 0.062 | 0.049 | 1.28 | 1 – 1.64 | 2.06 | 1.14E-07 | SNX20 | exonic | Locus Replication | 0 (A) | 0 (A) | 0.16 (A) | 0.01 (A) | |
| 5 | 40424426 | A | C | rs6896969 | 5.46E-04 | 0.333 | 0.387 | 1.26 | 1.11 – 1.44 | 4.22E-05 | 0.330 | 0.391 | 1.30 | 1.15 – 1.47 | 1.28 | 1.69E-07 | PTGER4 | intergenic | Locus Replication | 0.38 (A) | 0.14 (C) | 0.41 (A) | 0.45 (C) | ||
| 1 | 67665210 | A | G | rs1569923 | 2.91E-05 | 0.434 | 0.362 | 0.74 | 0.65 – 0.84 | 8.37E-04 | 0.425 | 0.389 | 0.86 | 0.76 – 0.97 | 0.78 | 1.88E-07 | IL23R | intronic | Locus Replication | 0.36 (A) | 0.39 (G) | 0.37 (G) | 0.37 (G) | ||
| 21 | 16799818 | G | A | rs7276764 | 7.76E-03 | 0.277 | 0.314 | 0.84 | 0.73 – 0.96 | 3.01E-05 | 0.276 | 0.320 | 0.81 | 0.71 – 0.92 | 0.76 | 2.59E-06 | USP25 | intergenic | 0.34 (A) | 0.24 (A) | 0.29 (A) | 0.23 (A) | |||
| 2 | 25145173 | T | A | rs59086897 | 9.54E-03 | 0.192 | 0.204 | 1.08 | 0.92 – 1.26 | 4.54E-05 | 0.199 | 0.259 | 1.41 | 1.22 – 1.63 | 1.33 | 4.00E-06 | ADCY3 | intergenic | 0.08 (T) | 0.44 (A) | 0.47 (A) | 0.48 (A) | |||
| 6 | 167541624 | T | C | rs41414848 * | 2.88E-02 | 0.069 | 0.089 | 0.76 | 0.6 – 0.96 | 7.09E-06 | 0.040 | 0.068 | 0.58 | 0.44 – 0.77 | 0.64 | 4.42E-06 | CCR6 | intronic | 0.09 (C) | 0 (C) | 0 (C) | 0 (C) | ESN - 0.12 (C) | ||
| 4 | 106934908 | C | A | rs6854424 * | 3.60E-05 | 0.055 | 0.033 | 1.71 | 1.27 – 2.3 | 2.75E-02 | 0.041 | 0.028 | 1.47 | 1.08 – 1.99 | 1.85 | 5.86E-06 | TBCK | intronic | 0.04 (A) | 0 (A) | 0 (A) | 0 (A) | MAG / MSL - 0.06 (A) | ||
|
| |||||||||||||||||||||||||
| IBD | 21 | 16800087 | C | T | rs7278277 | 1.60E-04 | 0.530 | 0.473 | 0.80 | 0.71 – 0.89 | 1.21E-05 | 0.513 | 0.476 | 0.86 | 0.78 – 0.96 | 0.77 | 1.99E-08 | USP25 | intergenic | GWS | 0.43 (C) | 0.24 (T) | 0.29 (T) | 0.23 (T) | |
| 5 | 40424426 | A | C | rs6896969 | 4.09E-04 | 0.340 | 0.387 | 1.22 | 1.09 – 1.37 | 2.85E-05 | 0.335 | 0.391 | 1.27 | 1.14 – 1.42 | 1.25 | 7.81E-08 | PTGER4 | intergenic | Locus Replication | 0.38 (A) | 0.14 (C) | 0.41 (A) | 0.45 (C) | ||
| 6 | 32594240 | T | A | rs139282044 * | 5.48E-05 | 0.050 | 0.031 | 1.67 | 1.26 – 2.21 | 1.74E-04 | 0.038 | 0.022 | 1.76 | 1.31 – 2.36 | 1.93 | 9.43E-08 | HLA-DQA1 | intergenic | Locus Replication | 0.04 (A) | 0 (A) | 0 (A) | 0 (A) | ESN - 0.08 (A) | |
| 9 | 117811778 | C | T | rs114032850 * | 3.14E-05 | 0.066 | 0.042 | 1.61 | 1.26 – 2.05 | 4.00E-04 | 0.063 | 0.041 | 1.56 | 1.24 – 1.95 | 1.65 | 9.59E-08 | TNC | intronic | Locus Replication | 0.06 (T) | 0 (T) | 0 (T) | 0 (T) | MAG - 0.09 (T) | |
| 5 | 158782684 | C | T | rs11749526 | 3.20E-05 | 0.020 | 0.034 | 0.58 | 0.41 – 0.83 | 9.64E-04 | 0.023 | 0.042 | 0.54 | 0.39 – 0.75 | 0.53 | 2.64E-07 | LOC285626: IL12B | intronic | Locus Replication | 0 (T) | 0.08 (T) | 0.14 (T) | 0.05 (T) | ||
| 3 | 45989578 | C | G | rs55698153 * | 1.82E-05 | 0.050 | 0.075 | 0.64 | 0.51 – 0.81 | 7.67E-03 | 0.037 | 0.049 | 0.75 | 0.57 – 0.98 | 0.60 | 6.94E-07 | CXCR6 | UTR3 | Locus Replication | 0.06 (G) | 0 (G) | 0 (G) | 0 (G) | YRI / ESN / LWK - 0.06 (G) | |
| 2 | 25145173 | T | A | rs59086897 | 3.58E-03 | 0.191 | 0.204 | 1.09 | 0.95 – 1.25 | 3.72E-05 | 0.204 | 0.259 | 1.36 | 1.2 – 1.55 | 1.31 | 1.18E-06 | ADCY3 | intergenic | Locus Replication | 0.08 (T) | 0.44 (A) | 0.47 (A) | 0.48 (A) | ||
| 1 | 67630341 | A | T | rs10889663 | 8.74E-03 | 0.299 | 0.331 | 0.86 | 0.77 – 0.97 | 3.46E-05 | 0.274 | 0.313 | 0.83 | 0.74 – 0.93 | 0.78 | 2.60E-06 | IL23R | intergenic | Observed variant without frequency or population information | ||||||
| 6 | 167541624 | T | C | rs41414848 * | 4.04E-02 | 0.071 | 0.089 | 0.78 | 0.64 – 0.96 | 1.50E-06 | 0.042 | 0.068 | 0.61 | 0.48 – 0.78 | 0.67 | 3.50E-06 | CCR6 | intronic | 0.09 (C) | 0 (C) | 0 (C) | 0 (C) | ESN - 0.12 (C) | ||
| 1 | 120553996 | C | T | rs7530844 | 3.63E-05 | 0.215 | 0.157 | 1.47 | 1.27 – 1.69 | 1.61E-02 | 0.230 | 0.233 | 0.99 | 0.87 – 1.12 | 1.33 | 8.32E-06 | NOTCH2 | intronic | 0.02 (T) | 0.23 (C) | 0.18 (C) | 0.4 (C) | |||
| 5 | 150596303 | C | T | rs13359932 | 1.01E-05 | 0.230 | 0.288 | 0.74 | 0.65 – 0.84 | 5.00E-02 | 0.238 | 0.248 | 0.95 | 0.84 – 1.07 | 0.80 | 9.06E-06 | CCDC69 | intronic | 0.31 (T) | 0 (T) | 0.01 (T) | 0 (T) | |||
Top associations per locus for UC, CD and IBD. The top 3 HLA associations are shown for illustrative purposes.
Chr: Chromosome; A1: tested allele; A2: Alternate allele; Freq: Tested allele frequency; OR: odds ratio; CI: Confidence interval; GWS: Genome-wide significant; MAF: Minor Allele Frequency.
"Locus replication" defined as p<1.5×10−6 at SNP within 250 kb of a maximal association of a previously established locus.
YRI: Yoruba in Ibadan, Nigeria; EAS: East Asian; EUR: European; SAS: South Asian; LWK: Luhya in Webuye, in Kenya; MAG: Mandinka in Gambia; ESN: Esan in Nigeria; MSL: Mende in Sierra Leone; (data from 1000 genome phase 3)
African Specific SNP (monomorphic in EAS, EUR ans SAS)
Figure 2.
Figure 2A, 2B and 2C. Meta-analysis Manhattan plots for UC (2A), CD (2A) and IBD phenotypes, respectively. All SNPs are plotted according to their position on each chromosome on x-axis, against their association on y-axis. The red and blue lines indicate the genome-wide significance (p≤5×10−8) and the suggestive significance threshold (p ≤ 1×10−5), respectively. Genome-wide significant signals are labeled with corresponding gene names. The inset QQ plots shows the observed (y-axis) against the expected (x-axis) distribution of p-values under the null hypothesis with and without MHC.
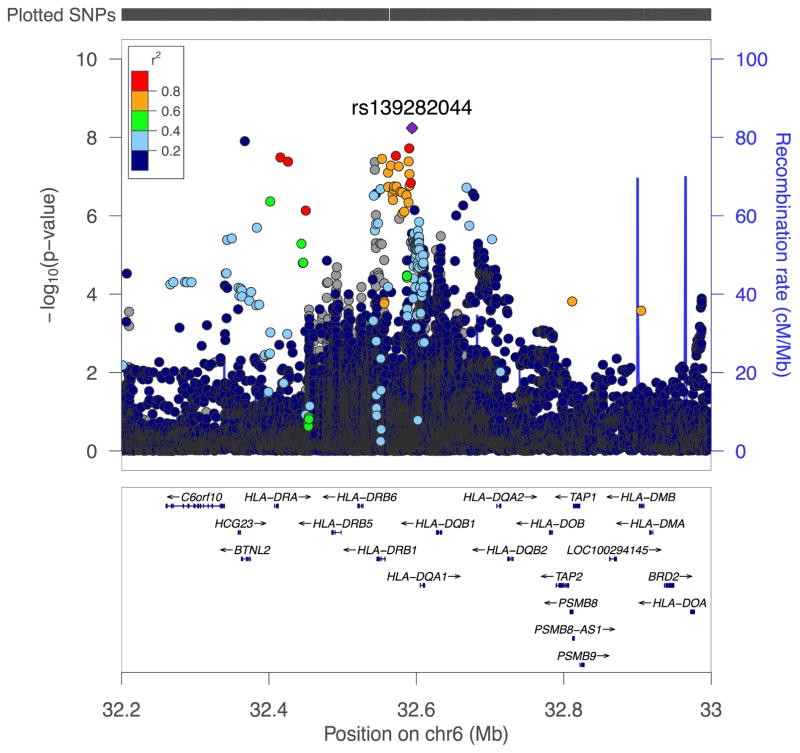
The 3 strongest UC associations are located within the HLA-DRB1 gene. We conditioned association in UC, CD and IBD on the second strongest association, a non-synonymous coding variant (rs9270299, c.179A>G, p.Ala29Thr), in an exploratory analysis to detect potential independent HLA signals (Table 2). Conditioning on rs9270299 in IBD revealed 9 SNPs with GWS for this phenotype located between BTNL2 and HLA-DQA1 (Table 2 and Figure 3), 6 of which are African specific (Table 2). Conditioning in CD and UC did not reveal additional significant associations.
Table 2.
Nine genome-wide significant (p≤5×10−8) variants on chromosome 6 are associated with IBD following conditional analysis on the HLA-DRB1 SNP rs9270299 (from the UC analysis in AA). A1: tested allele; A2: Alternate allele; OR: odds ratio.
| Phenotype | Chr | Position (hg19) | Gene | SNP_Name | A1 | A2 | Combined Freq | Unconditional | Conditional | MAF (Minor Allele) | African Sub-population with highest MAF | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||||||
| OR | Pvalue | OR | P-value | YRI | EAS | EUR | SAS | |||||||||
| IBD | 6 | 32594240 | HLA-DQA1 | rs139282044 * | T | A | 0.031 | 1.93 | 9.43E-08 | 2.08 | 5.78E-09 | 0.04 (A) | 0 (A) | 0 (A) | 0 (A) | ESN - 0.08 (A) |
| 6 | 32367297 | BTNL2 | rs144540865 * | A | G | 0.032 | 1.88 | 1.41E-07 | 2.01 | 1.25E-08 | 0 (G) | 0 (G) | 0 (G) | 0 (G) | LWK - 0.03 (G) | |
| 6 | 32590316 | HLA | rs75441240 * | C | T | 0.032 | 1.84 | 2.89E-07 | 1.97 | 1.91E-08 | 0.04 (T) | 0 (T) | 0(T) | 0 (T) | ESN - 0.08 (T) | |
| 6 | 32572195 | HLA | rs140435271 * | A | G | 0.033 | 1.82 | 4.51E-07 | 1.95 | 2.97E-08 | 0.04 (G) | 0 (G) | 0 (G) | 0 (G) | ESN - 0.08 (G) | |
| 6 | 32415572 | HLA | rs114243387 * | G | C | 0.038 | 1.69 | 4.92E-07 | 1.80 | 3.25E-08 | 0.04 (C) | 0 (C) | 0 (C) | 0 (C) | ESN - 0.08 (C) | |
| 6 | 32553401 | HLA | rs144404284 | G | A | 0.048 | 1.66 | 8.14E-07 | 1.79 | 3.54E-08 | 0.05 (A) | 0.02 (A) | 0.05 (A) | 0.01 (A) | ||
| 6 | 32589502 | HLA | rs114210446 | A | G | 0.033 | 1.80 | 5.73E-07 | 1.92 | 4.11E-08 | 0.05 (G) | 0 (G) | 0.01 (G) | 0 (G) | ||
| 6 | 32425875 | HLA | rs140228374 * | C | T | 0.036 | 1.75 | 6.46E-07 | 1.87 | 4.17E-08 | 0.04 (T) | 0 (T) | 0(T) | 0 (T) | ESN - 0.08 (T) | |
| 6 | 32543575 | HLA | rs41294271 | C | T | 0.210 | 1.36 | 2.72E-06 | 1.45 | 4.27E-08 | 0.09 (T) | Observed variant without frequency or population information | ||||
Genome-wide significant (<5.0×10−8) variants associated with IBD after analysis conditioned on the HLA-DRB1 SNP rs9270299 (from the UC analysis in AA). Nine SNPs on chromosome 6 exceeded genome-wide significance for association with IBD conditioned the HLA-DRB1 SNP rs9270299 (from the UC analysis in AA). Chr: Chromosome; A1: tested allele; A2: Alternate allele; Freq: frequency; OR: odds ratio; MAF: Minor Allele Frequency.
YRI: Yoruba in Ibadan, Nigeria; EAS: East Asian; EUR: European; SAS: South Asian; LWK: Luhya in Webuye, Kenya; ESN: Esan in Nigeria; (data from 1000 genome phase 3)
African Specific SNP (monomorphic in EAS, EUR ans SAS)
Figure 3.
LocusZoom plots of SNPs by chromosome position against –log10 p-value for their genetic associations with IBD phenotype. Conditional regional plot for the HLA locus (chr6p21) for the IBD phenotype shows SNPs reaching genome-wide significance conditioned on SNP rs9270299 from UC analysis. The top SNP is highlighted in purple. The surrounding SNPs, shown within 500kb of the top SNP are color-coded to reflect their linkage disequilibrium in r2 with the top SNP (see inset). Estimated recombination rates are plotted in pale blue to reflect local LD structure on secondary y-axis.
Meta-analysis of CD did not detect any GWS associations (Figure 2B). In IBD, meta-analysis detected two GWS SNPs (s73782531 located near ZNF608 and rs7278277 located near ubiquitin protease USP25, meta p= 8.6×10−9 and p=2.0×10−8, respectively [Figure 2C]. However, for ZNF608 association evidence only came from GWAS1 (p=1.6×10−16 with case and control allele frequencies 0.013 and 0.056, respectively) with no evidence from GWAS2 (p=0.63, and respective allele frequencies 0.043 and 0.44). Hence, for IBD, outside of the HLA signal observed on conditional analysis, only USP25 (Supplementary Figure 2C) met our minimal criteria of at least nominal evidence in both GWAS cohorts, and the ZNF608 association is more likely a false signal.
We detected numerous additional SNPs with significant locus replication (i.e. p<1.6×10−6 within 250 kb of maximal evidence of an established locus) at established European loci (Table 1) within or proximal to genes ADCY3, CXCR6, HLA-DRB1 to HLA-DQA2 (including rs139282044, GWS on conditioning), IL12B, PTGER4, and TNC for IBD; IL23R, PTGER4, and SNX20 (18kb from NOD2 and in LD with NOD2 R702W) for CD; and KCNQ2 (near TNFRSF6B) for UC. Several of these loci contained African specific variants (Table 1).
SNPs with strong evidence of association but below that of locus replication were also detected for multiple additional loci. In CD, African-specific SNPs were observed at rs6854424 (OR=1.85, p=5.86×10−6) 31Kb centromeric of TBCK, a gene involved with regulation of mTOR signaling. In contrast, multiple universal SNPs (i.e. polymorphic in all populations) maximal at rs141365838 (OR 1.67, p=3.90×10−6), were observed in UC at NFKBIZ, an inducible regulator of NF-kB, important for TH17 cell development.2 Interestingly, NFKBIZ was only recently identified as a UC gene via trans-ethnic meta-analysis in Europeans, East Asians, North Indians and Iranians; our association evidence was stronger than that reported for each of these separate study populations (p=9.34×10−6)3. Other locus associations of note included CCR6 for IBD and CD, and NOTCH2 and CCDC69 for IBD. We again observed African specific IBD associations at STAT3, but not as strong in the meta-analysis (i.e. rs12721583 GWAS2 p=5.0×10−6, meta p=8.2×10−5) as in our previous report4. All associations with p-values < 6.5×10−5 in either GWAS are shown in Supplementary Table 1.
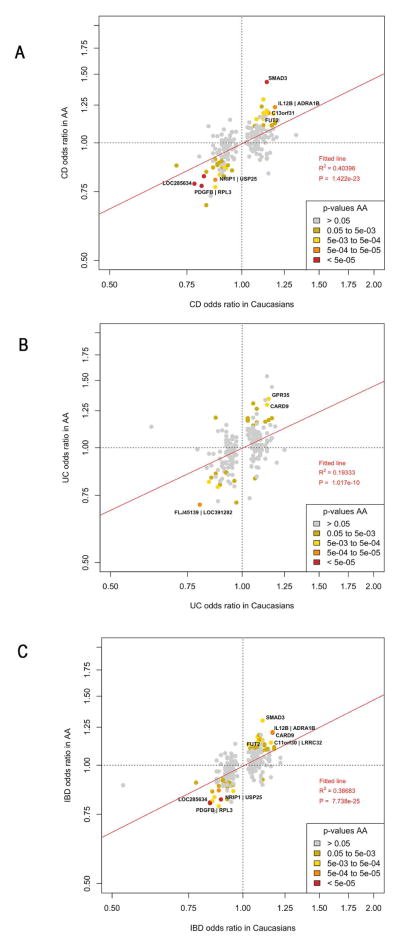
Among the SNPs reported in the updated European and trans-ethnic immunochip study3 we genotyped or successfully imputed 221 of 231 SNPs. Of these, 104 showed evidence of replication (p<0.05). ORs (direction and magnitude) observed in Europeans were excellent predictors for CD, UC and IBD in AA (Figure 4, A, B and C respectively). The full summary statistics of our analysis can be found using the following link: https://www.dropbox.com/sh/s653tyw3yxo4mcc/AADxhOZCvb9VCOn45vvsTic0a?dl=0
Figure 4.
Figure 4(A–C): Odds ratios of SNPs maximally associated in Caucasians versus AAs for CD (4A), UC (4B) and IBD (4C) phenotypes. Red line: best-fitting least-squares regression line.
DISCUSSION
In this first AA GWAS for IBD, we provide the first GWS evidence for AA IBD loci: we identified two novel and African specific GWS UC loci, and elevate our prior UC HLA association in the region of the DRB1 gene to certainty with GWS evidence. We provide GWS HLA association for IBD unrelated to the UC association and driven mostly by African specific polymorphisms; and establish the USP25 locus as GWS in AAs. We also found increased association evidence in AAs for NOD2 (via SNX20), IL23R and PTGER4 loci, above that from our prior, smaller sized immunochip study, and significant locus replication for 4 other loci, ADCY3, CXCR6, TNC, and KCNQ2 – the majority within or adjacent (e.g. KCNQ2) to immune regulatory genes.
ZNF649 acts as a transcriptional repressor and its overexpression suppresses transcriptional activities of the Serum Response Element (SRE) and the Activating protein-1 (AP-1) complex21. AP-1 upregulates pro-inflammatory responses, including TNF-α in IBD22. Among the 5 imputed GWS with internal replication on chromosome 19, 4 SNPs (rs78090981, rs116142355, rs149246580, and rs116148256) map within introns of ZNF649, while 1 (rs75075099) maps downstream of ZNF649 at 19q13.41 (Table 1 and Supplemental figure 3). All 5 SNPs, which are in perfect LD (r2~1), also map within ZNF649 antisense RNA1 (ZNF649-AS1) and are specific to Africans (i.e. present in African populations or in populations with African ancestry and monomorphic or unknown in other populations) and mostly found in Sub-Saharan Africa. The top ranked variant, rs78090981, resides in an intron that is retained in one transcript of the gene (ENST00000599671), and in addition, may alter a regulatory motif for basic leucine zipper ATF-like transcription factor (BATF). The SNP rs75075099 also potentially alters some transcription factor binding sites for Pou5f1, TCF11 and p30023, and is associated with a number of histone marks and DNase I hypersensitivity in a range of cell lines24. Also, as the antisense-RNA gene for ZNF649 overlaps with the ZNF649 gene itself, this is another possible mechanism for regulation. All of these pieces of evidence suggest that ZNF649 or its antisense might be causal genes for this novel susceptibility locus, and targeting ZNF649 may be a potential therapeutic approach.
LSAMP has been characterized as an axonal neuronal adhesion molecule. Its protein product is also expressed in colon and platelets. Interestingly, LSAMP somatic deletions were associated with prostate cancer specifically in AAs25. The top ranked variant, rs72947885, is located in the 3’UTR and also overlaps with several transcriptional regulator motifs and is also reported by Ensembl to be in an enhancer region (ENSR00001988698) in some cell lines including B cells and M1 macrophages from venous blood. However, this variant did not meet the threshold for GWS with internal replication and did not have additional regional variants with strong evidence for association (no variants in strong LD were testable in the meta-analysis) and therefore more evidence from additional studies is required to validate this locus and LSAMP as a candidate gene.
We also showed evidence for independent GWS associations in the HLA region for UC and IBD, as has been shown in other populations. The 3 most highly associated variants, including a non-synonymous coding variant (rs9270299), were located at HLA-DRB1, a gene previously associated with UC. These 3 SNPs are common to all major populations (Table 1) and they are in LD with the previously reported top UC association in our AA Immunochip study, rs92713664. However, SNP rs9271366 (also found as most highly associated in Asian UC) did not pass our QC. To determine if there is initial evidence for independent HLA signals in AA IBD, we conditioned association on the rs9270299 non-synonymous variant, and we detected multiple independent GWS SNP associations (Table 2) in the HLA region, the majority of which are African specific, for IBD. Given the great allelic and LD complexity of the HLA region, our conditional analysis is considered exploratory and far more in-depth analyses will be necessary to better define the independent associations in this region for IBD in AAs. Of note, the top IBD non-conditioned analysis HLA signal (rs139282044, p=9.4×10−8) is represented by the same African specific variant revealed in the conditional analysis. Conditioning on rs9270299 also revealed a separate signal at rs144540865, an uncharacterized rare variant (i.e. observed or found at frequencies <0.5% in non-African population) that resides in BTNL2, a gene associated with IBD in Caucasians26 and with CD in Koreans27 by deep sequencing only. BTNL2 is expressed in mice in the duodenum, ileum, cecum ascending and descending colon28. BTNL2 resides between the MHC class II and class III regions, but is class II associated and appears to regulate T-cell activation. These analyses support parallel observations in Europeans that multiple HLA alleles contribute independently to IBD29.
USP25 is a ubiquitously expressed gene and has been established as an IBD locus in Europeans and shown to be associated with CD in Koreans2, 7. The SNP rs7278277 was not internal to this gene but is in very high LD (r2>0.8) with a number of variants that correlate with a multitude of regulatory regions, including promotors, enhancers and transcription factor binding sites. Two of these SNPs, rs7278277 and rs2242830, showed suggestive evidence of association in GWAS2 for CD and IBD.
We also find evidence for multiple other IBD, CD and UC loci, several driven by African specific polymorphisms and some, like TNC, with unique location relative to those found in other populations. TNC codes for an extracellular matrix glycoprotein involved in epithelial cell migration, intestinal barrier function, arresting T-cell activation and is a marker of IBD activity30. It is located 100kb telomeric to TNFSF8 and 200kb telomeric to TNFSF15, the dominant CD locus in Asians and a significant locus for both CD and UC in Europeans31. The association patterns of diverse populations, as highlighted by the TNFSF15-TNC region, may allow for the fine dissection of the mechanisms of molecular genetic risk for all populations.
Owing to the smaller size of our AA cohort compared to the European cohorts used in previous studies2, 3, we had limited power to assess the extent of allele sharing between the two populations. However, we found evidence that disease variants for nearly half of European IBD loci have at least nominal evidence for an influence on IBD in AAs. This finding supports shared pathogenic mechanisms across different ethnicities.
One seemingly surprising finding is that we were able to identify 3 novel GWS loci for UC, whereas in the three-fold larger CD cases, no GWS loci were identified. Reasons include the dominance of HLA for UC, high risk (OR 3.41) of ZNF649 for UC with no comparable higher OR, single risk variants for CD, and potentially greater allelic and phenotypic heterogeneity underlying CD in AAs. We did however demonstrate significant locus replication for 6 IBD loci, 3 CD loci but only 1 UC loci. We are not aware of any evidence that AA UC is more genetic than CD (i.e. having a greater family history).
In summary, this first GWAS of AA IBD has demonstrated unique, African specific loci, as well as loci that are shared across multiple populations. While some of these shared loci contain unique association patterns and African specific risk variants, many contain universal risk variants (like HLA-DRB1) or risk variants that have arisen from European admixture (like NOD2). Given our results and the evolution of IBD genetics research in non-European populations, it is clear that further studies with larger sample sizes in the AA population are needed to identify additional population specific variants and novel loci, as well as more fully characterize the role of risk variants established in other populations on the development of IBD in AAs. Such research is paramount to allow for the future benefits of IBD genetics research, from risk prediction and family counseling to targeted therapies and eventually disease preventive strategies to be available for the understudied AA population.
Supplementary Material
Acknowledgments
We are grateful to the affected and unaffected individuals who participated in this study. We thank the coordinators and research assistants who helped in the recruitment of subjects. The National Institutes of Health (NIH) Grants DK062431 (S.R.B.), DK087694 (S.K.), DK062413 (D.P.B.M and K.T), DK046763-19, AI067068 and U54DE023789-01 (D.P.B.M.), DK062429, DK062422 (J.H.C.), DK062420 (R.H.D.), DK062432 (J.D.R.), and DK062423 (M.S.S.) supported this study. S.K. was supported by the endowed professorship from Marcus foundation; S.R.B. was supported by grant from Harvey M. and Lynn P. Meyerhoff Inflammatory Bowel Disease Center, the Morton Hyatt Family, the Buford and Linda Lewis family; D.P.B.M. was supported by the Joshua L and Lisa Z Greer Endowed Chair, grant 305479 from the European Union, and The Leona M. and Harry B. Helmsley Charitable Trust; C.L.S was supported in part by the intramural research program of the National Human Genome Research Institute (NIH).
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicts to declare.
AUTHOR CONTRIBUTIONS
S.R.B., D.J.C., J.H.C., D.P.B.M., and S.K. conceived and designed the study. S.R.B., D.T.O., C.L.S., D.J.C., T.H., J.P.B., P.C., F.B., M.E.Z., D.P.B.M. and S.K. performed analysis and interpretation of data. S.R.B., J.P., A.K., C.H., S.V., Z.W., K.T., L.H., J-M.A.K., A.J.Q. J.S., Z.L., J.S.A., R.N.B., S.D., R.K.C., T.D., T.A.D., G.T.D., J.S.H., J.K.H., S.Z.H., L.A.D., J.S.H., D.M., K.L.I., H.K., M.D.K., J.F., R.K., B.S.K., J.F.K., J.H.K., E.L., P.M., D.E.M., R.D.N., B.O.O., A.S.P., S.S., S.R.T., J.F.V., M-H.W., M.L. M.Z., J.D.R., R.H.D., M.S.S., H.H., and D.P.B.M. contributed material, reagents or acquired data. S.R.B., D.T.O., C.L.S., T.H., D.P.B.M. and S.K. wrote the manuscript. S.R.B., D.T.O., C.L.S., T.H., J.H.C., D.P.B.M. and S.K. provided critical revision of the manuscript for important intellectual content. All authors reviewed and approved the manuscript before submission.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
Author names in bold designate shared co-first authorship
- 1.Kappelman MD, Rifas-Shiman SL, Porter CQ, et al. Direct health care costs of Crohn's disease and ulcerative colitis in US children and adults. Gastroenterology. 2008;135:1907–13. doi: 10.1053/j.gastro.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jostins L, Ripke S, Weersma RK, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu JZ, van Sommeren S, Huang H, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47:979–86. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang C, Haritunians T, Okou DT, et al. Characterization of genetic loci that affect susceptibility to inflammatory bowel diseases in African Americans. Gastroenterology. 2015;149:1575–86. doi: 10.1053/j.gastro.2015.07.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Juyal G, Negi S, Sood A, et al. Genome-wide association scan in north Indians reveals three novel HLA-independent risk loci for ulcerative colitis. Gut. 2015;64:571–9. doi: 10.1136/gutjnl-2013-306625. [DOI] [PubMed] [Google Scholar]
- 6.Ye BD, Choi H, Hong M, et al. Identification of Ten Additional Susceptibility Loci for Ulcerative Colitis Through Immunochip Analysis in Koreans. Inflamm Bowel Dis. 2016;22:13–9. doi: 10.1097/MIB.0000000000000584. [DOI] [PubMed] [Google Scholar]
- 7.Yang SK, Hong M, Choi H, et al. Immunochip analysis identification of 6 additional susceptibility loci for Crohn's disease in Koreans. Inflamm Bowel Dis. 2015;21:1–7. doi: 10.1097/MIB.0000000000000268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Basson A, Swart R, Jordaan E, et al. The association between race and Crohn's disease phenotype in the Western Cape population of South Africa, defined by the Montreal Classification System. PLoS One. 2014;9:e104859. doi: 10.1371/journal.pone.0104859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Griglione N, Yarandi S, Srinivasan J, et al. A comparison of abdominal surgical outcomes between African-American and Caucasian Crohn's patients. Int J Colorectal Dis. 2014;29:917–22. doi: 10.1007/s00384-014-1902-2. [DOI] [PubMed] [Google Scholar]
- 10.Sofia MA, Rubin DT, Hou N, et al. Clinical presentation and disease course of inflammatory bowel disease differs by race in a large tertiary care hospital. Dig Dis Sci. 2014;59:2228–35. doi: 10.1007/s10620-014-3160-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malaty HM, Sansgiry S, Artinyan A, et al. Time Trends, Clinical Characteristics, and Risk Factors of Chronic Anal Fissure Among a National Cohort of Patients with Inflammatory Bowel Disease. Dig Dis Sci. 2015 doi: 10.1007/s10620-015-3930-3. [DOI] [PubMed] [Google Scholar]
- 12.Wang MH, Okazaki T, Kugathasan S, et al. Contribution of higher risk genes and European admixture to Crohn's disease in African Americans. Inflamm Bowel Dis. 2012;18:2277–87. doi: 10.1002/ibd.22931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cutler DJ, Zwick ME, Okou DT, et al. Dissecting Allele Architecture of Early Onset IBD Using High-Density Genotyping. PLoS One. 2015;10:e0128074. doi: 10.1371/journal.pone.0128074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abecasis GR, Cherny SS, Cookson WO, et al. GRR: graphical representation of relationship errors. Bioinformatics. 2001;17:742–3. doi: 10.1093/bioinformatics/17.8.742. [DOI] [PubMed] [Google Scholar]
- 15.Price A, Patterson N, Plenge R, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 16.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marchini J, Cutler D, Patterson N, et al. A comparison of phasing algorithms for trios and unrelated individuals. Am J Hum Genet. 2006;78:437–50. doi: 10.1086/500808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang H, Yuan W, Wang Y, et al. ZNF649, a novel Kruppel type zinc-finger protein, functions as a transcriptional suppressor. Biochem Biophys Res Commun. 2005;333:206–15. doi: 10.1016/j.bbrc.2005.05.101. [DOI] [PubMed] [Google Scholar]
- 22.Moriyama I, Ishihara S, Rumi MA, et al. Decoy oligodeoxynucleotide targeting activator protein-1 (AP-1) attenuates intestinal inflammation in murine experimental colitis. Lab Invest. 2008;88:652–63. doi: 10.1038/labinvest.2008.38. [DOI] [PubMed] [Google Scholar]
- 23.Kheradpour P, Kellis M. Systematic discovery and characterization of regulatory motifs in ENCODE TF binding experiments. Nucleic Acids Res. 2014;42:2976–87. doi: 10.1093/nar/gkt1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Romanoski CE, Glass CK, Stunnenberg HG, et al. Epigenomics: Roadmap for regulation. Nature. 2015;518:314–6. doi: 10.1038/518314a. [DOI] [PubMed] [Google Scholar]
- 25.Petrovics G, Li H, Stumpel T, et al. A novel genomic alteration of LSAMP associates with aggressive prostate cancer in African American men. EBioMedicine. 2015;2:1957–64. doi: 10.1016/j.ebiom.2015.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prescott NJ, Lehne B, Stone K, et al. Pooled sequencing of 531 genes in inflammatory bowel disease identifies an associated rare variant in BTNL2 and implicates other immune related genes. PLoS Genet. 2015;11:e1004955. doi: 10.1371/journal.pgen.1004955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hong SN, Park C, Park SJ, et al. Deep resequencing of 131 Crohn's disease associated genes in pooled DNA confirmed three reported variants and identified eight novel variants. Gut. 2016;65:788–96. doi: 10.1136/gutjnl-2014-308617. [DOI] [PubMed] [Google Scholar]
- 28.Stammers M, Rowen L, Rhodes D, et al. BTL-II: a polymorphic locus with homology to the butyrophilin gene family, located at the border of the major histocompatibility complex class II and class III regions in human and mouse. Immunogenetics. 2000;51:373–82. doi: 10.1007/s002510050633. [DOI] [PubMed] [Google Scholar]
- 29.Goyette P, Boucher G, Mallon D, et al. High-density mapping of the MHC identifies a shared role for HLA-DRB1*01:03 in inflammatory bowel diseases and heterozygous advantage in ulcerative colitis. Nat Genet. 2015;47:172–9. doi: 10.1038/ng.3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Riedl S, Tandara A, Reinshagen M, et al. Serum tenascin-C is an indicator of inflammatory bowel disease activity. Int J Colorectal Dis. 2001;16:285–91. doi: 10.1007/s003840100312. [DOI] [PubMed] [Google Scholar]
- 31.Yamazaki K, McGovern D, Ragoussis J, et al. Single nucleotide polymorphisms in TNFSF15 confer susceptibility to Crohn's disease. Hum Mol Genet. 2005;14:3499–506. doi: 10.1093/hmg/ddi379. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.