Abstract
Spectrins are multi-domain, elastic proteins that provide elasticity to the plasma membrane of erythrocytes and select nucleated cells. Spectrins have also been found in the nucleus of non-erythrocytes, but their function remains to be uncovered. It has been hypothesized that a spring-like spectrin network exists within the lamina nucleoskeleton, however, experiments testing a spectrin network’s mechanical impact on the nucleus are lacking. Here, we knock-down levels of nuclear αII-spectrin with the goal of disrupting this nucleoskeletal spectrin network. We mechanically test live cells with intranuclear particle tracking and compression assays to probe changes in nuclear mechanics with decreases in αII-spectrin. We show no changes in chromatin mechanics or in the stiffness of nuclei under compression. However, we do observe a reduction in the ability of nuclei with decreased αII-spectrin to recover after compression. These results establish spectrin as a nucleoskeletal component that specifically contributes to elastic recovery after compression.
Keywords: Nucleoskeleton mechanics, Mechanobiology, Lamina, Spectrins
1. Introduction
Spectrin proteins are known to have a dominant role in red blood cell mechanics, and despite their known association with nuclear structural proteins, their mechanical role within the nucleus remains to be determined. Spectrins are best studied in erythrocytes, where an αI spectrin forms the primary membrane-bound network that provides elasticity and resilience to the red blood cell plasma membrane (Boey et al., 1998; Li et al., 2007, 2005; Marchesi and Steers, 1968)). αII spectrin proteins are also found associated with the membrane and in the cytoskeleton of nucleated cells, wherein they help regulate membrane stability (Bennett and Healy, 2008; Metral et al., 2009; Zhong et al., 2011) and have been identified in various organelles such as the Golgi apparatus (Beck, 2005; Beck et al., 1994) and the nucleus (McMahon et al., 1999; Young and Kothary, 2005).
Nuclear αII-spectrin (αII-Sp) is associated with DNA damage repair. While there are no known human mutations of the αII-Sp gene SPTAN1, lymphoblastoid cells from patients with Fanconi anemia show a decrease in αII-Sp (Brois et al., 1999; McMahon et al., 1999). These cells, as well as HeLa cells with siRNA-induced α II-Sp reduction, show increased susceptibility to DNA interstrand cross-linking agents (Howlett et al., 2005; McMahon et al., 2009). Additionally, nuclear αII-Sp is critical for telomere stability after DNA damage, co-localizes with telomeric proteins after cells are treated with an interstrand cross-linking agent and is required for the DNA damage repair protein XPF to localize to telomeres after interstrand cross-links are induced (Zhang et al., 2013).
In addition to this role of DNA interstrand crosslink repair and chromosome stability in the nuclear interior, it is suspected that αII-Sp may serve a function in the nucleoskeleton as well (Holaska et al., 2004; Simon and Wilson, 2011; Sridharan et al., 2006). Within the nucleus, αII-Sp has been shown to associate with structural proteins including lamin A, actin, emerin and nuclear myosin via co-immunoprecipitation (Holaska and Wilson, 2007; Sridharan et al., 2006). These associations with structural proteins, along with spectrin’s understood role in erythrocyte membrane stabilization, lead to the hypothesis that spectrin proteins help to regulate the mechanics of the nucleus. However, a direct functional mechanical role has not been demonstrated for spectrins.
It is increasingly appreciated that nuclear structure and stiffness correlate with cell functionality (Charras and Sahai, 2014; Dahl et al., 2008; Martins et al., 2012). While lamins in the nucleoskeleton have been shown to be a primary component to nuclear stiffness (Lammerding et al., 2006; Shimi et al., 2015, 2008), a potential spectrin network may also play a critical role in regulating the mechanical properties of the nucleus (Simon and Wilson, 2011). In this paper we aim to address the mechanical role of nuclear spectrin. We reduce the level of αII-Sp via RNA interference and then probe mechanical changes of the chromatin using live cell intranuclear particle tracking and test the mechanics of the whole nucleus using a live cell compression assay. While the loss of αII-Sp is associated with inhibited DNA damage repair, we find that reduction αII-Sp has no impact on the rheology of the chromatin interior. Reduction in αII-Sp does not alter nuclear size, but αII-Sp does impact the resilience of the nucleus, as evident by a failure to return to initial nuclear area after being compressed; control cell nuclei return to their initial nuclear area after being compressed. Thus, the mechanical role of spectrins in the nucleus appears to be at the inner nuclear membrane, similar to their role at the plasma membrane. Unlike lamin filaments that provide stiffness, spectrins provide for resilience of the cell nucleus.
2. Materials and methods
Standard materials and methods can be found in Supplemental materials and methods including cell culture and transfection, western blot, immunofluorescence and cell imaging.
2.1. Live cell compression assay
HeLa cells were grown in glass bottom dishes (P35G-1.5–14-C, MatTek Corporation, Ashland, MA) and transfected with the appropriate vector. Cell nuclei were stained with Hoechst 33342, and propidium iodide (PI) (Invitrogen, Carlsbad, CA) was added to the media prior to imaging to reject cells whose membranes burst during compression. Hoechst 33342 intercalates into DNA and can influence mechanics of the nucleus, but this label was used for all samples experimental and control. Cells were imaged as described in Supplemental materials and methods. Cells were subjected to unconfined compression, and images were taken before application of weight, during compression, and after release of compression. A 50 g static mass was placed over an area of 0.5 cm2, with a silicon spacer placed between the stiff mass to evenly distribute the force over the given area. Trans-fected cells were detected by a green fluorescent protein (GFP) reporter protein. Only cells with high levels of GFP expression were used for knock-down spectrin (KDSp) or knock-down control (KDC) data. Cells not expressing GFP, in the same field of view as the KDSp cells were used as side-by-side controls. Cells not treated with any vector or transfection reagent were used as wild type (WT) controls. Projected nuclear area was measured based on the Hoechst nuclear stain using ImageJ. For analysis, nuclear area was normalized to the initial nuclear area of a given cell. A 1-way ANOVA followed by Tukey’s pairwise comparison test was performed to determine significant differences between normalized nuclear area increase for each condition. To obtain the side-by-side control puromycin was not used for compression experiments. For the compression assay, samples sizes were as follows: n=35 WT nuclei, n=13 side-by-side control nuclei, n=10 KDC nuclei, and n=12 KDSp nuclei.
2.2. Intranuclear particle tracking
HeLa cells were grown in glass bottom dishes (P35G-1.5–14-C, MatTek Corporation, Ashland, MA) and cells were transfected with RFP-TRF1, which is visualized as fluorescent speckles within the nucleus when expressed. The motion of these speckles were tracked over time, and mean square displacement (MSD) was calculated. Cells were imaged as described above. Images were gathered at 3-minute time intervals for 1 h, cells were imaged for an additional hour to assure cell viability, but only the first hour was used for analysis. Single point 2-D particle tracking analysis was performed on these images in MATLAB using custom Lap-track71 software developed by Ge Yang as previously published(Spagnol and Dahl, 2014; Yang et al., 2008). Using this software, nuclei were cropped and aligned at each frame to remove rigid body motion such that only intranuclear motion was captured. Only points which persisted for the entire hour of imaging were tracked and used for analysis. Student’s t-test was performed to test for statistical differences between MSD at each lag time. Sample sizes for intranuclear particle tracking analysis were as follows: n=12 WT cells, with 37 total points tracked and n=17 KDSp cells, with 65 total points tracked.
3. Results
3.1. Confirmation of αII-spectrin knock-down
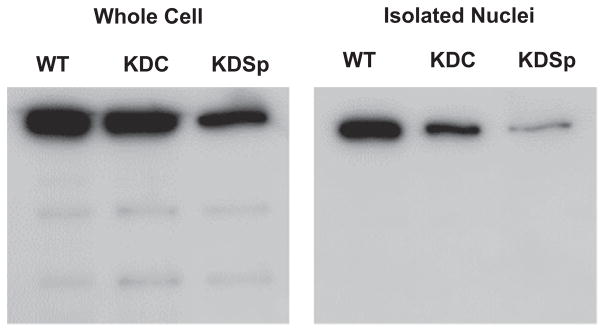
In order to determine the mechanical effects of nuclear spectrin proteins, we aimed to compare cells with decreased levels of nuclear αII-Sp to control cells. We investigated cells with reduced levels of αII-Sp since complete loss of αII-Sp is lethal to cells (McMahon et al., 2009). The gene SPTAN1, which codes for αII-Sp, was chosen as the knock-down target since it is the only gene which codes for α-spectrins in non-erythrocytes, while up to 5 genes code for β spectrins(Machnicka et al., 2012). Knockdown of αII-Sp was used to assess the mechanical impact of a potential nucleoskeletal spectrin network, since sufficient knockdown of αII-Sp would cause disruption of this network, and potentially alter the mechanical properties of the nucleus. Knockdown treatments were performed via transient transfection with a shRNA vector targeted against αII-Sp. While we aimed to isolate and identify the role of nuclear spectrin, αII-Sp is also present at the plasma membrane. As no nucleoskeletal-specific αII-Sp isoform is known, αII-Sp levels were decreased both in the cytoskeleton as well as in the nucleus with the shRNA, which we confirmed by western blot analysis, shown in Fig. 1 for both whole cell and isolated nuclear lysates. The level of αII-Sp in KDSp cells was compared to that in non-treated, WT cells, and KDC cells, which were treated with a scramble control vector with the same backbone as the αII-Sp knock-down vector. KDSp and KDC were treated with antibiotic (see Methods) to isolate cells which had taken up the vector. These results indicate that αII-Sp levels were decreased in the whole cell, as well as the nucleus of KDSp cells. With the shRNA vector, KDSp isolated nuclei had approximately a 68% reduction of αII-Sp compared KDC nuclei, as determined by western blot area analysis in ImageJ. By comparison, KDSp whole cells had a 51% reduction of αII-Sp compared to KDC whole cells. Interestingly, bands of lower molecular weight proteins were present in all of the whole cell lysates, but were not seen in the isolated nuclei lysates, even after prolonged exposure for chemiluminescence (Fig. 1, S1). This indicates that cytoskeleton-specific αII-Sp isoforms may exist, which are not present in the cell nucleus.
Fig. 1.
Western blot showing αII-spectrin protein level decrease in whole cells, and isolated nuclei. Bands below the primary band in whole cells demonstrate αII-spectrin isoforms may be present in the cytoskeleton which are not seen in the nucleus.
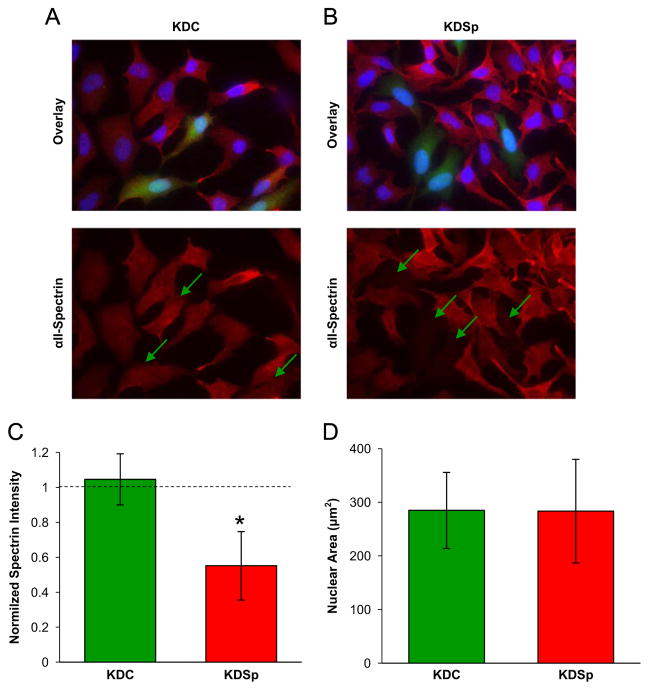
To further confirm decreased levels of αII-Sp, immunofluorescence experiments were run. While several antibodies against αII-Sp were tested, none were found to exclusively label nuclear αII-Sp. The αII-Sp knock-down vector and the scramble control vector contain a GFP reporter protein to visually determine which cells have taken up the plasmid. Fig. 2A and B, compare the normalized intensity of αII-Sp KDC cells to KDSp cells (no antibiotic). The αII-Sp IF intensity was decreased in KDSp (expressing GFP) cells, relative to cells which did not take up the vector in the same field of view. KDC (expressing GFP) cells had similar levels of αII-Sp to cells which did not take up the vector in the same field of view. The relative intensities of cells which took up a given vector compared to cells which did not take up the vector, are quantified in Fig. 2C. Based on this quantification, it is estimated that αII-Sp levels in KDSp cells were decreased to approximately 50% of those in KDC cells or in the side-by-side controls. These results, combined with the western blot results, show that cells treated with the αII-Sp knock-down vector, which express GFP, have decreased levels of nuclear αII-Sp, and therefore can be used for analysis in future experiments to determine the role of αII-Sp in nuclear mechanics.
Fig. 2.
Immunofluorescence (IF) and quantification of fluorescence of KDSp and KDC cells. (A) Shows IF of scramble control (KDC – green) cells which express control levels of αII-Sp. Green arrows show locations in the αII-Sp channel of cells which express the GFP-reporter, and thus have taken up the given vector. (B) Shows IF images of αII-Sp which demonstrates decreased αII-Sp levels in cells which have taken up the knock-down vector (KDSp – green). (C) Shows quantification of intensity of αII-spectrin channel from IF images. Average intensity of the αII-Sp channel was calculated for cells which had taken up a given vector, and divided by the average intensity of the αII-Sp channel of non-transfected cells in each field of view (n=12 fields of view for each condition). Error bars are standard deviation between fields of view. Asterisk denotes significant difference (p<0.05). (D) Shows a comparison of initial nuclear area (i.e. no compression) between KDC and KDSp cells. No significant difference in nuclear area was detected in these cells as measured by Student’s t-test. n=19 KDSp nuclei, and n=28 KDC nuclei. Error bars represent standard deviation. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
3.2. Intranuclear movement is unaffected in αII-spectrin depleted cells
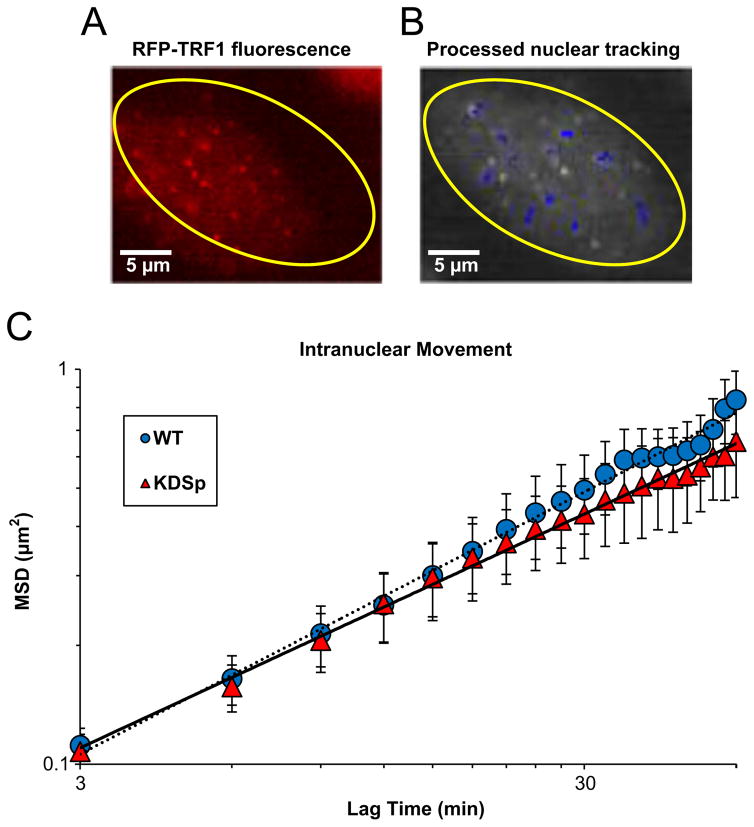
Previously, it was shown that loss of αII-Sp was associated with impaired DNA damage repair and telomere function following DNA interstrand crosslinks(McMahon et al., 2009; Zhang et al., 2013). Thus, we hypothesized that there may be a structural role of this large protein within the chromatin. Since chromatin structure and stability often influence mechanics, we hypothesized that reduction of αII-Sp would alter intranuclear rheology. To measure intranuclear movement, cells were transfected with a RFP-tagged telomeric protein TRF1 (RFP-TRF1). The fluorescent nuclear speckles of TRF1 were tracked over time and mean squared displacement (MSD) was calculated. Fig. 3A shows a nuclei transfected with RFP-TRF1. The tracks of high intensity speckles are overlaid on the fluorescent image in Fig. 3B. A plot of intranuclear MSD versus lag time comparing WT cells to KDSp cells is shown in Fig. 3C. KDSp cells show no change in MSD of intranuclear movement compared with WT cells. Thus, it appears that the decreased levels of αII-Sp have no effect on motion of the chromatin inside the nucleus.
Fig. 3.
Intranuclear particle tracking analysis. (A) Shows an example of a cell nucleus (yellow outline) expressing RFP-TRF1. This telomeric protein appears as punctate fluorescent speckles in the nucleus which are tracked at 3-minute time intervals for 1 hour. (B) Shows tracks (blue overlay) of these chromatin-bound proteins demonstrate Brownian motion. (C) Shows a plot of mean squared displacement (MSD) vs. lag time for αII-spectrin knock-down cells compared to wild type (non-treated) cells. Interestingly, no significant difference is seen in MSD between groups for any lag time, indicating no changes in intranuclear movement when spectrin levels are decreased. n=12 WT cells, with 37 total points tracked and n=17 KDSp cells, with 65 total points tracked.(For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
3.3. αII-spectrin depleted cell nuclei show decreased resilience to applied force
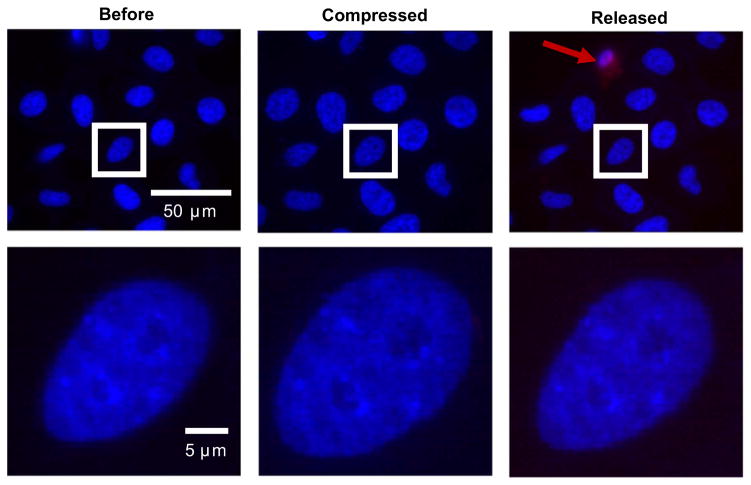
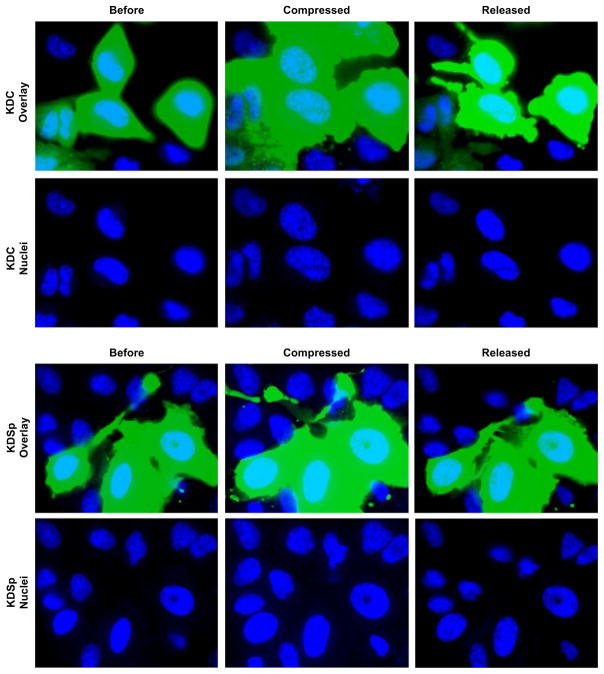
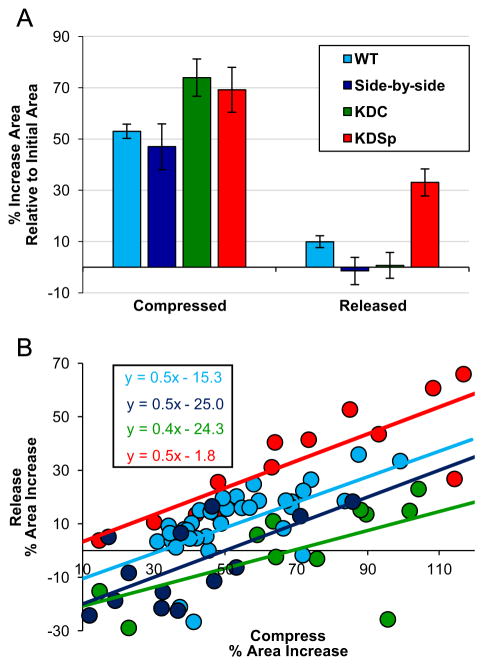
Once decreased levels of nuclear αII-Sp were confirmed, we aimed to determine the mechanical role of αII-Sp in the nucleus. First we investigated the impact of decreased αII-Sp on nuclear size in cultured cells. HeLa cells were transfected with the SPTAN1 shRNA, and no significant difference in projected nuclear area was detected between KDSp and KDC cells at the same confluency, as shown in Fig. 2D. We then compressed the cells and observed nuclear deformation inside the live cell. Briefly, a static load was placed on top of a population of cells, such that they underwent unconfined compression. Images were taken before compression, while being compressed (weight on), and once compression was released (weight removed), which were then used to determine nuclear area. Relative nuclear area was calculated by comparing nuclear area to the initial nuclear area before compression of a given cell. Propidium iodide (PI) was added to the media prior to compressing the cells to verify cell and cell membrane viability following compression. Only cells with intact membranes (i.e. those not stained by PI) upon removal of the weight were used for analysis. A representative image of WT control cell nuclei before, during, and after compression is shown in Fig. 4. This image shows the increase in nuclear area during compression, and return to initial area upon release. Additionally, a cell in the release frame is stained with PI, indicating a compromised membrane, and would not be used for analysis. A representative fluorescent image comparing KDSp to KDC treated cells is shown in Fig. 5. Cells expressing the GFP reporter are those which have taken up the vector. Cells which do not express GFP in the KDSp treated cells were used as side-by-side control cells, to assure that changes in nuclear area were not an artifact of changes in force distribution between experiments. Quantification of increase in nuclear area, normalized to initial nuclear area, for KDSp cells and 3 sets of control cells (WT, side-by-side, and KDC) under compression, followed by release of compression, is shown in Fig. 6. KDSp cells appear to increase in nuclear area the same extent as control cells, but upon release of compression, the KDSp cell nuclei do not return to their initial nuclear area, while control cells all return to their initial nuclear area. This suggests a potential spring-like role of αII-Sp, which allows nuclei to return to original area after deformation. However, given the similar extent of nuclear area increase during compression, αII-Sp appears to play a negligible role in regulating nuclear stiffness. We also investigated the role of the nucleoskeletal protein emerin in nuclear mechanics with the compression assays by treating cells with an emerin knock-down vector, and no significant difference was see in nuclear area between knock-down emerin and control cells (data not shown). Regression analysis comparing the deformation of each nucleus with its recovery shows the power of this facile compression imaging technique (Fig. 6B). For each sample (KDSp and all of the controls), larger deformed area correlated with larger area after release of compression. All samples had the same regression slope implying that dilation under compression and regression was not treatment-specific. However, the zero-strain intercept was very different between samples. Both the side-by-side control and KDC regressed to the same point, suggesting that the presence of the transfection agent may alter the cell or nucleus in some way. These controls were slightly different than the WT (no treatment control), but all controls showed a negative intercept suggesting that at a limit of no compression there is a nuclear contraction or pre-stress in cells that may be mediated by the spectrin network. In KDSp cells, the intercept is at zero.
Fig. 4.
Example of nuclear area increase, and subsequent decrease in wild type control cells after compression, and release of compression. Nuclei are stained with Hoechst. Propidium iodide (PI) was added to media for compression assays to assure cell membrane remained intact for the entire experiment. In the release frame, a PI stain is seen in one cell, indicated by a red arrow. This cell would not be used for analysis. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
Fig. 5.
Images of live cell compression assay comparing KDSp to KDC. Images are taken before, during, and after application of a 50 g static weight, and nuclear area was calculated. The GFP reporter is expressed in cells which have taken up the given vector, thus only these cells were used for analysis. Control cells which did not take up the knock-down vector are designated as side by side cells in Fig. 6. PI stain was added and only cells with intact membranes, which persisted for the entire assay were used for analysis. Nuclei are stained with Hoechst 33342.
Fig. 6.
Quantification of nuclear area for the various treatment groups after compression assay. (A) KDSp cells (red) fail to return to their initial nuclear area, in contrast to the various control groups (KDC, side-by-side, WT) which all return to their initial nuclear area upon removal of the static weight. Side-by-side cells are cells which did not take up the spectrin knock-down vector, but were in the same field of view as the KDSp cells. This control demonstrates that the failure to return to initial area was not an artifact of experimental variability. WT cells are control cells which did not receive any transfection treatment. Error bars show standard error of the mean. Asterisk denotes significant different (p<0.01). (B) Individual data points of each nucleus are shown for each sample comparing strains under compression (x-axis) to the strain of release (y-axis) normalized to the original size. In each case, high compression strain maps with larger strain after removal of the compressive force. In many control cases, removal of force in nuclei under small strains resulted in a shrinking of the nucleus (blue and green points). Compression of KDSp cells also resulted in the largest nuclear dilation. Regression analysis of the data (inset box) shows similar slopes for all samples, similar intercepts for transfected samples and largely divergent offset for KDSp cells. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
4. Discussion
Spectrin networks are known to be present at the lipid membranes of many biological systems, such as erythrocytes, the cell cytoplasm, and various intracellular organelles (Beck, 2005; Beck et al., 1994). Because of the association of spectrin with lipid bilayers, and the presence of spectrin proteins in the nucleus, it has been proposed that a spectrin network may exist in the nucleoskeleton (Simon and Wilson, 2011). Furthermore, this network may provide elasticity to the nucleus, playing a similar role as it does in the more well-characterized systems, which we sought to investigate. We too show the presence of spectrin in the nucleus (Fig. 1), and proceed to test the role of this potential nucleoskeletal spectrin network by knocking down αII-Sp. We demonstrate the potential presence of unique spectrin isoforms present in the cytoskeleton which do not reside in the nucleus. This can be seen in Fig. S1, where additional protein bands are seen in the western blot of whole cell lysates, but not in the isolated nuclear lysates. When nuclear spectrin levels are decreased, and thus this potential nucleoskeletal spectrin network is decreased, we see a change in the nuclear elastic restorative response to compression. Mainly, nuclei with decreased spectrin do not return to their initial nuclear area after compression, and release of compression. We attribute this effect to a nucleoskeletal spectrin network, the disruption of which limits nuclear restoration following compression, further suggesting a mechanical role of nuclear spectrins. As demonstrated by the compression experiments a disruption of this network causes a reduction in elasticity of the nucleus. Regression analysis of all of the data shows a net negative restoration force at a limit of zero compression, suggesting that nuclei are “primed” to have reduced area. This may be related to the nuclear shape change associated from the flat nuclei in spread cells to a more spherical phenotype with loss of cell adhesion or from a shape change associated with cytoskeletal-nuclear pre-stress. However this shape change priming is related to the αII-Sp network.
Proper nuclear mechanics are essential to cell function; as physical forces are known to alter genome expression. Generally, physical forces can be propagated from outside the cells, through the cytoskeleton, to the nucleus via the linker of the nucleoskeleton and cytoskeleton (LINC) complex (Wang et al., 2009). Any changes to nuclear mechanics can impact the effect that physical forces have on the nucleus, and ultimately gene expression based on the idea of mechanotransduction (Lammerding et al., 2004). Since spectrin appears to contribute to the elasticity of the nucleus, it may play a critical role in cells which undergo high levels of stress such as myocytes subjected to repeated contraction and relaxation cycles, endothelial cells subjected to shear stress and pressure changes, and cells which migrate through tight interstitial spaces which require drastic nuclear deformations. This recovery after deformation, referred to here as resilience but also called plasticity, has very recently been tested in numerous cell types (Bonakdar et al., 2016). Nucleoskeletal stability as well as resilience will be important during cell crawling through tight interstitial spaces (Denais et al., (2016)).
In addition to testing the changes in nuclear mechanics via live cell compression assays, we also probed changes in genome mechanics in cells with reduced levels of αII-Sp using live cell intranuclear particle tracking. Previous studies have demonstrated that cells with reduced levels of αII-Sp show increased susceptibility to DNA damage, and show a decrease in the ability to repair DNA damage (McMahon et al., 2009). Based on these studies, we suspected that αII-Sp reduced cells may show altered genome motion. Additionally, cells with decreased αII-Sp, and thus lacking a percolated nucleoskeletal spectrin network, may have changes in mechanotransduction arising from changes in nuclear mechanics. However, cells with decreased αII-Sp showed no difference in intranuclear movement relative to wild type control cells, and this reduction in αII-Sp appears to have no influence the intranuclear motion under the conditions measured. Since it is proposed that αII-Sp is involved in DNA interstrand crosslink repair it seems likely that in the absence of large-scale DNA damage induction the intranuclear motion is largely unchanged between wild type control and KDSp cells. It is possible that KDSp cells would not be able to repair the induced damage, thus leading to changes in intranuclear motion. Additionally, no external force was applied to cells during particle tracking experiments. In order to show changes in intranuclear movements due to mechanotransduction, KDSp cells may need to be exposed to forces such as shear flow in order for mechanotransduction related changes to be large enough to measure via particle tracking.
This work suggests the role of a nucleoskeletal spectrin network in providing elasticity to the non-erythrocyte cell nucleus. This was evident from data obtained via live cell compression assays, in which nuclei with reduced levels of αII-Sp fail to return to their initial nuclear area after release of compression, while control nuclei return to their initial area. αII-Sp may play an important role in mechanotransduction for cells undergoing high levels of stress, however, with no externally applied force, changes in intranuclear motion are minimal between control cells and αII-Sp reduced cells. The nucleoskeleton is composed of many interacting proteins, which play important roles in maintaining cell function. Here we begin to uncover a new mechanical phenotype of the cell nucleus provided by a lesser-studied nuclear protein, α II-Sp.
Supplementary Material
Acknowledgments
This work was funded by NSF-CMMI-1300476 (Dahl) and NIH-EB003392 (Armiger). We also acknowledge NIH-ES025606 to Opresko for the production of the TRF1 vector. The authors would like to acknowledge the lab of Ge Yang, at Carnegie Mellon University for providing the particle tracking software as well as insight on how to best utilize this technique. We would also like to acknowledge Patricia Opresko (University of Pittsburgh, Department of Environmental and Occupational Health) for providing the EYFP-TRF1 and for preparing the RFP-TRF1 vector used for particle tracking. Finally, we would like to acknowledge Katherine Wilson, Daniel Simon, Zhixia Zhong and Alexandre Ribeiro for intellectual contributions to this project.
Appendix A. Supporting information
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.jbiomech.2016.10.034i.
Footnotes
Conflict of interest
No competing interests declared.
Uncited references
(Alter and Kupfer, 2013; Boal, 2012; Butin-Israeli et al., 2012; Grum et al., 1999; Opresko et al., 2004).
References
- Alter B, Kupfer G. Fanconi anemia. GeneReviews 2013 [Google Scholar]
- Beck KA. Spectrins and the golgi. Biochim Et Biophys Acta. 2005;1744:374–382. doi: 10.1016/j.bbamcr.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Beck KA, Buchanan JA, Malhotra V, Nelson WJ. Golgi spectrin: identification of an erythroid beta-spectrin homolog associated with the Golgi complex. J Cell Biol. 1994;127:707–723. doi: 10.1083/jcb.127.3.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett V, Healy J. Organizing the fluid membrane bilayer: diseases linked to spectrin and ankyrin. Trends Mol Med. 2008;14:28–36. doi: 10.1016/j.molmed.2007.11.005. [DOI] [PubMed] [Google Scholar]
- Boal D. Mechanics of the Cell. 2. Cambridge University Press; Cambridge: 2012. [Google Scholar]
- Boey SK, Boal DH, Discher DE. Simulations of the erythrocyte cytoskeleton at large deformation. I Microscopic models Biophys J. 1998;75:1573–1583. doi: 10.1016/S0006-3495(98)74075-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonakdar N, Gerum R, Kuhn M, Spörrer M, Lippert A, Schneider W, Aifantis KE, Fabry B. Mechanical plasticity of cells. Nat Mater. 2016:1–6. doi: 10.1038/nmat4689. [DOI] [PubMed] [Google Scholar]
- Brois DW, McMahon LW, Ramos NI, Anglin LM, Walsh CE, Lambert MW. A deficiency in a 230 kDa DNA repair protein in Fanconi anemia complementation group A cells is corrected by the FANCA cDNA. Carcinogenesis. 1999;20:1845–1853. doi: 10.1093/carcin/20.9.1845. [DOI] [PubMed] [Google Scholar]
- Butin-Israeli V, Adam SA, Goldman AE, Goldman RD. Nuclear lamin function and disease. Trends Genet. 2012;28:464–471. doi: 10.1016/j.tig.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denais CM, Gilbert RM, Isermann P, McGregor AL, te Lindert M, Weigelin B, Davidson PM, Friedl P, Wolf JLK. Nuclear envelope rupture and repair during cancer cell migration. Science. 2016;352:353–358. doi: 10.1126/science.aad7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charras G, Sahai E. Physical influences of the extracellular environment on cell migration. Nat Rev Mol Cell Biol. 2014;15:813–824. doi: 10.1038/nrm3897. [DOI] [PubMed] [Google Scholar]
- Dahl KN, Ribeiro AJS, Lammerding J. Nuclear shape, mechanics, and mechanotransduction. Circ Res. 2008;102:1307–1318. doi: 10.1161/CIRCRESAHA.108.173989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grum VL, Li D, MacDonald RI, Mondragón A. Structures of two repeats of spectrin suggest models of flexibility. Cell. 1999;98:523–535. doi: 10.1016/s0092-8674(00)81980-7. [DOI] [PubMed] [Google Scholar]
- Holaska J, Wilson K. An emerin “proteome”: purification of distinct emerin-containing complexes from HeLa cells suggests molecular basis for diverse roles including gene regulation, mRNA splicing, signaling, mechanosensing and nuclear architecture. Biochemistry. 2007;46:8897–8908. doi: 10.1021/bi602636m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holaska JM, Kowalski AK, Wilson KL. Emerin caps the pointed end of actin filaments: evidence for an actin cortical network at the nuclear inner membrane. PLoS Biol. 2004;2:E231. doi: 10.1371/journal.pbio.0020231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlett NG, Taniguchi T, Durkin SG, D’Andrea AD, Glover TW. The Fanconi anemia pathway is required for the DNA replication stress response and for the regulation of common fragile site stability. Human Mol Genet. 2005;14:693–701. doi: 10.1093/hmg/ddi065. [DOI] [PubMed] [Google Scholar]
- Lammerding J, Fong LG, Ji JY, Reue K, Stewart CL, Young SG, Lee RT. Lamins A and C but not lamin B1 regulate nuclear mechanics. J Biol Chem. 2006;281:25768–25780. doi: 10.1074/jbc.M513511200. [DOI] [PubMed] [Google Scholar]
- Lammerding J, Schulze PC, Takahashi T, Kozlov S, Sullivan T, Kamm RD, Stewart CL, Lee RT. Lamin A/C deficiency causes defective nuclear mechanics and mechanotransduction. J Clin Investig. 2004;113:370–378. doi: 10.1172/JCI19670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Dao M, Lim CT, Suresh S. Spectrin-level modeling of the cytoskeleton and optical tweezers stretching of the erythrocyte. Biophys J. 2005;88:3707–3719. doi: 10.1529/biophysj.104.047332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Lykotrafitis G, Dao M, Suresh S. Cytoskeletal dynamics of human erythrocyte. Proc Natl Acad Sci U S A. 2007;104:4937–4942. doi: 10.1073/pnas.0700257104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machnicka B, Grochowalska R, Bogusławska DM, Sikorski aF, Lecomte MC. Spectrin-based skeleton as an actor in cell signaling. Cell Mol Life Sci: CMLS. 2012;69:191–201. doi: 10.1007/s00018-011-0804-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchesi VT, Steers E., Jr Selective solubilization of a protein component of the red cell. Membr Sci. 1968;159:203–204. doi: 10.1126/science.159.3811.203. [DOI] [PubMed] [Google Scholar]
- Martins RP, Finan JD, Guilak F, Lee DA. Mechanical regulation of nuclear structure and function. Annu Rev Biomed Eng. 2012;14:431–455. doi: 10.1146/annurev-bioeng-071910-124638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon LW, Walsh CE, Lambert MW. Human alpha Spectrin II and the Fanconi anemia proteins FANCA and FANCC interact to form a nuclear complex. J Biol Chem. 1999;274:32904–32908. doi: 10.1074/jbc.274.46.32904. [DOI] [PubMed] [Google Scholar]
- McMahon LW, Zhang P, Sridharan DM, Lefferts JA, Lambert MW. Knockdown of alphaII spectrin in normal human cells by siRNA leads to chromosomal instability and decreased DNA interstrand cross-link repair. Biochem Biophys Res Commun. 2009;381:288–293. doi: 10.1016/j.bbrc.2009.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metral S, Machnicka B, Bigot S, Colin Y, Dhermy D, Lecomte MC. AlphaII-Spectrin is critical for cell adhesion and cell cycle. J Biol Chem. 2009;284:2409–2418. doi: 10.1074/jbc.M801324200. [DOI] [PubMed] [Google Scholar]
- Opresko PL, Otterlei M, Graakjær J, Bruheim P, Dawut L, Kølvraa S, May A, Seidman MM, Bohr VA. The werner syndrome helicase and exonuclease cooperate to resolve telomeric D loops in a manner regulated by TRF1 and TRF2. Mol Cell. 2004;14:763–774. doi: 10.1016/j.molcel.2004.05.023. [DOI] [PubMed] [Google Scholar]
- Shimi T, Kittisopikul M, Tran J, Goldman AE, Adam SA. Structural organization of nuclear lamins A, C, B1 and B2 revealed by super-resolution microscopy. Mol Biol Cell. 2015;26:4075–4068. doi: 10.1091/mbc.E15-07-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimi T, Pfleghaar K, Kojima SI, Pack CG, Solovei I, Goldman AE, Adam SA, Shumaker DK, Kinjo M, Cremer T, Goldman RD. The A- and B-type nuclear lamin networks: microdomains involved in chromatin organization and transcription. Genes Dev. 2008;22:3409–3421. doi: 10.1101/gad.1735208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon DN, Wilson KL. The nucleoskeleton as a genome-associated dynamic “network of networks. Nat Rev Mol Cell Biol. 2011;12:695–708. doi: 10.1038/nrm3207. [DOI] [PubMed] [Google Scholar]
- Spagnol ST, Dahl KN. Active cytoskeletal force and chromatin condensation independently modulate intranuclear network fluctuations. Integr Biol. 2014;6:523–531. doi: 10.1039/c3ib40226f. [DOI] [PubMed] [Google Scholar]
- Sridharan DM, McMahon LW, Lambert MW. AlphaII-Spectrin interacts with five groups of functionally important proteins in the nucleus. Cell Biol Int. 2006;30:866–878. doi: 10.1016/j.cellbi.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Wang N, Tytell JD, Ingber DE. Mechanotransduction at a distance: mechanically coupling the extracellular matrix with the nucleus. Nat Rev Mol Cell Biol. 2009;10:75–82. doi: 10.1038/nrm2594. [DOI] [PubMed] [Google Scholar]
- Yang G, Cameron La, Maddox PS, Salmon ED, Danuser G. Regional variation of microtubule flux reveals microtubule organization in the metaphase meiotic spindle. J Cell Biol. 2008;182:631–639. doi: 10.1083/jcb.200801105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young KG, Kothary R. Spectrin repeat proteins in the nucleus. BioEssays. 2005;27:144–152. doi: 10.1002/bies.20177. [DOI] [PubMed] [Google Scholar]
- Zhang P, Herbig U, Coffman F, Lambert MW. Non-erythroid alpha spectrin prevents telomere fragility after DNA interstrand cross-link damage. Nucleic Acids Res. 2013;41:5321–5340. doi: 10.1093/nar/gkt235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Zhang C, Zhao Q, Li D. Spectrin: structure, function and disease. Sci China Life Sci. 2013;56:1076–1085. doi: 10.1007/s11427-013-4575-0. [DOI] [PubMed] [Google Scholar]
- Zhong Z, Booth-Gauthier EA, Dahl KN. αII Spectrin stabilizes stress fibers and actin–membrane interactions. Cell Mol Bioeng. 2011;4:106–115. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.