Abstract
Prions are infectious protein conformations that are generally ordered protein aggregates. In the absence of prions, newly synthesized molecules of these same proteins usually maintain a conventional soluble conformation. However, prions occasionally arise even without a homologous prion template. The conformational switch that results in the de novo appearance of yeast prions with glutamine/aspargine (Q/N)-rich prion domains (e.g., [PSI+]), is promoted by heterologous prions with a similar domain (e.g., [RNQ+], also known as [PIN+]), or by overexpression of proteins with prion-like Q-, N-, or Q/N-rich domains. This finding led to the hypothesis that aggregates of heterologous proteins provide an imperfect template on which the new prion is seeded. Indeed, we show that newly forming Sup35 and preexisting Rnq1 aggregates always colocalize when [PSI+] appearance is facilitated by the [RNQ+] prion, and that Rnq1 fibers enhance the in vitro formation of fibers by the prion domain of Sup35 (NM). The proteins do not however form mixed, interdigitated aggregates. We also demonstrate that aggregating variants of the polyQ-containing domain of huntingtin promote the de novo conversion of Sup35 into [PSI+]; whereas nonaggregating variants of huntingtin and aggregates of non-polyQ amyloidogenic proteins, transthyretin, α-synuclein, and synphilin do not. Furthermore, transthyretin and α-synuclein amyloids do not facilitate NM aggregation in vitro, even though in [PSI+] cells NM and transthyretin aggregates also occasionally colocalize. Our data, especially the in vitro reproduction of the highly specific heterologous seeding effect, provide strong support for the hypothesis of cross-seeding in the spontaneous initiation of prion states.
The term “prion” and the hypothesis of a self-replicating protein were originally introduced to explain the unusual nature of the infectious agent that causes spongiform encephalopathies (1, 2). In 1994, Wickner (3) expanded the prion concept to explain the inheritance of two non-Mendelian traits, [PSI+] and [URE3], that occur naturally in laboratory strains of Saccharomyces cerevisiae, and now more yeast prions are known (see ref. 4). During prion propagation, molecules already in a prion state template the conversion of other molecules of the same amino acid sequence into the prion conformation. How prions arise de novo is unknown. Presumably, the spontaneous folding of a protein into the prion shape or a chance interaction of two or more non-prion molecules leads to the formation of a prion seed. Judging from the low rates of spontaneous prion appearance, this is an inefficient process often assumed to be nontemplated and therefore quite distinct from prion propagation.
The [PSI+] prion (reviewed in refs. 4–7) results from a self-propagating change of conformation of Sup35 associated with its aggregation. Because Sup35 is a component of the release factor, its prion aggregation inhibits translational termination. The N-terminal (or prion) domain of Sup35 is dispensable for the translation function, but is required for the formation and propagation of [PSI+]. Purified Sup35 forms protease-resistant aggregates (amyloid fibers). Modeling prion propagation, the lag time preceding aggregation can be eliminated by the addition of a small amount of premade fibers or cell lysate from a [PSI+] culture (8, 9). Other yeast prions, [RNQ+], [NU+], and [URE3], also form aggregates in vivo and fibers in vitro (reviewed in refs. 4 and 7; however, see ref. 10).
As predicted by the prion model, [PSI+] can appear spontaneously, and the frequency of its appearance can be enhanced >1,000-fold by overproduction of Sup35 or its prion domain (11, 12). However, the presence of an epigenetic element named [PIN+], for [PSI+] inducibility, facilitates both the spontaneous appearance of [PSI+] or its induction with excess Sup35 (13). Although [PIN+] is required for efficient [PSI+] appearance, it is not needed for [PSI+] propagation (14). Recently, by using the strains in which [PIN+] was first characterized, the Pin+ phenotype (i.e., the possibility of de novo formation of [PSI+]), was shown to be due to a prion form of Rnq1 (15), a previously described prion (16). Thus, [PIN+] is synonymous to [RNQ+] and is the cause of the Pin+ phenotype in these strains. However, [URE3], [NU+], or expression of any of 11 proteins from high-copy plasmids can also cause the Pin+ phenotype (15, 17). Two of these proteins were previously known or proposed to be able to become prions themselves (Ure2 and New1). The other nine have Q- or Q/N-rich domains similar to those present in Sup35 and other yeast prions.
Protein aggregation is a hallmark of many degenerative human diseases. For example, Huntington's and Machado–Joseph diseases are familial disorders caused by polyQ expansions in the huntingtin (Ht) and Machado–Joseph disease proteins, respectively, and are characterized by the accumulation of aggregates composed of these proteins in neurons (see ref. 18). In healthy individuals, the polyQ region of these proteins is generally composed of 12–40 Q's, when the number of Q's is greater than this the diseases occur with increased frequency, corresponding to the increase in polyQ region length. Amyloidogenic proteins not carrying Q- or Q/N-rich domains have been linked to Parkinson's disease (PD), Alzheimer's disease (AD) and systemic amyloidoses (see refs. 19 and 20), and recent data suggest that interactions of different amyloidogenic proteins play a role in PD and AD pathogenesis (21).
We previously hypothesized that various heterogeneous prions and prion-like aggregates help initiate the de novo formation of [PSI+], and that the Q- or Q/N-rich domains found in Sup35, as well as in heterologous aggregate-forming proteins, are likely to be important for epigenetic protein-based inheritance as well as for protein folding diseases (15). Here, we study the effects of polyQ and non-polyQ aggregates on [PSI+] induction and the acceleration of Sup35 fiber formation by Q/N-rich and non-Q/N amyloids.
Materials and Methods
Yeast Strains and Plasmids. Yeast strains were [psi–][rnq–], [psi–][RNQ+], [PSI+][RNQ+], and [PSI+][rnq–] derivatives of 74-D694 (MATa ade1–14 his3-Δ200 ura3–52 leu2–3,112 trp1–289; see Supporting Materials and Methods, which is published as supporting information on the PNAS web site). Unless specified, growth was at 30°C in media selective for plasmid maintenance.
Centromeric pNM-GFP plasmids, identical except for URA3 (22), HIS3 (15), or LEU2 (this work) markers and made from pRS316, pRS413, and pRS415, respectively, carry a CUP1-driven fusion of amino acids 1–254 of Sup35 with GFP. Similar plasmids with LEU2 and HIS3 markers carrying a Rnq1-GFP fusion were made based on pRnq1-GFP, which was described (16). Otherwise identical pRnq-CFP and pNM-YFP constructs were made by replacing GFP with appropriate cyan fluorescent protein (CFP) or yellow fluorescent protein (YFP) amplicons (see Supporting Materials and Methods for details on these and other vectors). The sequence of the RNQ1 ORF in all of the constructs is identical to that of WT Rnq1, except for a 3-aa N-terminal extension (MGS) and a 5-aa deletion (NNGNQN) at the extreme C terminus. Constructs carrying this amplicon fused to either GFP or Sup35MC maintain Pin+ activity in the absence of WT Rnq1 (I.L.D. and S.W.L., unpublished work).
Human amyloidogenic proteins or their fragments were fused with YFP or CFP and expressed with the strong constitutive GPD promoter (for basic vectors, see ref. 23). Centromeric URA3 and multicopy LEU2 versions of the GPD-HtQ(n)-GFP plasmids for expression of Ht exon 1 with different lengths of the polyQ tract (n = 25, 47, 72, and 103) were provided by S. Krobitsch (University of Chicago; ref. 24). Non-Q-rich amyloidogenic sequences were expressed from multicopy vectors. Synphilin-1 (Synph-1) and WT α-synuclein (αSyn-WT) and mutant α-Syn (αSynA30P) were expressed as YFP fusions (ref. 25 and this work) in LEU2 vectors. WT transthyretin (TTR-WT) and mutant D-strand-deleted TTR lacking the signal sequence (TTRd) were fused to CFP in URA3 vectors.
Scoring for the de Novo Formation of [PSI+] in Vivo. Induction and scoring of [PSI+] were as described (see ref. 26). The assay for de novo formation of [PSI+] in 74-D694 derivatives (11, 27) uses a premature UGA stop in ADE1, ade1–14. In [psi–] cells, translation is terminated at this UGA and cells are Ade– and are red on complex YPD media. In [PSI+] cells, Sup35 is partially inactivated, causing readthrough of the UGA (nonsense suppression) and allowing colonies to grow on adenineless media (–Ade) and to be white or pink (instead of red) on YPD. The de novo formation of [PSI+] was induced by NM-GFP(YFP) overproduction. [psi–][RNQ+] and [psi–][rnq–] cells were cotransformed with pNM-GFP or pNM-YFP, and with constructs expressing HtQ(n)-GFP, Synph-1, TTR-WT, TTRd, αSyn-WT, or αSynA30P. Transformants grown for ≈7 or 14 generations to allow for overproduction of Ht or non-polyQ amyloidogenic sequences were replica-plated once or twice to media containing 70 μM of CuSO4 to induce NM-GFP and then to different types of –Ade media (2% glucose or ethanol) where [PSI+] appearance was scored as growth at 30°Cor20°C. To distinguish [PSI+] colonies from chromosomal suppressor mutations, Ade+ colonies were subjected to the guanidine hydrochloride (GuHCl) test (see ref. 28 and the legend to Fig. 1B).
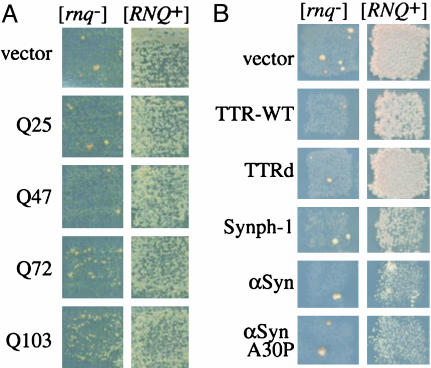
Fig. 1.
High levels of proteins with expanded polyQ tracts, but not non-polyQ proteins, permit the de novo induction of [PSI+] in the absence of [RNQ+]. [PSI+] suppressed a nonsense mutation in the ADE1 gene allowing growth on glucose –Ade medium (scored after 2 weeks at 20°C). All cells contained a [PSI+]-inducing construct and either an empty vector or one of the constitutive Ht-expressing multicopy vectors with polyQ stretches of different length (A) or one of the multicopy vectors expressing the indicated proteins (B). [psi–][rnq–] carrying an empty vector and transformants of the [psi–][RNQ+] derivative served as negative and positive controls. Note that growth on –Ade in [RNQ+] cells in B reflects growth inhibition by TTR-WT, Synph-1, and αSyn overproduction (data not shown) and not reduced [PSI+] induction.
The fluorescence assay for the de novo formation of [PSI+] uses NM-GFP (29) or NM-YFP reporters. In [psi–] cells, Sup35 is soluble, and fluorescence is evenly distributed throughout the cell. [PSI+] is associated with Sup35 aggregation and the appearance of bright fluorescent foci (29). Furthermore, in cultures where [PSI+] is forming de novo, but not in established [PSI+] derivatives, characteristic ring- and line-shaped aggregates appear (22), which are easily distinguished from similarly tagged αSyn-WT and Synph-1 aggregates that are exclusively dot-like.
Analysis of Colocalization of [PSI+] Aggregates with [RNQ+] or TTR Aggregates. For [PSI+]/[RNQ+] colocalization, fusion proteins were overexpressed in pNM-YFP/pRnq1-CFP cotransformants of [psi–][RNQ+] in late-log phase (OD595 = 2.2–2.5) by adding CuSO4 to 50 μM. At least four cotransformants were analyzed for each derivative. For [PSI+]/TTR colocalization, NM-YFP expression was induced by 50 μM CuSO4 for 3 h in late-log-phase cultures of pNM-YFP/p425GPD-TTR-CFP cotransformants of [psi–][RNQ+], [PSI+][RNQ+], or [PSI+][rnq–]. See Supporting Materials and Methods for microscopy details.
In Vitro Conversion and Amyloid Detection. Recombinant 0.2, 0.5, or 1.0 μM NM was incubated in 1× PBS, pH 7.4, with or without other proteins. See Supporting Materials and Methods for methods of protein preparation. Conversion occurred at room temperature without agitation and was monitored by fluorescence emission of thioflavin T (100-fold molar excess over NM; Sigma) using a Cary Varian Eclipse (bandpass 5-nm excitation and 10-nm emission) or a Jobin Yvon Horiba Fluoromax-3 (bandpass 3-nm excitation and emission) fluorescence spectrophotometer. The excitation and emission wavelengths were 450 nm and either 481 or 485 nm, respectively.
Results
Facilitation of the de Novo Induction of [PSI+] by Aggregation-Prone Alleles of Ht. To investigate the importance of Q- or Q/N-rich sequences for initiating prion seeding, we tested whether constructs carrying the first exon of Ht with an uninterrupted polyQ sequence of variable length could facilitate the de novo appearance of [PSI+]. When expressed in yeast, the constructs encompassing Ht exon 1 with an expanded polyQ region associated with early disease onset (Q53, Q72, and Q103) form cytoplasmic aggregates (and aggregation increases with the length of the polyQ tract, as well as with increased levels of expression), whereas WT Ht (Q25) remains soluble (24, 30). Some constructs of Ht with expanded Q tracts are toxic (31, 32); however, we used a nontoxic version that retains Q-length-dependent aggregation (M. Duennwald, S. Jagadish, S. Willingham, S.L.L., and P. Muchowski, unpublished work and ref. 24).
Expression of Ht variants containing polyQ tracts rendered some cells phenotypically Pin+, i.e., enabled overexpression of the Sup35 prion domain to induce the de novo appearance of [PSI+] in [rnq–] cells (Fig. 1 A and Table 1). Furthermore, the efficiency with which Ht constructs promoted [PSI+] induction correlated with their tendency to aggregate (Table 2). Curiously, Ht aggregates in [rnq–] cells were distinct from those in [RNQ+] cells (two vs. one dot per cell; data not shown). The level of Ht expression and the polyQ tract length affected the brightness of the aggregates and the percent of cells with aggregates, but not the number of aggregates per cell. However, more cells had very large aggregates when Q103 was expressed and Q103 inhibited growth slightly.
Table 1. The GuHCl test confirms facilitation of [PSI+] induction by Ht exon 1.
| No. of Ade+ colonies
|
|||
|---|---|---|---|
| Construct | Total tested | [PSI+] | [PSI+], % |
| Vector | 17 | 8 | 47 |
| Q25 | 17 | 9 | 53 |
| Q47 | 14 | 9 | 64 |
| Q72 | 8 | 7 | 88 |
| Q103 | 28 | 27 | 96 |
Ade+ colonies from [rnq-] transformants shown in Fig. 1 were transiently grown on YPD plus 5 mM GuHCl and were then scored for retention of suppression on media without GuHCl. Because some suppressors cause a transient change of color in GuHCl (53), cells were considered to have been [PSI+] only if they remained red following growth on GuHCl. Note that occasional [PSI+] colonies found among the rare Ade+ colonies in cultures carrying the empty vector, Q25, or Q47 constructs (see Fig. 1) probably reflect the spontaneous appearance of [RNQ+] (see ref. 15).
Table 2. Ability of Ht to facilitate [PSI+] induction correlates with polyQ aggregation.
| Moderate polyQ expression
|
High polyQ expression
|
|||
|---|---|---|---|---|
| PolyQ insert | Cells with aggregates*, % | [PSI+] induction† | Cells with aggregates*, % | [PSI+] induction† |
| None | 0 | - | 0 | - |
| Q25 | 0 | - | 0 | - |
| Q47 | 0 | - | 0 | - |
| Q72 | 0 | - | 1 | + |
| Q103 | 1 | ± | 10 | ++ |
The [psi-][rnq-] transformed with centromeric (moderate polyQ expression) or multicopy (high polyQ expression) Ht-GFP expressing vectors were grown for 7-14 generations. Fluorescent foci indicated Ht aggregation. Four or more transformants were analyzed for each plasmid, and the fraction of cells with aggregates was estimated from at least 1,000 cells.
The above transformants also carried the [PSI+]-inducing construct NM-GFP. The ±, +, and ++, levels of growth on -Ade after transient expression of NM-GFP correspond to very weak, weak, and moderate [PSI+] induction, respectively. Nine transformants were analyzed for each plasmid combination. The induction seen here is low in comparison with that seen in the presence of a high [RNQ+] variant that causes high-level [PSI+] induction (44); see Fig. 1A.
Non-Q-Rich Amyloidogenic Proteins Do Not Promote [PSI+] Induction in the Absence of [RNQ+]. We next asked whether the formation of [PSI+] could be facilitated by aggregates of TTR-WT, TTRd, αSyn-WT, αSynA30P, or Synph-1, none of which contain Q- or N-rich regions. Amyloid fibrils of TTR-WT cause senile systemic amyloidosis. The TTRd mutant in the region encoding β-strand D destabilizes TTR tetramers facilitating amyloid formation (33). αSyn plays a central role in PD, forming cytoplasmic inclusions called Lewy bodies, and is implicated in several other neurodegenerative disorders: the point mutation αSynA30P is associated with early onset of PD (34). Synph-1, an αSyninteracting protein (35), is found in Lewy bodies in brains of patients with sporadic PD (36).
Each protein, fused to a fluorescent tag to visualize aggregates, was constitutively expressed. TTR-WT, TTRd, and Synph-1 aggregates were easily detected, and their frequency and shape were not affected by the presence of [RNQ+] (Fig. 2A). Recently, αSyn-WT, but not αSynA30P, was found to aggregate in yeast (25). Here, we found these aggregation properties to be independent of the presence of [RNQ+] (Fig. 2 A and data not shown).
Fig. 2.
Colocalization of amyloidogenic aggregates. (A) Non-polyQ amyloidogenic proteins form aggregates in [RNQ+] and [rnq–] cells. (B) Occasional colocalization of TTR-WT (columns 1 and 4) and TTRd (columns 2 and 3) aggregates with [PSI+]. Cells shown originate from [psi–][RNQ+] (column 1), [PSI+][rnq–] (column 2), and [PSI+][RNQ+] (columns 3 and 4). (C) Coaggregation of [RNQ+] and newly forming [PSI+] aggregates in [psi–][RNQ+] cells during [PSI+] induction. For the first two samples, an array of focal planes 1 μm apart (Z-stack) is shown. DIC, differential interference contrast.
When TTR-WT, TTRd, αSyn-WT, αSynA30P, and Synph-1 were coexpressed with an NM-YFP [PSI+]-inducing construct, no [PSI+] induction was observed in the absence of [RNQ+] (Fig. 1B). Even when rare potentially [PSI+] (Ade+) colonies were recovered, further testing revealed that they were not [PSI+] (see Materials and Methods, and Table 4, which is published as supporting information on the PNAS web site). Furthermore, when a fluorescence assay was used to score for [PSI+] formation, the ring- and line-shaped YFP aggregates associated with de novo induction of [PSI+] (22) were absent in a [rnq–] background. Such aggregates were present in [RNQ+] transformants, where their frequency was not influenced by the heterologous aggregation-prone proteins (data not shown). Thus, the de novo formation of [PSI+] was facilitated in the presence of Ht polyQ aggregates, but not by the presence of various non-polyQ aggregates.
Sup35NM Aggregates Colocalize with Rnq1. We tested whether Sup35 aggregates that appear during [PSI+] induction colocalize with preexisting [RNQ+] aggregates. Rnq1 aggregates were visualized in a [RNQ+][psi–] strain by expressing a Rnq1-CFP fusion; the newly forming [PSI+] aggregates were induced and visualized by overexpressing NM-YFP. The fusion proteins were expressed when the cells were in late-log phase when [PSI+] can be efficiently induced (22), and expression continued for 2–6 h, long enough for some new [PSI+] aggregates to appear, but not to be propagated to daughter cells. CFP fluorescent aggregates, indicative of [RNQ+], were detected in most cells; YFP aggregates, indicative of [PSI+], were detected in <1% of the cells. Every newly appearing NM-YFP aggregate colocalized (completely or partially) with a Rnq1-CFP aggregate (Fig. 2C and Table 3). This finding is consistent with the idea that [RNQ+] can directly seed the formation of [PSI+].
Table 3. Newly appearing NM-YFP aggregates are detected in close proximity to preexisting [RNQ+] aggregates.
| [PSI+] induction, h | No. of cells examined with NM-YFP aggregates | No. of cells with/without overlapping Rnq-CFP and NM aggregates* |
|---|---|---|
| 2 | 12 | 11/0 |
| 4.5 | 30 | 12/0 |
| 6 | 57 | 41/0 |
All cells in which only NM-YFP aggregates but not Rnq-CFP aggregates were detected had no CFP fluorescence, indicative of the loss of the plasmid bearing the Rnq-CFP construct.
Colocalization of [PSI+] and [RNQ+] aggregates was also frequently observed in cultures propagating established forms of both prions. Here however, unlike in cultures where [PSI+] was appearing de novo, colocalization of NM-YFP with Rnq1-CFP aggregates was not always seen (Fig. 4, which is published as supporting information on the PNAS web site). Furthermore, CFP-tagged TTR-WT or TTRd and NM-YFP exhibited frequent but not obligatory colocalization (Fig. 2B). Thus, even amyloidogenic non-polyQ aggregates can sometimes colocalize with Sup35 aggregates.
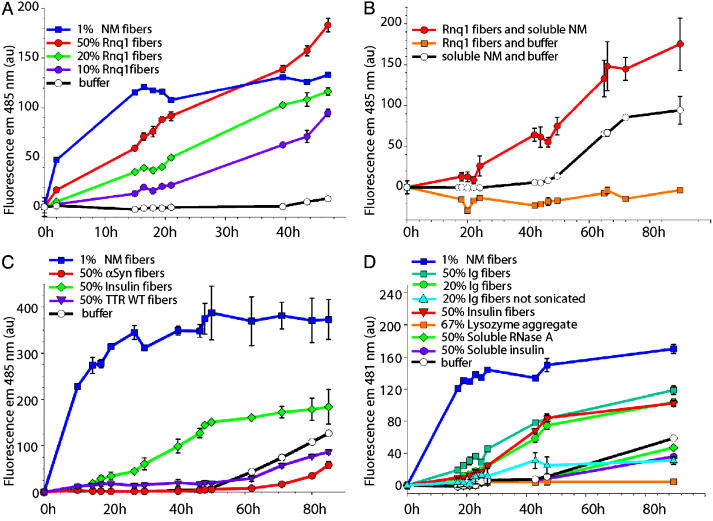
Stimulation of Conversion of NM to Amyloid Fibers by Heterologous Amyloids. We tested whether preparations of Rnq1 fibers would stimulate conversion of NM into amyloid in vitro. The formation of such amyloids closely mimics the propagation of the prion and has recently been shown to provide bona fide seed that transforms cells to the prion state (37, 38). Fiber formation was monitored by fluorescence emission of thioflavin T (Fig. 3 A and B) and confirmed by atomic force microscopy (G. J. Sawicki, S.M.U., and S.L.L., unpublished work). Conversion of soluble NM into amyloid occurred when Rnq1 preformed sonicated fibers were added at a value equal to 10% mol/mol, and the rate of NM conversion increased as the ratio of Rnq1 to NM increased (Fig. 3A). Clearly, Rnq1 aggregates stimulated conversion of NM into amyloid, although much less efficiently than NM stimulated its own conversion.
Fig. 3.
Effect of amyloidic, aggregated, or soluble proteins on NM conversion kinetics in vitro. The proportion of proteins is given as percent mol/mol. (A) Effect of increasing amounts of sonicated Rnq1 fibers on soluble NM conversion. (B) Control for A. The addition of sonicated Rnq1 fibers converts soluble NM to fibers, because thioflavin T emission did not increase when Rnq1 fibers at the same concentration were incubated without NM. (C) Effect of TTR-WT and αSyn-sonicated fibers on NM conversion. (D) Effect of Ig fibers, insulin fibers, and nonspecifically aggregated and soluble proteins on NM conversion.
On the contrary, TTR-WT and αSyn amyloids did not seed NM conversion even when added at 50% mol/mol ratio (Fig. 3C). This result in vitro is in agreement with the inability of TTR and αSyn aggregates to facilitate [PSI+] induction in vivo. However, bovine pancreas insulin and human Ig light-chain amyloids, which lack Q/N-rich regions, did stimulate NM conversion (Fig. 3D), and the degree of stimulation decreased when less amyloid was added. Also, sonication of these amyloids before their addition to soluble NM further increased the efficiency of conversion, as expected, if the amyloid fibers were seeding NM at their ends. The ability to stimulate NM conversion was specific to the amyloid form: 50% soluble insulin failed to stimulate NM conversion (Fig. 3D). Moreover, other proteins that were β-sheet-rich but not in an aggregated form (RNase A, Fig. 3D), or that were nonspecifically aggregated (lysozyme, Fig. 3D), did not stimulate NM conversion. Thus, neither β-sheet structure nor aggregation alone was sufficient to augment conversion, nor does the lack of a Q/N-rich domain necessarily preclude it.
Discussion
A seeding model was proposed to explain how heterologous prions, e.g., [RNQ+], facilitate the de novo appearance of [PSI+]. According to this model, a heterologous preexisting protein in the prion conformation templates the conversion of Sup35 into its prion form. The alternative titration model postulates that preexisting heterologous prions or prion-like aggregates capture and inactivate an inhibitor that prevents conversion of Sup35 into a prion (15, 17). The observations in this paper do not disprove the titration model, but they strongly support the seeding model.
[PSI+] aggregates appear to be amyloid (39), and the behavior of a short N-terminal Sup35 peptide in vitro suggests that Sup35 amyloid is composed of densely packed parallel β-sheets (40) that are likely similar to the parallel β-sheet structure proposed for ataxin-3, which has a polyQ domain (41). The Q-, N-, and Q/N-rich domains from two different proteins might form “polar zippers” (42), enabling heterologous protein aggregates to seed [PSI+]. Amyloidogenic sequences that are not Q- or N-rich may not be as likely to interact with the Sup35 prion domain. Indeed, we found that overexpression of the aggregation-prone, polyQ-containing domain of Ht supplied Pin+ activity, whereas several amyloid forming proteins that were not Q- or N-rich did not.
The seeding model postulates a direct interaction between newly forming prions and preexisting heterologous prion or prion-like aggregates. Reported antagonistic interactions between prions also suggest a direct interaction (43, 44). Indeed, binding of a Q/N-rich domain to the growth tip of a heterologous prion seed that could occasionally lead to the formation of a novel prion might also block the propagation of the heterologous seed. One argument against the seeding model is that visible new Sup35-YFP aggregates induced in [psi–] cells did not colocalize with heterologous New1-CFP aggregates that were shown to provide Pin+ activity (17). We revisited this issue by looking for colocalization of Rnq1-CFP and NM-YFP in [RNQ+] cells with either newly induced or already established [PSI+]. As might be predicted from the seeding model, the newly induced NM-YFP aggregates always colocalized, completely or partially, with preexisting Rnq1-CFP aggregates. In cells with established [PSI+], which can propagate in the absence of [RNQ+], we also found colocalization of NM-YFP and Rnq1-CFP aggregates, but the degree of colocalization varied considerably between experiments (I.L.D. and S.W.L., unpublished work). Likewise, some colocalization of Sup35 from different yeasts with divergent prion domains has been observed (45). Although the colocalization of Rnq1 and newly forming Sup35 aggregates supports the seeding model, our results suggest that once seeding has occurred Rnq1 and Sup35 do not coassemble, but, rather, each protein has a much greater capacity to convert itself than to cross convert. Also, colocalization without extensive coaggregation was reported (46) for an aggregation-prone rhodopsin mutant and a construct with an expanded polyQ stretch.
We also observed occasional colocalization of [PSI+] and TTR aggregates. Because TTR aggregates did not promote [PSI+] formation, colocalization of existing aggregates does not necessarily mean that these proteins can promote prion formation. More likely, it reflects the tendency of certain types of aggregates to colocalize in common sites (47).
The relationship between naturally occurring prion aggregates and visible aggregates detected upon overexpression of tagged constructs is unclear. Several groups recently proposed that [PSI+] and [URE3] propagons are low oligomeric weight aggregates and larger aggregates may represent dead-end products (48–50). Still, during the de novo induction of [PSI+], cells with visible aggregates gave rise to colonies that contained 20–100% [PSI+] cells, whereas cells without NM-GFP aggregates gave only [psi–] progeny (22).
In agreement with in vivo data, preformed Rnq1 amyloid fibers shorten the lag time for Sup35 fiber formation, but TTR and αSyn fibers do not. This finding strongly supports the seeding model. Also, in vivo, [PSI+] propagates very efficiently, but appears de novo only rarely, even in the presence of [RNQ+] (<1:105). Likewise, in vitro, Rnq1 fibers are much less efficient than Sup35 fibers at shortening the lag phase for conversion of soluble Sup35 into amyloid. The simplicity of the in vitro system, where Sup35 and Rnq1 aggregates are the only components, assures that Sup35 fiber formation was indeed seeded by Rnq1 fibers. However, although soluble Sup35 forms fibers in the absence of other proteins in vitro,[PSI+] does not appear without [RNQ+] or its substitutes in vivo (see ref. 14). Clearly, other factors present in yeast cells can affect prion conversion (9, 51).
Two other non-Q/N-rich amyloids tested, insulin and Ig, weakly augmented Sup35 fiber formation in vitro, perhaps through other β-strand interactions. It is unknown whether any non Q/N rich amyloids can support [PSI+] conversion in vivo, nor whether the assay used here would be sensitive enough to detect it.
Recently, aggregation-prone proteins have been detected together in disease-associated aggregates and have been shown to facilitate each other's polymerization into amyloid lesions (52). A link between the conformational changes associated with Q-length-dependent Huntington's disease and yeast prions was established by demonstrating enhanced aggregation and toxicity of polyQ-expanded proteins in the presence of [RNQ+]or[NU+] (17, 31) and a possible interaction of Rnq1 and polyQ (32). Our results further support the concept that common protein-folding mechanisms underlie processes as diverse as epigenetic protein-based inheritance and neurodegenerative diseases.
Supplementary Material
Acknowledgments
We thank R. Craig-Schapiro, G. J. Sawicki, S. Zadorsky, and J. Hong for assistance with experiments; M. J. Saraiva (Universidade do Porto, Porto, Portugal) for TTR cDNA; B. Hyman (Massachusetts General Hospital, Charlestown, MA) for Synph-1 cDNA; K. Sciarretta and Y. Argon (University of Chicago) for Ig light-chain amyloid (SMA); J. W. Kelly (The Scripps Research Institute, La Jolla, CA) for TTR and αSyn amyloids; N. Sondheimer (University of Chicago) for purified Rnq1; A. Manogaran and J. Shorter for comments on the manuscript; and C. Potenski, J. G. Smyth, and B. Bevis for assistance with the figures and the manuscript. This work was partially supported by National Institutes of Health Grants GM56350 (to S.W.L.), GM57840 [to M. Arnsdorf (University of Chicago) with partial support to S.M.U.], a Howard Hughes Medical Institute Award (to S.L.L.), an American Cancer Society postdoctoral fellowship (to S.M.U.), and Programa Praxis XXI, Fundacao para a Ciencia e Tecnologia, Portugal (to T.F.O.).
Abbreviations: Ht, huntingtin; PD, Parkinson's disease; CFP, cyan fluorescent protein; YFP, yellow fluorescent protein; TTR, transthyretin; TTRd, mutant D-strand-deleted TTR; αSyn, α-synuclein; Synph-1, synphilin-1; GuHCl, guanidine hydrochloride.
References
- 1.Griffith, J. S. (1967) Nature 215, 1043–1044. [DOI] [PubMed] [Google Scholar]
- 2.Prusiner, S. B. (1982) Science 216, 136–144. [DOI] [PubMed] [Google Scholar]
- 3.Wickner, R. B. (1994) Science 264, 566–569. [DOI] [PubMed] [Google Scholar]
- 4.Uptain, S. M. & Lindquist, S. (2002) Annu. Rev. Microbiol. 56, 703–741. [DOI] [PubMed] [Google Scholar]
- 5.Serio, T. R. & Lindquist, S. L. (2001) Adv. Protein Chem. 59, 391–412. [DOI] [PubMed] [Google Scholar]
- 6.Tuite, M. F. & Cox, B. S. (2003) Nat. Rev. Mol. Cell Biol. 4, 878–890. [DOI] [PubMed] [Google Scholar]
- 7.Wickner, R. B., Liebman, S. W. & Saupe, S. J. (2004) in Prion Biology and Diseases, ed. Prusiner, S. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 305–372.
- 8.Glover, J. R., Kowal, A. S., Schirmer, E. C., Patino, M. M., Liu, J. J. & Lindquist, S. (1997) Cell 89, 811–819. [DOI] [PubMed] [Google Scholar]
- 9.Uptain, S. M., Sawicki, G. J., Caughey, B. & Lindquist, S. (2001) EMBO J. 20, 6236–6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernandez-Bellot, E., Guillemet, E., Ness, F., Baudin-Baillieu, A., Ripaud, L., Tuite, M. & Cullin, C. (2002) EMBO Rep. 3, 76–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Derkatch, I. L., Chernoff, Y. O., Kushnirov, V. V., Inge-Vechtomov, S. G. & Liebman, S. W. (1996) Genetics 144, 1375–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chernoff, Y. O., Derkach, I. L. & Inge-Vechtomov, S. G. (1993) Curr. Genet. 24, 268–270. [DOI] [PubMed] [Google Scholar]
- 13.Derkatch, I. L., Bradley, M. E., Zhou, P., Chernoff, Y. O. & Liebman, S. W. (1997) Genetics 147, 507–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Derkatch, I. L., Bradley, M. E., Masse, S. V., Zadorsky, S. P., Polozkov, G. V., Inge-Vechtomov, S. G. & Liebman, S. W. (2000) EMBO J. 19, 1942–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Derkatch, I. L., Bradley, M. E., Hong, J. Y. & Liebman, S. W. (2001) Cell 106, 171–182. [DOI] [PubMed] [Google Scholar]
- 16.Sondheimer, N. & Lindquist, S. (2000) Mol. Cell 5, 163–172. [DOI] [PubMed] [Google Scholar]
- 17.Osherovich, L. Z. & Weissman, J. S. (2001) Cell 106, 183–194. [DOI] [PubMed] [Google Scholar]
- 18.Perutz, M. F. (1999) Trends Biochem. Sci. 24, 58–63. [DOI] [PubMed] [Google Scholar]
- 19.Saraiva, M. J. (2001) FEBS Lett. 498, 201–203. [DOI] [PubMed] [Google Scholar]
- 20.Shastry, B. S. (2003) Neurochem. Int. 43, 1–7. [DOI] [PubMed] [Google Scholar]
- 21.Kurosinski, P., Guggisberg, M. & Gotz, J. (2002) Trends Mol. Med. 8, 3–5. [DOI] [PubMed] [Google Scholar]
- 22.Zhou, P., Derkatch, I. L. & Liebman, S. W. (2001) Mol. Microbiol. 39, 37–46. [DOI] [PubMed] [Google Scholar]
- 23.Mumberg, D., Muller, R. & Funk, M. (1995) Gene 156, 119–122. [DOI] [PubMed] [Google Scholar]
- 24.Krobitsch, S. & Lindquist, S. (2000) Proc. Natl. Acad. Sci. USA 97, 1589–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Outeiro, T. F. & Lindquist, S. (2003) Science 302, 1772–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chernoff, Y. O., Uptain, S. M. & Lindquist, S. L. (2002) Methods Enzymol. 351, 499–538. [DOI] [PubMed] [Google Scholar]
- 27.Chernoff, Y. O., Lindquist, S. L., Ono, B., Inge-Vechtomov, S. G. & Liebman, S. W. (1995) Science 268, 880–884. [DOI] [PubMed] [Google Scholar]
- 28.Tuite, M. F., Mundy, C. R. & Cox, B. S. (1981) Genetics 98, 691–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patino, M. M., Liu, J. J., Glover, J. R. & Lindquist, S. (1996) Science 273, 622–626. [DOI] [PubMed] [Google Scholar]
- 30.Muchowski, P. J., Schaffar, G., Sittler, A., Wanker, E. E., Hayer-Hartl, M. K. & Hartl, F. U. (2000) Proc. Natl. Acad. Sci. USA 97, 7841–7846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meriin, A. B., Zhang, X., He, X., Newnam, G. P., Chernoff, Y. O. & Sherman, M. Y. (2002) J. Cell Biol. 157, 997–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meriin, A. B., Zhang, X., Miliaras, N. B., Kazantsev, A., Chernoff, Y. O., McCaffery, J. M., Wendland, B. & Sherman, M. Y. (2003) Mol. Cell. Biol. 23, 7554–7565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goldsteins, G., Andersson, K., Olofsson, A., Dacklin, I., Edvinsson, A., Baranov, V., Sandgren, O., Thylen, C., Hammarstrom, S. & Lundgren, E. (1997) Biochemistry 36, 5346–5352. [DOI] [PubMed] [Google Scholar]
- 34.Trojanowski, J. Q. & Lee, V. M. (2003) Ann. N.Y. Acad. Sci. 991, 107–110. [DOI] [PubMed] [Google Scholar]
- 35.Engelender, S., Kaminsky, Z., Guo, X., Sharp, A. H., Amaravi, R. K., Kleiderlein, J. J., Margolis, R. L., Troncoso, J. C., Lanahan, A. A., Worley, P. F., et al. (1999) Nat. Genet. 22, 110–114. [DOI] [PubMed] [Google Scholar]
- 36.Wakabayashi, K., Engelender, S., Yoshimoto, M., Tsuji, S., Ross, C. A. & Takahashi, H. (2000) Ann. Neurol. 47, 521–523. [PubMed] [Google Scholar]
- 37.King, C. Y. & Diaz-Avalos, R. (2004) Nature 428, 319–323. [DOI] [PubMed] [Google Scholar]
- 38.Tanaka, M., Chien, P., Naber, N., Cooke, R. & Weissman, J. S. (2004) Nature 428, 323–328. [DOI] [PubMed] [Google Scholar]
- 39.Kimura, Y., Koitabashi, S. & Fujita, T. (2003) Cell Struct. Funct. 28, 187–193. [DOI] [PubMed] [Google Scholar]
- 40.Balbirnie, M., Grothe, R. & Eisenberg, D. S. (2001) Proc. Natl. Acad. Sci. USA 98, 2375–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bevivino, A. E. & Loll, P. J. (2001) Proc. Natl. Acad. Sci. USA 98, 11955–11969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perutz, M. F., Pope, B. J., Owen, D., Wanker, E. E. & Scherzinger, E. (2002) Proc. Natl. Acad. Sci. USA 99, 5596–5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schwimmer, C. & Masison, D. C. (2002) Mol. Cell. Biol. 22, 3590–3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bradley, M. E., Edskes, H. K., Hong, J. Y., Wickner, R. B. & Liebman, S. W. (2002) Proc. Natl. Acad. Sci. USA 99, Suppl. 4, 16392–16399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakayashiki, T., Ebihara, K., Bannai, H. & Nakamura, Y. (2001) Mol. Cell 7, 1121–1130. [DOI] [PubMed] [Google Scholar]
- 46.Rajan, R. S., Illing, M. E., Bence, N. F. & Kopito, R. R. (2001) Proc. Natl. Acad. Sci. USA 98, 13060–13065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnston, J. A., Ward, C. L. & Kopito, R. R. (1998) J. Cell Biol. 143, 1883–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ness, F., Ferreira, P., Cox, B. S. & Tuite, M. F. (2002) Mol. Cell. Biol. 22, 5593–5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Narayanan, S., Bosl, B., Walter, S. & Reif, B. (2003) Proc. Natl. Acad. Sci. USA 100, 9286–9291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ripaud, L., Maillet, L. & Cullin, C. (2003) EMBO J. 22, 5251–5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shorter, J. & Lindquist, S. (2004) Science 304, 1793–1797. [DOI] [PubMed] [Google Scholar]
- 52.Giasson, B. I., Lee, V. M. & Trojanowski, J. Q. (2003) Neuromolecular Med. 4, 49–58. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.