Abstract
Background
Examining transcriptional regulation by antidepressants in key neural circuits implicated in depression, and understanding the relationship to transcriptional mechanisms of susceptibility and natural resilience, may help in the search for new therapeutics. Given the heterogeneity of treatment response in human populations, examining both treatment response and non-response is critical.
Methods
We compared the effects of a conventional monoamine-based tricyclic antidepressant, imipramine, and a rapidly acting, non-monoamine-based antidepressant, ketamine, in mice subjected to chronic social defeat stress, a validated depression model, and used RNA-sequencing to analyze transcriptional profiles associated with susceptibility, resilience and antidepressant response and non-response in prefrontal cortex (PFC), nucleus accumbens, hippocampus, and amygdala.
Results
We identified similar numbers of responders and non-responders following ketamine or imipramine treatment. Ketamine induced more expression changes in hippocampus; imipramine induced more expression changes in nucleus accumbens and amygdala. Transcriptional profiles in treatment responders were most similar in PFC. Non-response reflected both the lack of response-associated gene expression changes and unique gene regulation. In responders, both drugs reversed susceptibility-associated transcriptional changes as well as induced resilience-associated transcription in PFC.
Conclusions
We generated a uniquely large resource of gene expression data in four inter-connected limbic brain regions implicated in depression and its treatment with imipramine or ketamine. Our analyses highlight the PFC as a key site of common transcriptional regulation by both antidepressant drugs and in both reversing susceptibility- and inducing resilience-associated molecular adaptations. In addition, we found region-specific effects of each drug suggesting both common and unique effects of imipramine versus ketamine.
Keywords: depression, RNA-seq, susceptibility, resilience, ketamine, imipramine
Introduction
Depression is a complex and heterogeneous disorder and a leading cause of disability worldwide, yet existing pharmacotherapies have limited efficacy (1). Virtually all drugs used to treat depression today target the same basic mechanisms identified more than 60 years ago, inducing full remission in fewer than 50% of affected individuals (2). Earlier treatments, such as tricyclic antidepressants (e.g. imipramine), target multiple neurotransmitter systems. Specifically, imipramine inhibits reuptake of serotonin and norepinephrine (thought to mediate its therapeutic actions), and influences numerous monoaminergic and cholinergic receptors. More recently developed antidepressants have greater selectivity at inhibiting serotonin and/or norepinephrine transporters, but roughly the same intrinsic efficacy as older tricyclic medications. Moreover, the therapeutic actions of both tricyclics and more selective reuptake inhibitors require weeks or months of treatment. While the initial target of these drugs is known, the slowly developing drug-induced adaptations that mediate antidepressant outcomes remain unknown (3, 4). There is a great unmet need to develop more effective and more rapidly acting treatments for depression, ideally guided by an improved understanding of the pathophysiology of the syndrome.
Several groups have shown that ketamine, a dissociative anesthetic, induces rapid antidepressant effects in ~50% of patients who are resistant to available tricyclic and reuptake inhibitor antidepressants (5, 6). While ketamine’s mechanism of action as a non-competitive NMDA glutamate receptor antagonist has been studied with regard to its anesthetic and recreational use at high doses, the functional and molecular underpinnings of ketamine’s antidepressant action at lower doses are a matter of ongoing study, with several attractive models of altered synaptic and structural changes proposed (7–9). Unbiased genome-wide transcriptional profiling may shed new light on the molecular mechanisms targeted by both established and experimental pharmacotherapies, thereby facilitating the development of novel antidepressant treatments.
A key challenge in understanding the mechanism of action of existing pharmacotherapies for depression is to identify the brain regions in which antidepressant treatments exert their effects. Neuroimaging studies of depressed patients, and findings in animal models, show that depression is a circuit-level disorder in which several functionally inter-connected brain regions are affected (10–13). One involved circuit is the highly studied cortico-mesolimbic reward system consisting of several limbic brain regions, including nucleus accumbens (NAC), prefrontal cortex (PFC), hippocampus (HIP) and amygdala (AMY). The NAC integrates information from glutamatergic inputs from PFC, AMY and HIP, among other regions (14). Structural, functional and transcriptional changes in each of these brain regions have been reported in both rodent depression models and depressed humans (12, 15–25). Thus, examining how antidepressant drugs regulate transcriptional profiles in these functionally interconnected brain regions may offer important mechanistic insights into their therapeutic actions.
In studying the mechanism of action of antidepressant drugs, it is important to address both the individual receiving the treatment and the heterogeneity of treatment response. Antidepressants do not elevate mood in non-depressed individuals, suggesting that unique responses may occur in depressed patients. Likewise, analyzing drug-induced transcriptional changes in both responders and non-responders may be particularly informative in distinguishing drug-induced therapeutic changes from off-target effects. A key question is whether lack of response reflects simply the lack of drug-induced therapeutic changes, or induction of aberrant transcriptional programs that mask antidepressant actions.
Here, we compared imipramine and ketamine action in mice subjected to chronic social defeat stress (CSDS), an ethologically validated model of depression and social stress-related disorders (26, 27). Chronic, but not acute, administration of imipramine or other standard antidepressants has been shown to reverse a range of behavioral abnormalities in roughly 60% of mice (26, 28). Recently, single doses of ketamine were shown to induce roughly equivalent treatment responses (29). We used RNA-sequencing (RNA-seq) to characterize transcriptomic responses genome-wide to either chronic imipramine or acute ketamine within the limbic circuitry noted above: NAC, PFC, HIP and AMY. Our findings demonstrate fundamental differences in the molecular and brain region targets of these two medications in responders and non-responders, results which have important implications for antidepressant drug discovery efforts.
Methods & Materials
(See also Supplementary Methods)
CSDS, behavioral testing and drug treatment
An established CSDS protocol was used to induce depressive-like behaviors in mice (26, 27). C57BL/6J mice were subjected to 10 daily, 5-min defeats by a novel CD1 aggressor and social-avoidance behavior was assessed in a two-stage social-interaction (SI) test 24h after the final defeat. In the ‘no target’ test mice freely explored an arena containing an empty enclosure. In the ‘target’ test mice were returned to the arena with a novel CD1 mouse in the enclosure. Time spent in the ‘interaction zone’ (IZ) surrounding the enclosure was measured. Resilient mice spent more time in IZ in target than no target and total time in IZ in target >60s. Susceptible mice spent less time in IZ with target than no target and total time in IZ in target <60s.
Susceptible mice were treated with either saline, ketamine or imipramine. 24h following the final injection, mice were subjected to a second SI test (SI2). Mice were defined as “responders” to imipramine or ketamine treatment if they spent more time in IZ in target following antidepressant treatment and had an increase of >20s in IZ in target from SI1 to SI2. Mice were defined as “non-responders” if they spent less time in IZ in target following treatment or had an increase of <10s in IZ in target from SI1 to SI2. Saline-treated resilient and susceptible animals were included in transcriptome-wide analyses if they continued to meet the SI1 criteria in SI2. All control animals were included in downstream analysis.
RNA isolation, library preparation and RNA-sequencing
Mice were killed 2 days following SI2 and NAC, PFC, HIP and AMY tissues rapidly dissected and frozen on dry ice. Tissue from 2 mice were pooled for each sample for n=3–5 biological replicates for each brain region and phenotype. RNA isolation, qPCR and data analyses were performed as described (12). Libraries were prepared using the TruSeq RNA Sample Prep Kit v2 protocol (Illumina, San Diego, CA) and sequenced with 50 base pair paired-end reads. (See Supplementary Methods)
Statistical and bioinformatic data analysis
Differential expression analyses
Pair-wise differential expression comparisons were performed using Voom Limma (34) and a nominal significance threshold of fold change >1.3 and p<0.05. (See Supplementary Methods)
Enrichment analyses
Enrichment between gene lists was analyzed using the GeneOverlap R package (www.bioconductor.org/packages/release/bioc/html/GeneOverlap.html).
Results
Differential expression signatures of susceptibility vs. resilience to CSDS and treatment response vs. non-response
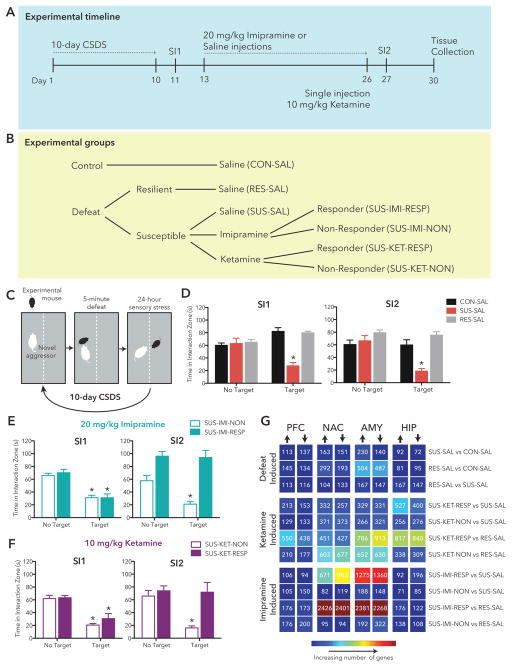
C57BL/6J mice were exposed to CSDS and (Figure 1A,C) 24 hr after the final defeat, underwent initial social interaction testing (SI1) to screen for susceptibility vs. resilience (Figure 1D–F). Previous work has established that CSDS induces two phenotypes: mice that are susceptible to stress (~67%) exhibiting profound and enduring social avoidance, and a resilient population (~33%) that continue to show a preference for social interaction similar to control mice (27). The mechanisms underlying such different responses to stress among inbred mice raised under identical conditions remain unknown. Our data showed a similar split with 55 susceptible animals and 22 resilient animals (Figure S1). Figure 1D–F shows group averages for animals included in downstream sequencing analysis (highlighted in Figure S1).
Figure 1. Study Overview.
(a) Schematic outlining study design and experimental manipulations. (b) Social interaction data 24h post CSDS and again following drug treatment. (c) The number of DEGs in each pair-wise comparison (p<0.05) is displayed in the matrix with warmer colors indicating increasing numbers of DEGs. Time spent in the interaction zone in the absence (No Target) or presence (Target) of a novel mouse 24h after CSDS (SI1) and 24h following 14 daily injections (SI2) in: (d) saline (SAL) treated control (CON), susceptible (SUS) and resilient (RES) mice, (e) imipramine (IMI) treated susceptible responders (RESP) and non-responders (NON) and (f) ketamine (KET) treated susceptible responders (RESP) and non-responders (NON). (g) Table summarizes number of differentially expressed genes (p<0.05, FC>1.3; DEGs) in each pair-wise comparison in each brain region with warmer colors representing increasing numbers of DEGs and text indicating exact number.
Control and resilient animals were treated with saline for 14d (control = 10, resilient = 8; Figure 1B). Groups of susceptible mice were treated chronically (14d) with saline, 20 mg/kg imipramine, or saline followed by acute treatment with 10 mg/kg ketamine (saline = 6 animals, imipramine = 14, ketamine = 12; Figure 1B). Following treatment, all mice were re-tested in a second social interaction test (SI2). Repeated measures ANOVA analysis of time in interaction zone (IZ) found a phenotype x test type (no target, target) interaction in both SI1 and SI2 (Figure 1D; SI1:F(2,21) = 26.09, p < 0.0001; SI2: F(2,21) = 13.31, p = 0.0002). Susceptible animals spent significantly less time in IZ when target was present than when the enclosure was empty (Bonferroni post hoc SI1:t(5) = 5.596, p = 0.0025; SI2: t(5) = 6.427, p = 0.0014), and in the presence of the social target, both control and resilient mice spent more time in IZ than susceptible mice in both SI1 and SI2 (SI1:t(13) = 6.910, p < 0.0001; SI2: t(13) = 5.882, p < 0.0001) or control (SI1:t(15) = 7.392, p < 0.0001; SI2: t(15) = 4.372, p = 0.0005). Mice in which imipramine or ketamine treatment increased social interaction (responders) and those in which drug treatment did not alter social interaction (non-responders) were identified and included in downstream sequencing analysis (post hoc analysis imipramine responders vs. non-responders w/target present, SI2: t(13) = 6.590, p < 0.0001; ketamine responders vs. non-responders w/target present, SI2: t(11) = 4.049, p = 0.0019; Figure 1E–F). Each treatment reversed social interaction deficits in approximately 50% of susceptible mice—similar to previous studies (26, 28, 29). To generate circuit-wide transcriptional profiles we used RNA-seq to analyze the NAC, PFC, HIP and AMY from seven groups of mice—control, susceptible, and resilient saline-treated mice as well as ketamine and imipramine responders and non-responders (control = 10 animals; resilient, imipramine non-responders = 8; susceptible, ketamine responders, ketamine non-responders, imipramine responders = 6).
In each brain region, we profiled differential gene expression in susceptible-saline mice vs. control-saline mice (SUS-SAL vs. CON-SAL) and resilient-saline vs. control-saline mice (RES-SAL vs. CON-SAL) We also directly compared resilient and susceptible mice (RES-SAL vs. SUS-SAL)to identify transcriptional changes associated uniquely with either condition. Additionally, we examined differential gene expression in ketamine and imipramine responders (SUS-KET-RESP, SUS-IMI-RESP) and non-responders (SUS-KET-NON, SUS-IMI-NON) relative to SUS-SAL and RES-SAL to examine how treatment response and non-response relate to natural processes of susceptibility and resilience to chronic stress. Figure 1G summarizes the number of upregulated and downregulated differentially expressed genes (DEGs) in each comparison. The largest number of DEGs in ketamine responders compared to saline-treated susceptible mice (SUS-KET-RESP vs. SUS-SAL) were observed in HIP and AMY. In contrast, more DEGs were detected in NAC and AMY in imipramine responders compared to saline-treated susceptible mice (SUS-IMI-RESP vs. SUS-SAL). Intriguingly, the largest number of DEGs across all comparisons was observed in NAC and AMY comparing imipramine responders to resilient mice (SUS-IMI-RESP vs. RES-SAL), and a large number of DEGs were observed across all brain regions in comparing ketamine responders to resilient mice (SUS-KET-RESP vs. RES-SAL). These initial observations raise interesting questions about how transcriptional profiles associated with ketamine or imipramine response relate to natural processes of resilience and susceptibility.
Comparison of ketamine and imipramine treatment response and non-response
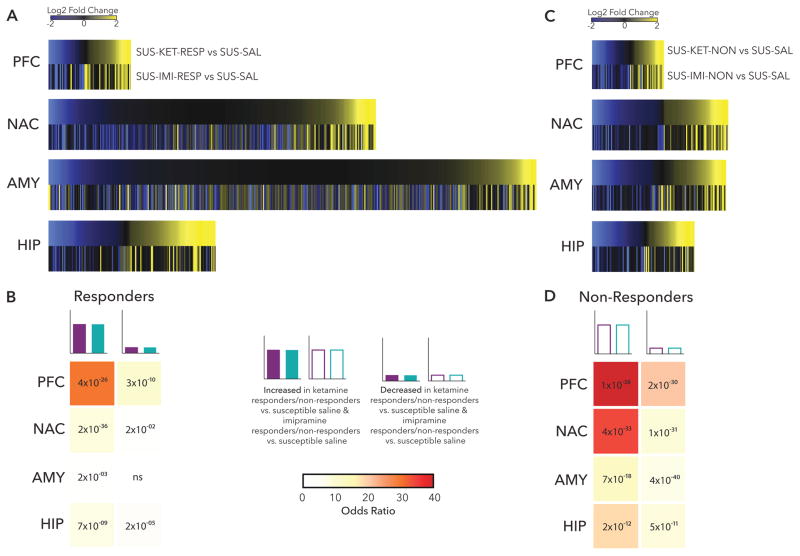
To identify similarities between transcriptional profiles associated with effective treatment response to either ketamine or imipramine, we plotted union heatmaps of log2 fold change of all significant DEGs in either SUS-KET-RESP vs. SUS-SAL or SUS-IMI-RESP vs. SUS-SAL in each brain region (Figure 2A). Visual inspection reveals the greatest similarity between treatment responses in up- and downregulated DEGs in PFC, the region in which the fewest DEGs were detected for both drug responses (Figure 1G). In contrast, transcriptional profiles were most distinct in AMY (Figure 2A), where many more DEGs were associated with imipramine response than ketamine response (Figure 1G). These observations were confirmed by Fisher’s exact tests of enrichment which identified a highly significant overlap of DEGs upregulated (29.96×, p= 4×10−26) and downregulated (10.07×, p=3×10−10) in both SUS-KET-RESP vs. SUS-SAL and SUS-IMI-RESP vs. SUS-SAL in PFC (Figure 2B). In contrast, the overlap of ketamine and imipramine response was non-significant for DEGs downregulated in AMY (1.09×, p>0.05), with only modest overlap observed for DEGs upregulated in this region (1.74×, p= 2×10−03) (Figure 2B).
Figure 2. Characterization of Treatment Response & Non-Response.
(a) Heatmaps show the union of ketamine response (SUS-KET-RESP vs. SUS-SAL) and imipramine response (SUS-IMI-RESP vs. SUS-SAL) DEGs rank ordered by log2 fold changes of ketamine response and scaled by relative number of DEGs. (b) Table of p-value (text) and odds ratio (warmer colors indicating increasing odds ratio) for Fisher’s exact test for enrichment of ketamine response DEGs in imipramine response DEGs. (c) Heatmaps show the union of ketamine non-response (SUS-KET-NON vs. SUS-SAL) and imipramine non-response (SUS-IMI-NON vs. SUS-SAL) DEGs rank ordered by log2 fold changes of ketamine non-response and scaled by relative number of DEGs. (d) Table of p-value (text) and odds ratio (warmer colors indicating increasing odds ratio) for Fisher’s exact test for enrichment of ketamine non-response DEGs in imipramine non-response DEGs. *p<0.05
Contrasting with the large number of DEGs and limited overlap between ketamine and imipramine response across brain regions, relatively fewer DEGs associated with non-response to the two drugs (Figure 1G) and there was a much greater similarity in non-response transcriptional profiles across all brain regions (Figure 2C). Fisher’s exact tests identified the largest overlap in DEGs upregulated in both SUS-KET-NON vs. SUS-SAL and SUS-IMI-NON vs. SUS-SAL in PFC (69.92×, p= 1× 10−38) and in NAC (34.22×, p= 4× 10−33), with substantial overlap observed across DEGs in all brain regions (Figure 2D). The increased overlap in DEGs of non-response to ketamine and imipramine points to greater specificity in transcriptional profiles associated with a therapeutic-like response to each drug than non-response, which may reflect that, in part, non-responders to either drug remain more similar to susceptible mice.
Characteristics of DEGs in treatment responders and non-responders
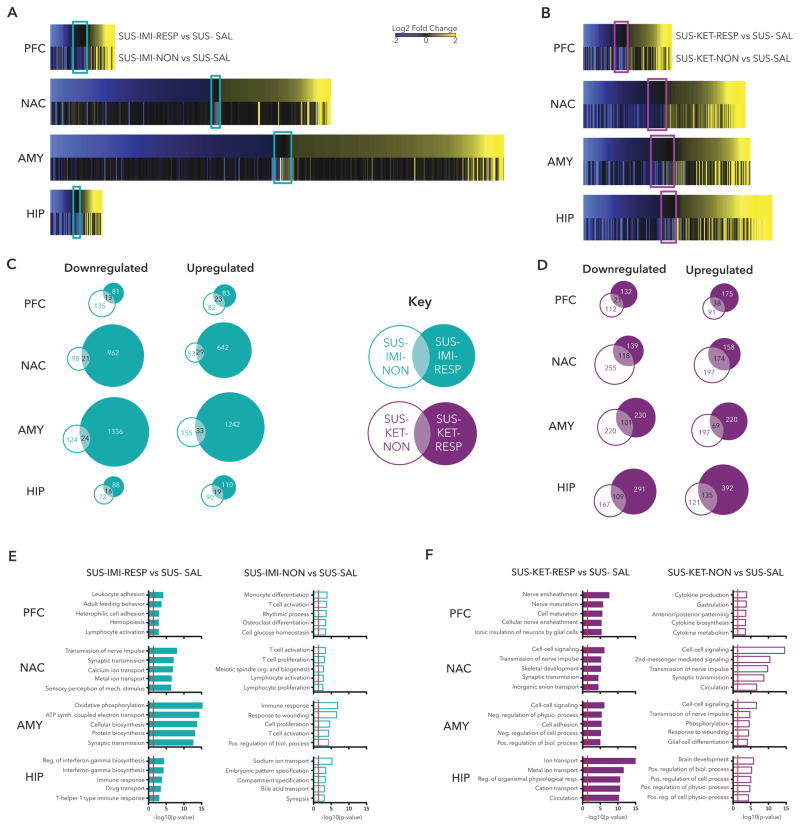
Direct comparison of ketamine responders vs. non-responders, and of imipramine responders vs. non-responders, revealed that, while non-response is associated with the lack of some transcriptional changes seen in responders, non-response is also associated with transcriptional regulation unique to this condition (Figure 3A–D). In imipramine treated mice, AMY had the largest number of genes uniquely associated with response and the largest number of genes uniquely associated with non-response (Figure 3C). Gene ontology analysis revealed that genes regulated in AMY in imipramine responders enriched for biological processes including oxidative phosphorylation (5.65×, p=3.91×10−17) and synaptic transmission (2.14×, p=2.111×10−13), whereas genes regulated in non-responders were modestly enriched in biological processes such as immune response (2.35×, p=1.56×10−07) and response to wounding (2.73×, p=2.81×10−07; Figure 3E). In ketamine treated mice, HIP had the largest number of genes uniquely associated with response, whereas NAC had the largest number of genes associated with non-response (Figure 3F). Gene ontology analysis revealed that genes regulated in HIP in ketamine responders enriched for biological processes of ion transport (2.72×, p=8.24×10−17) and circulation (3.78×, p=5.05×10−11). Genes regulated in non-responders in NAC strongly enriched for biological processes including cell-cell signalling (2.76×, p=2.56×10−15), transmission of nerve impulse (2.98×, p=1.65×10−10) and circulation (3.49×, p=2.10×10−07).
Figure 3. Comparison of Treatment Response and Non-Response.
(a) Heatmaps show the union of ketamine response DEGs (SUS-KET-RESP vs. SUS-SAL) and ketamine non-response DEGs (SUS-KET-NON vs. SUS-SAL) in each brain region rank ordered by log2 fold changes of ketamine response and scaled by relative number of DEGs. (b) Heatmaps show the union of imipramine response DEGs (SUS-IMI-RESP vs. SUS-SAL) and imipramine non-response DEGs (SUS-IMI-NON vs. SUS-SAL) in each brain region, rank ordered by log2 fold changes of imipramine response and scaled by relative number of DEGs. Colored rectangles highlight DEGs significantly regulated exclusively in non-responders and not in responders. (c) Venn diagrams represent the number of common and unique downregulated (left panel) and upregulated (right panel) DEGs in imipramine responders and non-responders in each brain region. (d) Venn diagrams represent the number of common and unique downregulated (left panel) and upregulated (right panel) DEGs in ketamine responders and non-responders in each brain region. (e) Bar graphs show biological pathways enriched in DEGs in imipramine responders (left panel) and non-responders (right panel). (f) Bar graphs show biological pathways enriched in DEGs in ketamine responders (left panel) and non-responders (right panel). Red line indicates p=0.05.
Treatment-response is associated with induction of transcriptional profiles of resilience
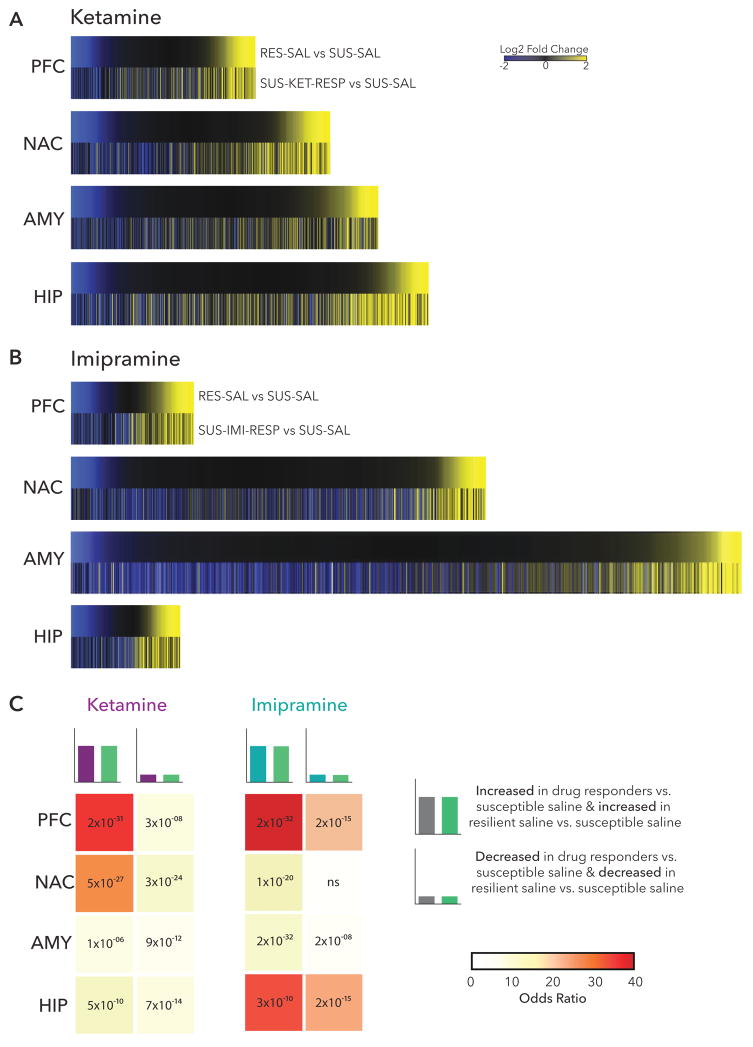
A growing body of literature demonstrates that resilience to CSDS or other forms of stress reflects active processes and not merely the absence of susceptibility (27, 35–38). We therefore examined the possibility that ketamine and imipramine induce—in responders—transcriptional profiles associated with natural resilience. Plotting union heatmaps of log2 fold changes of all significant DEGs regulated in either RES-SAL vs. SUS-SAL and SUS-KET-RESP vs. SUS-SAL and separately RES-SAL vs. SUS-SAL and SUS-IMI-RESP vs. SUS-SAL in each brain region (Figure 4A,B) revealed considerable overlap across all brain regions with both drug responses, although the magnitude of overlap differed by brain region and drug. In ketamine responders, DEGs upregulated in PFC (36.05×, p= 2×10−31), or those upregulated in NAC (24.36×, p= 5×10−27), showed the largest amount of overlap with resilient-specific DEGs (Figure 4C). (Note that SUS-SAL, not CON-SAL, was used as the reference condition for these analyses since many gene expression changes seen in resilience also occur in susceptibility (27) and we sought to isolate those associated uniquely with resilience.)
Figure 4. Induction of Resilience DEGs with Treatment Response.
(a) Heatmaps show the union of ketamine response (SUS-KET-RESP vs. SUS-SAL) and resilience (RES-SAL vs. SUS-SAL) DEGs in each brain region rank ordered by log2 fold changes of resilience and scaled by relative number of DEGs. (b) Heatmaps show the union of imipramine response (SUS-IMI-RESP vs. SUS-SAL) and resilience (RES-SAL vs. SUS-SAL) DEGs in each brain region rank ordered by log2 fold changes of resilience and scaled by relative number of DEGs. (c) Table of p-value (text) and odds ratio (warmer colors indicating increasing odds ratio) for Fisher’s exact test for enrichment of ketamine response and imipramine response DEGs with resilience DEGs.
Similarly, the largest overlap between resilient-specific DEGs and imipramine response DEGs occurred in PFC (64.31×, p= 2×10−32). While less overlap was seen in NAC between resilient-specific and imipramine response DEGs (12.39×, p= 1×10−20) than with ketamine response, there was a large overlap in upregulated HIP DEGs (33.24, p= 3×10−10).
Taken together, these findings suggest that both ketamine and imipramine exert antidepressant effects in part through regulating expression of genes associated with natural resilience. However, interestingly, while both drugs induce resilient-specific transcriptional profiles in PFC, this effect of ketamine is also seen in NAC whereas imipramine’s effect is also seen in HIP, indicating important circuit-level specificity of these two different antidepressant drugs.
Treatment-response is also associated with reversal of transcriptional profiles of susceptibility
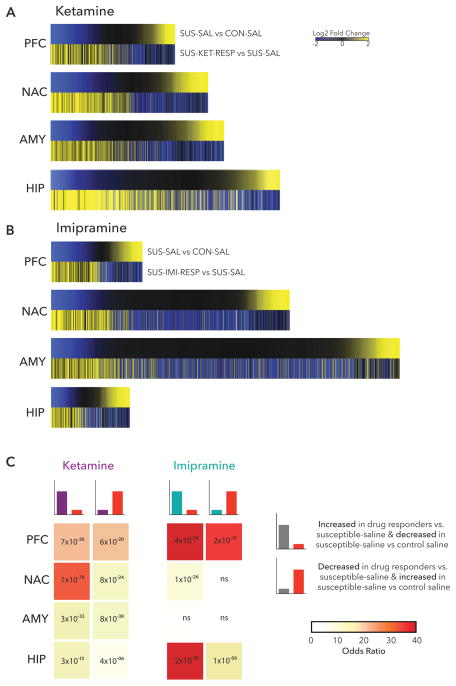
We next examined the possibility that antidepressant-like effects of ketamine and imipramine reverse transcriptional profiles associated with susceptibility. Plotting log2 fold changes of all significant DEGs regulated in either SUS-SAL vs. CON-SAL and SUS-KET-RESP vs. SUS-SAL in each brain region in union heatmaps revealed substantial opposing regulation between ketamine response and susceptibility in all brain regions (Figure 5A). As with induction of resilient-specific DEGs, the largest overlap was observed in NAC (31.28×, p=1×10−78) and PFC (p=6×10−20) with genes increased in drug responders overlapping with genes decreased in susceptibility (Figure 5C).
Figure 5. Reversal of Susceptibility DEGs with Treatment Response.
(a) Heatmaps show the union of ketamine response (SUS-KET-RESP vs. SUS-SAL) and susceptibility (SUS-SAL vs. CON-SAL) DEGs in each brain region rank ordered by log2 fold changes of susceptibility and scaled by relative number of DEGs. (b) Heatmaps show the union of imipramine response (SUS-IMI-RESP vs. SUS-SAL) and susceptibility (SUS-SAL vs. CON-SAL) DEGs in each brain region rank ordered by log2 fold changes of susceptibility and scaled by relative number of DEGs. (c) Table of p-value (text) and odds ratio (warmer colors indicating increasing odds ratio) for Fisher’s exact test for enrichment of ketamine response and imipramine response DEGs with susceptible DEGs.
The union of log2 fold changes of all significant DEGs regulated in either SUS-SAL vs. CON-SAL and SUS-IMI-NON vs. SUS-SAL revealed more region-specific reversal of susceptible-specific transcriptional profiles by imipramine than for ketamine (Figure 5B). Genes decreased in drug responders also overlapped considerably with genes increased in susceptibility in PFC (18.93×, p=19.35×, 6×10−20). Again, as observed with induction of resilient-specific DEGs by imipramine, the strongest effects in reversing susceptibility occurred in PFC and HIP. Genes increased in drug responders were greatly enriched amongst genes decreased in susceptibility in both PFC (44.80×, p= 4×10−34) and HIP (47.77×, p=2×10−18). Genes decreased in drug responders also strongly overlapped with genes increased in susceptibility in PFC (35.45×, p=2×10−19). In contrast, only limited opposing regulation was observed in NAC (7.60×, p=8×10−24) and no significant regulation in AMY (1.36× p>0.05) (Figure 5C).
Discussion
We generated a uniquely large resource of genome-wide gene expression data (publicly available in GEO) in four inter-connected limbic brain regions implicated in depression and its treatment to extend our understanding of transcriptional mechanisms of antidepressant response vs. non-response with a conventional monoamine-based tricyclic antidepressant (imipramine) and a rapidly acting, non-monoamine-based antidepressant (ketamine). Importantly, our study examines drug responses in stressed mice exhibiting behavioral susceptibility, as opposed to naive mice, and encompasses the heterogeneity of response—responders vs. non-responders—to antidepressant treatment. By independently analyzing transcriptional profiles in brains of responders (mice in which treatment reversed the deleterious behavioral effects of stress) and non-responders (mice in which treatment failed to induce this positive behavioral change) our data differentiate between transcriptional regulation associated with the therapeutic-like effects of each drug and off-target drug effects that either do not contribute to or may even antagonize antidepressant actions. Our data show that, at the transcriptional level, treatment response is characterized by both the reversal of some susceptibility-associated changes in gene expression plus the induction of some resilience-specific gene regulation. Likewise, our data reveal that non-response is more than simply the lack of response to treatment, but additionally reflects some aberrant regulation of gene expression. These features of treatment response and non-response differ for ketamine vs. imipramine, with clear differences observed across the four brain regions examined. Interestingly, the brain regions associated with the greatest number of DEGs with drug response were distinct from the regions in which the most statistically significant enrichment of pro-resilience and anti-susceptible transcriptional profiles was detected. One interpretation of this observation is that antidepressant response encompasses unique transcriptional regulation above and beyond both the normalization of aberrant transcription associated with susceptibility and induction of transcriptional programs of resilience.
Our findings suggest that antidepressant effects can be achieved through different circuit-level mechanisms. The greatest similarity between ketamine and imipramine responders was observed in PFC. Interestingly, the most overlap in ketamine and imipramine non-responders was also found in PFC. This suggests that PFC is a key locus of stress-induced transcriptional changes relevant to depression and a common site of action for these two very distinct antidepressant drugs. Both drugs exerted robust pro-resilience effects in PFC, inducing patterns of gene expression significantly enriched for resilient-specific DEGs. Imipramine, and to a lesser extent, ketamine, also powerfully reversed susceptibility in PFC, inducing patterns of gene expression that strongly opposed susceptible DEGs. These transcriptional level findings with imipramine are consistent with an earlier report, where we also showed similarities in genome-wide patterns of phosphoCREB binding and in repressive histone methylation in NAC of resilient vs. imipramine-treated mice after CSDS (36). At the functional level, the importance of normalization of disrupted PFC activity is well established in mouse models and humans (7, 12, 15, 18, 39). Altered activity in PFC is observed in human depression, where deep brain stimulation induces an antidepressant response in treatment resistant depression and normalizes depression-associated metabolic changes (39). Additionally, successful antidepressant response with either pharmacological or cognitive-behavioral treatments is associated with regulation of PFC activity (18, 40–42).
Beyond similarities in PFC, our findings identify interesting circuit-level differences in the sites of action of ketamine and imipramine. We identified NAC as a key site of action for both the pro-resilience and anti-susceptibility effects of ketamine although the NAC appeared far less important than HIP in the case of imipramine. Antidepressant response was most disparate in AMY, with many more genes regulated in imipramine responders than ketamine responders. Interestingly, although imipramine response was associated with a very large number of genes in AMY, these DEGs were not highly enriched for either pro-resilience or anti-susceptible DEG signatures suggesting that the antidepressant effects of imipramine do not relate to natural processes of susceptibility and resilience in this brain region. Together, our findings identify the PFC as a common, and potentially essential, target of antidepressant drugs in addition to which more drug-specific antidepressant effects are mediated by other brain regions in this functionally interconnected circuit.
The rich datasets presented in this study provide an invaluable resource to aid the development of new antidepressant compounds that selectively target transcriptional changes associated with stress susceptibility, resilience or treatment responsiveness. Of interest, a number of genes identified in key signatures of reversal of susceptibility or induction of resilience in association with drug response have been previously studied in depression. In PFC, ketamine normalized the reduced levels of Dusp1 associated with susceptibility. DUSP1 is a regulator of MAP-kinase signalling that was found to be increased in postmortem HIP of depressed humans and is regulated by antidepressant treatment in mice (43). Ketamine also reversed the reduction of Arc observed in PFC of susceptible mice, which we have implicated in depression in previous studies (12, 15). Fgf23, a member of the fibroblast growth factor family of genes known to regulate affective behavior, was induced in PFC by both ketamine and imipramine similar to effects in resilient mice (44, 45). Likewise, in NAC, among the susceptibility genes reversed by ketamine was another family member, Fgf3. Interestingly, ketamine also reversed decreased levels of Htr1b, which encodes the serotonin 1b receptor, seen in NAC of susceptible mice. Single nucleotide polymorphisms in HTR1B have been reported to regulate response to serotonin-selective reuptake inhibitor antidepressants and manipulation of this gene in mouse models regulates emotional behaviour (46, 47). In HIP, imipramine induced expression of Ctla4 similar to effects in resilient mice. Polymorphisms in CTLA4, a member of the immunoglobulin superfamily expressed on helper T cells, have been reported in both Korean and Han Chinese populations in association with depression (48, 49). Finally, our discovery of unique changes that occur in non-responders raises the interesting possibility that a subset of these changes may oppose therapeutic efficacy, something that now warrants direct examination. If this proves to be true, it might be possible to enhance therapeutic efficacy of available antidepressant treatments by developing ways of opposing these non-response associated transcriptional changes.
As the impact of depression on humanity increases, we need to develop a far broader range of antidepressant treatments with more rapid onset of action that also effectively treat patients failed by existing medications. Ketamine is an extremely promising new agent, however, it is not fully efficacious in all treatment-resistant individuals; also its antidepressant effects are transient and the safety of long-term ketamine treatment needs to be established. The genome-wide transcriptome mapping utilized here offers a template of several strategies to identify—in unbiased ways—novel drug targets which alone or in combination can be exploited to develop improved treatments for depression.
Supplementary Material
Acknowledgments
The authors would like to acknowledge James Palmer and Guang Chen for valuable discussions on the experimental design.This work was supported by P50 MH096890, R01 MH051399, a grant from Janssen Research and Development, and a Hope for Depression Research Foundation grant to E.J.N and a 2014 NARSAD Young Investigator Award 22713 from the Brain & Behavior Research Foundation to R.C.B.
Footnotes
Financial Disclosures
E.J.N reports consulting income from Janssen Research and Development, and G.M.W and L.Y. are employees of Janssen Research and Development, LLC. R.C.B., H.M.C., I.P., V.V., E.A.H., B.L., C.J.P, L.S. reported no biomedical financial interests or potential conflicts of interest.
Accession Numbers
Raw and processed RNA-seq gene expression data are available via the Gene Expression Omnibus data (accession number GEO:GSE81672).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Greenberg PE, Fournier AA, Sisitsky T, Pike CT, Kessler RC. The economic burden of adults with major depressive disorder in the United States (2005 and 2010) The Journal of clinical psychiatry. 2015;76:155–162. doi: 10.4088/JCP.14m09298. [DOI] [PubMed] [Google Scholar]
- 2.Block SG, Nemeroff CB. Emerging antidepressants to treat major depressive disorder. Asian journal of psychiatry. 2014;12:7–16. doi: 10.1016/j.ajp.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 3.Krishnan V, Nestler EJ. The molecular neurobiology of depression. Nature. 2008;455:894–902. doi: 10.1038/nature07455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duric V, Duman RS. Depression and treatment response: dynamic interplay of signaling pathways and altered neural processes. Cellular and molecular life sciences : CMLS. 2013;70:39–53. doi: 10.1007/s00018-012-1020-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murrough JW, Iosifescu DV, Chang LC, Al Jurdi RK, Green CE, Perez AM, et al. Antidepressant efficacy of ketamine in treatment-resistant major depression: a two-site randomized controlled trial. The American journal of psychiatry. 2013;170:1134–1142. doi: 10.1176/appi.ajp.2013.13030392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zarate CA, Jr, Singh JB, Carlson PJ, Brutsche NE, Ameli R, Luckenbaugh DA, et al. A randomized trial of an N-methyl-D-aspartate antagonist in treatment-resistant major depression. Archives of general psychiatry. 2006;63:856–864. doi: 10.1001/archpsyc.63.8.856. [DOI] [PubMed] [Google Scholar]
- 7.Li N, Lee B, Liu RJ, Banasr M, Dwyer JM, Iwata M, et al. mTOR-dependent synapse formation underlies the rapid antidepressant effects of NMDA antagonists. Science. 2010;329:959–964. doi: 10.1126/science.1190287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Autry AE, Adachi M, Nosyreva E, Na ES, Los MF, Cheng PF, et al. NMDA receptor blockade at rest triggers rapid behavioural antidepressant responses. Nature. 2011;475:91–95. doi: 10.1038/nature10130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Monteggia LM, Zarate C., Jr Antidepressant actions of ketamine: from molecular mechanisms to clinical practice. Current opinion in neurobiology. 2015;30:139–143. doi: 10.1016/j.conb.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pandya M, Altinay M, Malone DA, Jr, Anand A. Where in the brain is depression? Current psychiatry reports. 2012;14:634–642. doi: 10.1007/s11920-012-0322-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ressler KJ, Mayberg HS. Targeting abnormal neural circuits in mood and anxiety disorders: from the laboratory to the clinic. Nature neuroscience. 2007;10:1116–1124. doi: 10.1038/nn1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bagot RC, Parise EM, Pena CJ, Zhang HX, Maze I, Chaudhury D, et al. Ventral hippocampal afferents to the nucleus accumbens regulate susceptibility to depression. Nature communications. 2015;6:7062. doi: 10.1038/ncomms8062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christoffel DJ, Golden SA, Walsh JJ, Guise KG, Heshmati M, Friedman AK, et al. Excitatory transmission at thalamo-striatal synapses mediates susceptibility to social stress. Nature neuroscience. 2015;18:962–964. doi: 10.1038/nn.4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goto Y, Grace AA. Limbic and cortical information processing in the nucleus accumbens. Trends in neurosciences. 2008;31:552–558. doi: 10.1016/j.tins.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Covington HE, 3rd, Lobo MK, Maze I, Vialou V, Hyman JM, Zaman S, et al. Antidepressant effect of optogenetic stimulation of the medial prefrontal cortex. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2010;30:16082–16090. doi: 10.1523/JNEUROSCI.1731-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vialou V, Bagot RC, Cahill ME, Ferguson D, Robison AJ, Dietz DM, et al. Prefrontal cortical circuit for depression- and anxiety-related behaviors mediated by cholecystokinin: role of DeltaFosB. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2014;34:3878–3887. doi: 10.1523/JNEUROSCI.1787-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kennedy SH, Evans KR, Kruger S, Mayberg HS, Meyer JH, McCann S, et al. Changes in regional brain glucose metabolism measured with positron emission tomography after paroxetine treatment of major depression. The American journal of psychiatry. 2001;158:899–905. doi: 10.1176/appi.ajp.158.6.899. [DOI] [PubMed] [Google Scholar]
- 18.Mayberg HS, Brannan SK, Tekell JL, Silva JA, Mahurin RK, McGinnis S, et al. Regional metabolic effects of fluoxetine in major depression: serial changes and relationship to clinical response. Biological psychiatry. 2000;48:830–843. doi: 10.1016/s0006-3223(00)01036-2. [DOI] [PubMed] [Google Scholar]
- 19.Jaworska N, Yang XR, Knott V, Macqueen G. A review of fMRI studies during visual emotive processing in major depressive disorder. The world journal of biological psychiatry : the official journal of the World Federation of Societies of Biological Psychiatry. 2014 doi: 10.3109/15622975.2014.885659. [DOI] [PubMed] [Google Scholar]
- 20.Sequeira A, Mamdani F, Ernst C, Vawter MP, Bunney WE, Lebel V, et al. Global brain gene expression analysis links glutamatergic and GABAergic alterations to suicide and major depression. PloS one. 2009;4:e6585. doi: 10.1371/journal.pone.0006585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guilloux JP, Douillard-Guilloux G, Kota R, Wang X, Gardier AM, Martinowich K, et al. Molecular evidence for BDNF- and GABA-related dysfunctions in the amygdala of female subjects with major depression. Molecular psychiatry. 2012;17:1130–1142. doi: 10.1038/mp.2011.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang LC, Jamain S, Lin CW, Rujescu D, Tseng GC, Sibille E. A conserved BDNF, glutamate- and GABA-enriched gene module related to human depression identified by coexpression meta-analysis and DNA variant genome-wide association studies. PloS one. 2014;9:e90980. doi: 10.1371/journal.pone.0090980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ding Y, Chang LC, Wang X, Guilloux JP, Parrish J, Oh H, et al. Molecular and Genetic Characterization of Depression: Overlap with other Psychiatric Disorders and Aging. Molecular neuropsychiatry. 2015;1:1–12. doi: 10.1159/000369974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nasca C, Zelli D, Bigio B, Piccinin S, Scaccianoce S, Nistico R, et al. Stress dynamically regulates behavior and glutamatergic gene expression in hippocampus by opening a window of epigenetic plasticity. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:14960–14965. doi: 10.1073/pnas.1516016112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McEwen BS, Nasca C, Gray JD. Stress Effects on Neuronal Structure: Hippocampus, Amygdala, and Prefrontal Cortex. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2016;41:3–23. doi: 10.1038/npp.2015.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berton O, McClung CA, Dileone RJ, Krishnan V, Renthal W, Russo SJ, et al. Essential role of BDNF in the mesolimbic dopamine pathway in social defeat stress. Science. 2006;311:864–868. doi: 10.1126/science.1120972. [DOI] [PubMed] [Google Scholar]
- 27.Krishnan V, Han MH, Graham DL, Berton O, Renthal W, Russo SJ, et al. Molecular adaptations underlying susceptibility and resistance to social defeat in brain reward regions. Cell. 2007;131:391–404. doi: 10.1016/j.cell.2007.09.018. [DOI] [PubMed] [Google Scholar]
- 28.Tsankova NM, Berton O, Renthal W, Kumar A, Neve RL, Nestler EJ. Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nature neuroscience. 2006;9:519–525. doi: 10.1038/nn1659. [DOI] [PubMed] [Google Scholar]
- 29.Donahue RJ, Muschamp JW, Russo SJ, Nestler EJ, Carlezon WA., Jr Effects of striatal DeltaFosB overexpression and ketamine on social defeat stress-induced anhedonia in mice. Biological psychiatry. 2014;76:550–558. doi: 10.1016/j.biopsych.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang JC, Yao W, Dong C, Yang C, Ren Q, Ma M, et al. Comparison of ketamine, 7,8-dihydroxyflavone, and ANA-12 antidepressant effects in the social defeat stress model of depression. Psychopharmacology. 2015;232:4325–4335. doi: 10.1007/s00213-015-4062-3. [DOI] [PubMed] [Google Scholar]
- 31.Ichikawa J, Kuroki T, Meltzer HY. Differential effects of chronic imipramine and fluoxetine on basal and amphetamine-induced extracellular dopamine levels in rat nucleus accumbens. European journal of pharmacology. 1998;350:159–164. doi: 10.1016/s0014-2999(98)00247-7. [DOI] [PubMed] [Google Scholar]
- 32.Papp M, Gruca P, Lason-Tyburkiewicz M, Willner P. Antidepressant, anxiolytic and procognitive effects of rivastigmine and donepezil in the chronic mild stress model in rats. Psychopharmacology. 2016;233:1235–1243. doi: 10.1007/s00213-016-4206-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome biology. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Friedman AK, Walsh JJ, Juarez B, Ku SM, Chaudhury D, Wang J, et al. Enhancing depression mechanisms in midbrain dopamine neurons achieves homeostatic resilience. Science. 2014;344:313–319. doi: 10.1126/science.1249240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilkinson MB, Xiao G, Kumar A, LaPlant Q, Renthal W, Sikder D, et al. Imipramine treatment and resiliency exhibit similar chromatin regulation in the mouse nucleus accumbens in depression models. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2009;29:7820–7832. doi: 10.1523/JNEUROSCI.0932-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dias C, Feng J, Sun H, Shao NY, Mazei-Robison MS, Damez-Werno D, et al. beta-catenin mediates stress resilience through Dicer1/microRNA regulation. Nature. 2014;516:51–55. doi: 10.1038/nature13976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duclot F, Kabbaj M. Individual differences in novelty seeking predict subsequent vulnerability to social defeat through a differential epigenetic regulation of brain-derived neurotrophic factor expression. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2013;33:11048–11060. doi: 10.1523/JNEUROSCI.0199-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mayberg HS, Lozano AM, Voon V, McNeely HE, Seminowicz D, Hamani C, et al. Deep brain stimulation for treatment-resistant depression. Neuron. 2005;45:651–660. doi: 10.1016/j.neuron.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 40.Brody AL, Saxena S, Silverman DH, Alborzian S, Fairbanks LA, Phelps ME, et al. Brain metabolic changes in major depressive disorder from pre- to post-treatment with paroxetine. Psychiatry research. 1999;91:127–139. doi: 10.1016/s0925-4927(99)00034-7. [DOI] [PubMed] [Google Scholar]
- 41.Brody AL, Saxena S, Stoessel P, Gillies LA, Fairbanks LA, Alborzian S, et al. Regional brain metabolic changes in patients with major depression treated with either paroxetine or interpersonal therapy: preliminary findings. Archives of general psychiatry. 2001;58:631–640. doi: 10.1001/archpsyc.58.7.631. [DOI] [PubMed] [Google Scholar]
- 42.Haertzen CA, Hooks NT., Jr Dictionary of drug associations to heroin, benzedrine, alcohol, barbiturates and marijuana. Journal of clinical psychology. 1973;29:115–164. doi: 10.1002/1097-4679(197304)29:2<115::aid-jclp2270290202>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 43.Duric V, Banasr M, Licznerski P, Schmidt HD, Stockmeier CA, Simen AA, et al. A negative regulator of MAP kinase causes depressive behavior. Nature medicine. 2010;16:1328–1332. doi: 10.1038/nm.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Turner CA, Watson SJ, Akil H. The fibroblast growth factor family: neuromodulation of affective behavior. Neuron. 2012;76:160–174. doi: 10.1016/j.neuron.2012.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Elsayed M, Banasr M, Duric V, Fournier NM, Licznerski P, Duman RS. Antidepressant effects of fibroblast growth factor-2 in behavioral and cellular models of depression. Biological psychiatry. 2012;72:258–265. doi: 10.1016/j.biopsych.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Villafuerte SM, Vallabhaneni K, Sliwerska E, McMahon FJ, Young EA, Burmeister M. SSRI response in depression may be influenced by SNPs in HTR1B and HTR1A. Psychiatric genetics. 2009;19:281–291. doi: 10.1097/YPG.0b013e32832a506e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhuang X, Gross C, Santarelli L, Compan V, Trillat AC, Hen R. Altered emotional states in knockout mice lacking 5-HT1A or 5-HT1B receptors. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 1999;21:52S–60S. doi: 10.1016/S0893-133X(99)00047-0. [DOI] [PubMed] [Google Scholar]
- 48.Jun TY, Pae CU, Chae JH, Bahk WM, Kim KS. Polymorphism of CTLA-4 gene for major depression in the Korean population. Psychiatry and clinical neurosciences. 2001;55:533–537. doi: 10.1046/j.1440-1819.2001.00901.x. [DOI] [PubMed] [Google Scholar]
- 49.Liu J, Li J, Li T, Wang T, Li Y, Zeng Z, et al. CTLA-4 confers a risk of recurrent schizophrenia, major depressive disorder and bipolar disorder in the Chinese Han population. Brain, behavior, and immunity. 2011;25:429–433. doi: 10.1016/j.bbi.2010.10.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.