Abstract
Molecular genetic studies of model plants in the past few decades have identified many key genes and pathways controlling development, metabolism and environmental responses. Recent technological and informatics advances have led to unprecedented volumes of data that may uncover underlying principles of plants as biological systems. The newly emerged discipline of synthetic biology and related molecular engineering approaches is built on this strong foundation. Today, plant regulatory pathways can be reconstituted in heterologous organisms to identify and manipulate parameters influencing signalling outputs. Moreover, regulatory circuits that include receptors, ligands, signal transduction components, epigenetic machinery and molecular motors can be engineered and introduced into plants to create novel traits in a predictive manner. Here, we provide a brief history of plant synthetic biology and significant recent examples of this approach, focusing on how knowledge generated by the reference plant Arabidopsis thaliana has contributed to the rapid rise of this new discipline, and discuss potential future directions.
In the 15 years since the first plant genome was fully sequenced1, plant biology has been at the forefront of developing tools to connect genotype to phenotype. It is a significant technical challenge to make the leap from a sequenced genome to cellular signalling architecture to forecasting systems-level outputs. Among the greatest challenges is the fact that several pervasive features of biological networks, such as redundancy, convergence on shared signalling components and feedback, are not easily resolved by molecular genetics and systems approaches. Synthetic biology, as a complementary bottom-up approach, offers an opportunity to significantly accelerate our understanding of normal plant growth and development.
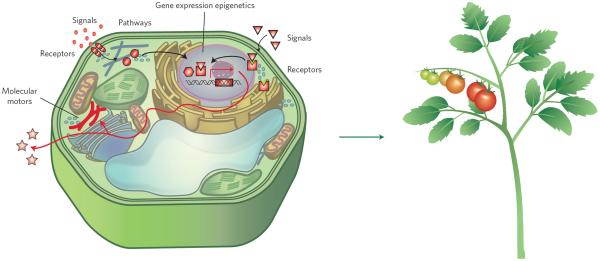
In this Review, we define `synthetic biology' as an engineering approach to design, build and analyse dynamic molecular devices and/or pathways from biological components to produce cells and organisms with customized functionality (Fig. 1). This broad definition builds bridges to approaches with natural affinity to synthetic biology (for example, chemical biology) and spans work anywhere along the spectrum, from solving explicit engineering problems to answering fundamental biological questions. A hallmark of synthetic biology is the incorporation of precise models and computational predictions of the properties of the engineered systems2. Although quantitative predictability is already crucial for rewiring or designing pathways and networks, precise prediction of outcomes is quite daunting, given the complex nature of higher plants as multicellular organisms and multiscale levels of regulation. Such approaches have been successful for producing simple genetic switches in bacteria3 and are now being applied to re-engineering photosynthesis in cyanobacteria4,5. Recent major advances in plant synthetic biology incorporate engineered variants of receptors, ligands, signal transduction components, epigenetic regulatory machinery and cytoskeletal motors with a diverse array of potential applications (Fig. 1).
Figure 1. A schematic diagram of an idealized plant cell with synthetic engineered pathways to produce a plant with ideal traits and functionality.
Each synthetic engineered component is coloured in red. Figure courtesy of Haruko Hirukawa (ITbM, Japan).
Identification and remediation of toxins
Long before the term synthetic biology was introduced and recognized in the plant biology community, the first generation of these approaches was already being implemented. Although here we focus primarily on the engineering of plant parts drawn from signalling and cell biology, there is significant research activity in synthetic metabolic engineering6,7, with many recent headlines stemming from successful porting of opiate8 and cannabinoid9 production into yeast.
Phytoremediation was the intended application of some of the earliest examples of successful metabolic engineering. For instance, engineered transgenic tobacco plants expressing human cytochrome P450 2E1 showed substantially enhanced ability to oxidize the toxic halogenic environmental pollutants trichloroethylene (TCE) and ethylene dibromide (EDB)10. Transgenic hybrid poplar expressing P450 2E1 effectively removes TCE, chloroform and even the gaseous form of benzene11.
The classic concept of directed protein evolution has also been incorporated into synthetic biology to boost the performance of transgenic organisms. Traditionally, mutagenesis and iterative cycles of selection have been used to produce proteins or pathways with enhanced activity12. Together with structure-guided rational design and high-throughput cloning/screening methods, the directed evolution approach could accelerate future protein engineering13–16. By applying such techniques to enzymes such as the P450 proteins involved in phytoremediation approaches, it may be possible in the future to design and optimize new functionalities.
The ground-up design of a complete, artificial signalling pathway to detect environmental pollutants was reported recently using an engineered bacterial two-component system17. This system consists of membrane-bound sensor histidine kinases and response regulators, which trigger conformational changes following phosphorylation, resulting in target gene expression18. One study17 introduced into Arabidopsis and tobacco a complete set of signalling components, from upstream ligand binding to downstream gene expression: (i) modified bacterial periplasmic binding protein (PBP) in the plant apoplast that could bind trinitrotoluene (TNT) as a ligand; (ii) a chimaeric bacterial two-component receptor histidine kinase (Trg:PhoR) that would bind TNT–PBP; (iii) adapted response regulator with synthetic transcriptional activation domain (PhoB–VP64); and (iv) transcriptional signalling readout using the modified PhoB promoter driving a reporter enzyme (β-glucuronidase, or GUS) or a suite of `degreening' genes that interfere with chlorophyll biosynthesis17,19.
The two-component system, although lost in animals, is retained in yeasts and plants, where the conserved circuits were adopted into hormone signalling pathways20–24. For example, the cytokinin-sensing pathway retains the basic framework of two-component circuitry. Using yeast, a synthetic signal transduction pathway was created, replacing the yeast osmosensor SLN1 with the Arabidopsis cytokinin receptor CRE1. This synthetic pathway rescued the lethal phenotype of the sln1D mutant only when cytokinin was exogenously applied23. The result implicated that the two-component systems can be introduced or exchanged between the kingdoms of life, providing a compelling rationale for repurposing this module for diverse applications. Future work can probably address some limitations of the engineered two-component systems, including leaky (background) activation, crosstalk with the endogenous two-component signalling pathways, and efficacy of synthetic circuit in the contexts of feedback regulation (tissue-specific and plant phase/age-related) and diurnal fluctuations of transgene expression and responsiveness.
Receptors
Engineered plant receptors could be powerful tools to hijack or bolster endogenous signal transduction pathways to manipulate plant behaviours. Such an approach does not require introduction of a complete set of synthetic signalling components. Rather, structural information on receptors at an atomic resolution, as well as computational structural modelling and simulation, are essential. Crystal structures of several key plant receptors have been solved recently, including receptors for auxin25, gibberellins26 (GA), abscisic acid (ABA)27–29, jasmonates (JA)30, brassinosteroids (BR)31,32 and peptides33. This deep structural knowledge has been instrumental in the engineering of new hormone biosensors, including DII–VENUS34 and its ratiometric variant R2D235 for auxin, Jas9–VENUS for JA36, and ABACUS37 and ABAleons38 for ABA.
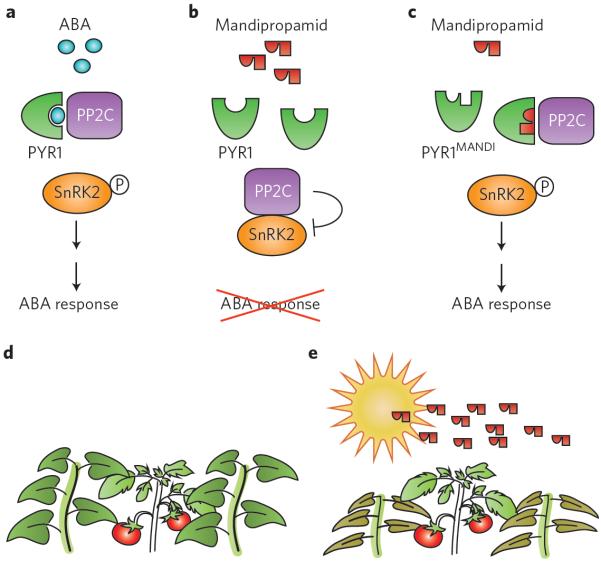
Molecular structural information on plant hormone–receptor interaction and signal activation can be used directly to engineer receptors to confer novel recognition specificity. Among the ongoing efforts, engineering of the ABA receptor PYRABACTIN RESISTANCE 1 (PYR1) has been particularly noteworthy. PYR1 was identified through chemical genetic screen using pyrabactin, a selective ABA agonist that preferentially interacts with PYR139. Structure-guided receptor engineering has been used to increase pyrabactin selectivity between PYR1 and PYR2, illustrating a path to manipulating signalling outputs via receptor modification40. One study41 further engineered PYR1 to perceive mandipropamid, an agrochemical compound used to fight blight pathogens (Fig. 2). A wide mutational search followed by directed mutagenesis yielded a receptor variant called PYR1mandi that essentially piggybacks on the ABA response in transgenic Arabidopsis or tomato plants (Fig. 2b,c). The synthetic pyrabactin–PYR1mandi ligand–receptor system has important implications in both basic and translational research for plant biology. For instance, cell-type-specific expression of PYR1mandi probably triggers ABA responses in the specific cell type of interest following mandipropamid spraying. The cell-type-specific roles of ABA signalling could be investigated without being hampered by the complexity owing to redundancies among PYR/PYR-like (PYL) proteins or systemic ABA transport. In the crop fields, mandipropamid spraying could enhance the drought tolerance of transgenic plants expressing PYR1mandi, but not of non-transgenic weeds, thereby selectively protecting the crops from abiotic stress (Fig. 2d,e).
Figure 2. An engineered ABA receptor can perceive a fungicide and trigger an ABA response.
a, Endogenous ABA signalling. Binding of ABA to the PYR1 receptor promotes PYR1–PP2C association, which in turn activates the downstream protein kinase SnRK2. Active SnRK2 triggers ABA responses, including stomatal closure and drought tolerance. b, Mandipropamid treatment does not elicit an ABA response in wild-type plants. c, In a plant expressing the engineered PYR1MANDI receptor, mandipropamid treatment triggers ABA response and, consequently, drought tolerance. d,e, Potential and idealized translational application of a synthetic ligand–receptor system in a crop field. Here, a transgenic tomato crop plant expressing PYR1MANDI is outcompeted by nearby weeds (d). During drought season, mandipropamid application triggers ABA response to the transgenic tomato plant, thereby boosting its drought tolerance. Surrounding non-transgenic weeds do not respond to the chemical spray. Mandipropamid has already been approved by the Environmental Protection Agency as a fungicide (reg. no. 100-1281) for field application.
Receptors can also be expressed in heterologous settings, making it possible to test the function of potential orthologues across a vast range of species. These head-to-head comparative studies in a common, evolutionarily distant background could lead to a more sophisticated assessment of how gene function evolves. Porting orthologous pathways from divergent lineages into a `blank slate' background could be combined with other synthetic approaches, such as ancestral sequence reconstruction (ASR)42. ASR aligns extant protein sequences within phylogenies and then uses statistical models of amino acid substitution rates to determine the maximum-likelihood sequence at an ancestral node. Such studies have helped resolve apparent paradoxes where co-evolution of complex components leave a `chicken or egg' dilemma, such as in the diversification of animal hormone receptors43. ASR and related approaches could aid in the transfer of knowledge from model plants to other plants of interest. For example, evolutionary and molecular phylogenetic studies coupled with developmental and physiological analysis may shed light on the specialization and diversification of plant receptors for strigolactones (SL) (which are butenolide hormones for branching and also used as a cue for parasitic plants and arbuscular mycorrhizal fungi) and karrikins (chemical compounds found in smoke)44–46. Engineering receptors using an ASR approach could be applied for manipulating plant growth and symbiotic relationships while limiting parasitism.
Signals
Small chemical analogues represent a powerful tool to discover, visualize and manipulate plant signalling pathways47,48. Here again, structural resolutions of small chemical hormone binding sites to corresponding receptors enable the rational design of analogues that interfere with endogenous hormone signalling. For instance, a series of auxin agonists and antagonists were created by extending alkyl chains to the α-position of indole-3-acetic acid (IAA)49. The crystallographic and molecular-docking analyses revealed that introduction of butyl or longer alkyl chains at this position blocks access of the Aux/IAA degron to the TRANSPORT INHIBITOR RESPONSE 1 (TIR1) auxin-binding pocket, thereby acting as an antiauxin49. Similarly, ABA analogues have been engineered with a long alkyl chain that interferes with PYL–protein phosphatase 2C (PP2C) interactions, thus acting as ABA antagonists50.
Fluorescent analogues of hormones and other signalling molecules are a major advance for probing in vivo events in real time and across different spatial scales. Perhaps the greatest advantage of these compounds is that they can be used in any plant species, even those without established transformation protocols. This overcomes a significant limitation of the fluorescence resonance energy transfer (FRET)-based biosensors, which require the introduction and expression of recombinant, synthetic protein fusions in the whole plant or in cell/tissue types of interest. Ideally, both methods can be used to validate each other, as data from chemical analogues may not always accurately reflect the activities or dynamics of endogenous chemical signals.
Some recent examples include fluorescently labelled bioactive GA (GA–FI) and BR analogues (Alexa Fluor 647-castasterone; AFCS)51,52. Treatment of Arabidopsis roots with GA–FI revealed the selective accumulation of signals in elongating endodermal cells, suggesting the presence of active GA transport and specific tissues as GA sinks51. AFCS enabled visualization of the internalization of BRI1–AFCS ligand–receptor complexes at the plasma membrane52. In these cases, conjugation of a large fluorescent dye with the introduction of a long alkyl chain reduces the binding affinity to the receptor. This inevitably makes such analogues less competitive against the endogenous, non-tagged counterparts. Similarly, the fluorescent auxin analogues 7-nitro-2,1,3-benzoxadiazole (NBD)–α-naphthalene acetic acid (NAA) and NBD–IAA allow for visualization of auxin transport, but are unable to mediate signalling through TIR1–Aux/IAA53,54. To improve the design, each plant hormone needs to be carefully investigated to elegantly replace the non-essential backbone with a fluorescent moiety. Recently, a fluorescent SL analogue, CISA-1 (cyano-isoindole strigolactone analogue-1), was created on the basis of such a design principle55.
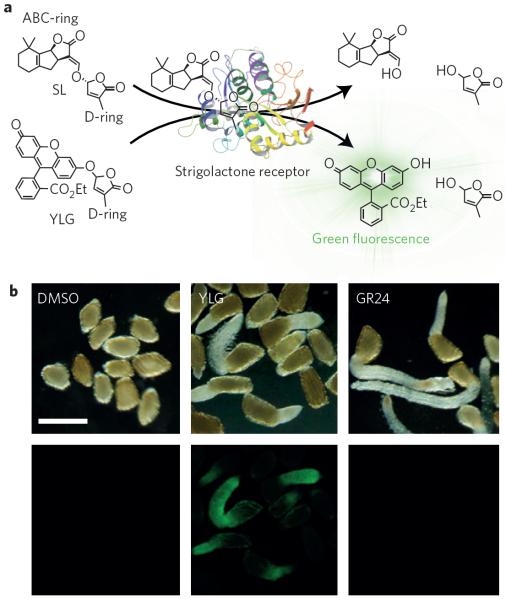
The creative and effective design of synthetic analogues could enable the visualization of the actual receptor perception and signal transduction with fluorescence. One study recently reported the synthesis and application of such an innovative synthetic chemical, Yoshimulactone Green (YLG), which can visualize the SL receptor activity in real time with high spatial resolution56. The SL receptor's α/β-hydrolase activity cleaves SLs into two pieces: the ABC-ring, which is structurally free, and the D-ring, which is required for bioactivity (Fig. 3a). In YLG, the ABC-ring is replaced by a fluorescein derivative, which emits fluorescence only if the D-ring is hydrolysed (Fig. 3a,b). Hence, YLG retains SL bioactivity. YLG and its variant with a higher on/off ratio, YLG-double (YLGW), trigger the germination of Striga seeds (Fig. 3b)56. Strikingly, the long-term time-lapse imaging of Striga germination by YLGW revealed the biphasic response of SL perception and signal activation: the first `wake up wave' sharply illuminates the root tip of Striga embryos within 20 minutes of YLGW application, diffuses towards cotyledons and then disappears after six hours56. This initial strong reaction is specific to Striga embryos and is not observed in Arabidopsis embryos, whose germination is not SL-specific. The second `elongation tide' of fluorescent wave coincided with germination and root elongation56.
Figure 3. Mode of action of YLG.
a, Perception of SL (top, left) by the SL receptor results in hydrolysis of SL, releasing the D-ring (top, right). YLG (bottom, left) is also recognized by the SL receptor with high affinity. The receptor perception cleaves off the D-ring of YLG, releasing a fluorescein derivative, which emits green fluorescence. b, Visualization of YLG perception in germinating Striga seeds. Although both YLG and the synthetic SL analogue GR24 trigger germination, strong green fluorescence is visible in the roots of only YLG-treated seedlings. Scale bar, 0.5 mm; DMSO, dimethylsulfoxide. Figure courtesy of Shinya Hagihara, Kenichiro Itami and Masahiko Yoshimura (ITbM, Japan).
As molecular structures of ligand–receptor associations are increasingly being resolved at atomic resolutions25,30,31, it will be possible to design diverse ligand analogues with specific properties to visualize and manipulate signalling. Although the new chemical approach offers great promise, there are many potential pitfalls. For instance, conjugating fluorescent dyes to hormones inevitably affects receptor binding affinity and diffusion rates, as well as degradation and transport kinetics. As a result of these differential effects, synthetic analogues may not accurately reflect the full activity of their endogenous counterparts. In addition, synthetic hormone agonists, antagonists and fluorescent probes are added exogenously to plants, rather than synthesized endogenously. Exogenous application may lead to a highly artificial spatial distribution of pathway activation, perturb the effect of endogenous hormones and provoke feedback regulation of endogenous hormone biosynthesis, signalling and transport pathways. Careful, multifaceted approaches that combine genetic tools, biosensors and mathematical modelling57–59 could moderate the impact of these limitations.
Combining engineering approaches with metabolic and signalling pathways is the obvious next step in synthetic signalling, as this would allow implementation of entire synthetic networks. The network motifs needed for complex dynamic output functions would be greatly facilitated by plants that could synthesize multiple synthetic signalling molecules that are, in turn, selectively perceived by engineered receptors and downstream components (Fig. 1). New approaches for rapid prototyping of synthetic parts in plants57 are key to developing the library of components needed to scale-up synthetic pathway engineering. Implementation of engineered networks in basal plant lineages may also be a means to accelerate the design–build–test engineering cycle, as these organisms often have smaller families of competing signalling components and more streamlined genome editing58,60.
Epigenetics
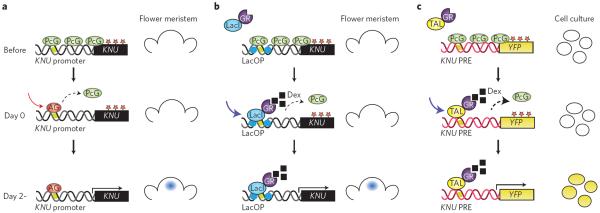
A synthetic approach has also been effective at manipulating plant gene expression. One powerful example makes use of the C-function homeotic transcription factor AGAMOUS (AG), which acts during floral development to turn on the transcription factor KNUCKLE (KNU; Fig. 4)61. KNU represses the expression of the stem cell gene WUSCHEL (WUS) to terminate stem cells in flower primordia. Interestingly, KNU induction by AG requires two days, and this involves displacement of Polycomb group (PcG) proteins by AG at the KNU locus (Fig. 4a)61. To unravel the mechanism underlying the time lag, one study62 generated an artificial DNA-binding protein, LacI–GR, which binds to the bacterial lactose repressor, coexpressed with a KNU reporter whose AG-binding domains within the promoter region were replaced by a lac operator sequence. Furthermore, this promoter region contains a Polycomb response element (PRE)-like domain from other plant genes, so that the KNU reporter is silenced unless the PcG proteins are removed from the system. Notably, following nuclear localization of LacI–GR, this synthetic system recapitulated the KNU expression with a two-day time lag (Fig. 4b).
Figure 4. A synthetic epigenetic timer for gene expression.
a, The endogenous timer. During flower development, KNU is covered with H3K27me2 repressive marks (red stars). The PcG complex maintains such marks (top). Binding of AG to the KNU promoter elements triggers eviction of PcG (middle). After two days, cell-division-dependent loss of repressive histone marks facilitates KNU gene expression in the flower meristem (bottom; right). b, A partially synthetic timer. On dexamethasone (Dex) treatment, LacI–GR, a synthetic DNA-binding protein consisting of the LacI DNA-binding domain fused to the glucocorticoid receptor (GR), can trigger the eviction of the PcG complex from the KNU promoter region that contains Lac operator (LacOp) sequences (middle). This leads to KNU gene expression after two days, thus mimicking the AG action (bottom, right). c, A completely synthetic timer. YFP driven by an unrelated promoter can be silenced with repressive histone marks (asterisks) if a Polycomb response element from KNU (KNU PRE) is inserted (top). A synthetic DNA-binding protein consisting of GR fused with TAL that is designed to bind near the AG-binding site, TAL–GR, can trigger the eviction of PcG (middle). This leads to YFP reporter gene expression in cultured cells (bottom; right).
This work was extended with a second synthetic epigenetic timer, consisting of a synthetic promoter-driven YFP construct and an artificial transcription activator-like (TAL)–GR DNA-binding protein, which was designed to bind the DNA sequence around the AG-binding site62. The culture cells expressing these constructs beautifully recapitulated the two-day lag of gene expression (Fig. 4c). These results indicate that any DNA-binding protein at the KNU promoter region that physically and competitively interferes with the association of the PcG protein complex would be sufficient to induce KNU gene expression. Moreover, the work shows the power and promise of a synthetic approach to controlling timing of gene expression using artificially designed epigenetic timers. Such an approach could be expanded and coupled with feedback/feedforward circuits and genetic switches to control artificial gene expression programs in plants.
A synthetic transcription factor can also be used to uncouple chromatin architecture and chemical-stimulus-induced gene expression. Very recently, a study reported that an auxin-regulated chromatin switch, mediated by MONOPTEROS (MP)/ARF5 (AUXIN RESPONSE FACTOR 5), triggers epigenetic reprogramming for floral primordial initiation63. Auxin perception normally frees MP/ARF5 from repression by degradation of Aux/IAA co-repressors. In this new work, the authors propose that degradation of Aux/IAAs allows MP/ARF5 to recruit a chromatin-remodelling complex containing SPLAYED (SYD) or BRAHMA (BRM) to open up nearby chromatin. In support of this model, an artificial transcription factor that fuses the MP/ARF5 DNA-binding domain and BUSHY, a protein that recruits SYD/BRM complexes, was able to mimic MP function and rescue the mp `pin' inflorescence phenotype63. These findings provide a blueprint for engineering artificial DNA-binding proteins with the capacity to directly recruit the SYD/BRM complex to any known cis-regulatory elements and precisely manipulate the chromatin landscape.
Molecular motors
Plant cytoskeleton and motor proteins are intimately coupled with plant cell division, cell elongation, cell shape and polarity specification64–66. For example, cortical microtubules position the cellulose synthase complex, and thereby determine the site of cellulose microfibril deposition67,68. As such, understanding and manipulating their dynamics in a controlled manner could have a huge impact on engineering cell wall biosynthesis and plant biomass.
Myosin is a motor protein that uses energy from ATP hydrolysis to `walk' along actin filaments. Researchers have engineered myosin VI and performed single-molecule imaging to investigate its behaviour in vitro, and compared this with predictions from mathematical models. Three- and four-headed myosins with various arm lengths, instead of natural myosins with two heads, have been created and their processivity and behaviours were tested in vitro15. These researchers tactically combined four-head myosin with Chara myosin XI, the fastest myosin known69, and created the fastest synthetic myosin known to date15. The next and most important question is how such engineered motor proteins perform in living cells. The plant myosin (myosin XI) was engineered to manipulate its velocity and processivity. Faster myosin, and thus faster cytoplasmic streaming, led to bigger Arabidopsis plants, whereas slower myosin resulted in smaller plants70. This work revealed a surprising connection between myosin velocity and plant size control. If such an approach can be expanded to also engineer actins and tubulins, it would allow precise control of the cytoskeleton, as well as cortical microtubule density and dynamics. This would allow researchers to manipulate cell growth and shape in a predictable manner.
Engineering other organisms with plant-derived pathways
Plant proteins and small molecules are increasingly being used to engineer heterologous systems. For example, the field of optogenetics has harvested a number of plant proteins involved in light perception that work effectively to trigger activation of neurons in living animals with exquisite spatial resolution71. A similar logic and related plant components were exploited to develop tools for light-induced gene expression72–74, protein splicing75,76 and nuclear localization77. Plant hormone pathways have been engineered to allow small-molecule regulation of target proteins, including triggering their turnover78–80 or relocalization81. Recently, an enzyme for auxin biosynthesis has been combined with components from the auxin response pathway to produce a robust sender–receiver system in yeast82. A sender strain expresses an enzyme from bacteria that converts a precursor called indole-3-acetamide into auxin. A library of auxin-degradable CRISPR transcription factors coexpressed with an auxin receptor translates the auxin signal in receiver cells.
In addition to producing new tools for heterologous systems, synthetic recapitulation of entire pathways or networks can generate new hypotheses about plant function. A suite of auxin response circuits encompassing signal perception to gene expression has been analysed in yeast83. Sensitivity analysis of the isolated pathway guided studies in plants by generating the hypothesis that auxin sets an Aux/IAA-regulated timer, coordinating progression through development84. In this way, parallel analysis of synthetic and natural systems can be synergistic. In a recent review outlining plans for re-engineering photosynthesis to meet the urgent need for increased crop yields, the authors highlighted the critical role of synthetic biology in realizing the most ambitious, and probably most effective, of these ideas85.
Perspective
Synthetic biology is rapidly becoming as essential to molecular biology as crystallography, in vitro biochemistry, genetics and the many `omics'. When combined with high-fidelity genomic engineering86,87, higher efficiency transformation88, high-throughput phenotyping platforms89 and improved in silico tools for pathway design and testing90–94, the scale of reasonable plant engineering projects expands dramatically. Detailed knowledge of a genome, specifically the genotype-to-phenotype map, is essential for targeting and rapidly prototyping the optimal candidates for engineering. Synthetic biology, as a multiscale and cross-disciplinary approach, offers a deeper understanding of plant development and signalling. This new perspective will make it possible to unleash the full potential of plants for the benefit of our health and environment.
Acknowledgements
We thank D. Wagner (Univ. Pennsylvania, USA) for sharing unpublished materials; S. Hagihara, M. Yoshimura and K. Itami (Institute of Transformative Biomolecules (ITbM), Nagoya Univ., Japan) for providing diagrams and unpublished Striga seedling images for Fig. 3; H. Hirukawa and S. Hagihara (ITbM) for the illustrations for Figs 1 and 3; and M. Maes (Univ. Washington, USA) for proofreading. Funding for synthetic biology research in J.L.N.'s laboratory is provided by the National Institute of Health (R01 GM107084) and the National Science Foundation (MCB-1411949). K.U.T. is an investigator of Howard Hughes Medical Institute and Gordon and Betty Moore Foundation (HHMI-GBMF), and her group is supported by a grant (GBMF3035).
Footnotes
Competing interests The authors declare no competing financial interests.
References
- 1.The Arabidopsis Genome Initiative Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 2.Medford JI, Prasad A. Plant synthetic biology takes root. Science. 2014;346:162–163. doi: 10.1126/science.1261140. [DOI] [PubMed] [Google Scholar]
- 3.Purnick PE, Weiss R. The second wave of synthetic biology: from modules to systems. Nature Rev. Mol. Cell Biol. 2009;10:410–422. doi: 10.1038/nrm2698. [DOI] [PubMed] [Google Scholar]
- 4.Huang HH, Camsund D, Lindblad P, Heidorn T. Design and characterization of molecular tools for a synthetic biology approach towards developing cyanobacterial biotechnology. Nucleic Acids Res. 2010;38:2577–2593. doi: 10.1093/nar/gkq164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jensen PE, Leister D. Cyanobacteria as an experimental platform for modifying bacterial and plant photosynthesis. Front. Bioeng. Biotechnol. 2014;2:7. doi: 10.3389/fbioe.2014.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu W, Stewart CN., Jr Plant synthetic biology. Trends Plant Sci. 2015;20:309–317. doi: 10.1016/j.tplants.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 7.Farre G, Twyman RM, Christou P, Capell T, Zhu C. Knowledge-driven approaches for engineering complex metabolic pathways in plants. Curr. Opin. Biotechnol. 2015;32:54–60. doi: 10.1016/j.copbio.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 8.Thodey K, Galanie S, Smolke CD. A microbial biomanufacturing platform for natural and semisynthetic opioids. Nature Chem. Biol. 2014;10:837–844. doi: 10.1038/nchembio.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zirpel B, Stehle F, Kayser O. Production of Δ9-tetrahydrocannabinolic acid from cannabigerolic acid by whole cells of Pichia (Komagataella) pastoris expressing Δ9-tetrahydrocannabinolic acid synthase from Cannabis sativa L. Biotechnol. Lett. 2015;37:1869–1875. doi: 10.1007/s10529-015-1853-x. [DOI] [PubMed] [Google Scholar]
- 10.Doty SL, et al. Enhanced metabolism of halogenated hydrocarbons in transgenic plants containing mammalian cytochrome P450 2E1. Proc. Natl Acad. Sci. USA. 2000;97:6287–6291. doi: 10.1073/pnas.97.12.6287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doty SL, et al. Enhanced phytoremediation of volatile environmental pollutants with transgenic trees. Proc. Natl Acad. Sci. USA. 2007;104:16816–16821. doi: 10.1073/pnas.0703276104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuan L, Kurek I, English J, Keenan R. Laboratory-directed protein evolution. Microbiol. Mol. Biol. Rev. 2005;69:373–392. doi: 10.1128/MMBR.69.3.373-392.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lehmann M, Wyss M. Engineering proteins for thermostability: the use of sequence alignments versus rational design and directed evolution. Curr. Opin. Biotechnol. 2001;12:371–375. doi: 10.1016/s0958-1669(00)00229-9. [DOI] [PubMed] [Google Scholar]
- 14.Bornscheuer UT, Pohl M. Improved biocatalysts by directed evolution and rational protein design. Curr. Opin. Chem. Biol. 2001;5:137–143. doi: 10.1016/s1367-5931(00)00182-4. [DOI] [PubMed] [Google Scholar]
- 15.Schindler TD, Chen L, Lebel P, Nakamura M, Bryant Z. Engineering myosins for long-range transport on actin filaments. Nature Nanotechnol. 2014;9:33–38. doi: 10.1038/nnano.2013.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Voigt CA, Mayo SL, Arnold FH, Wang ZG. Computational method to reduce the search space for directed protein evolution. Proc. Natl Acad. Sci. USA. 2001;98:3778–3783. doi: 10.1073/pnas.051614498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Antunes MS, et al. Programmable ligand detection system in plants through a synthetic signal transduction pathway. PLOS ONE. 2011;6:e16292. doi: 10.1371/journal.pone.0016292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu. Rev. Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 19.Antunes MS, et al. A synthetic de-greening gene circuit provides a reporting system that is remotely detectable and has a re-set capacity. Plant Biotechnol. J. 2006;4:605–622. doi: 10.1111/j.1467-7652.2006.00205.x. [DOI] [PubMed] [Google Scholar]
- 20.Chang C, Kwok SF, Bleecker AB, Meyerowitz EM. Arabidopsis ethylene-response gene ETR1: similarity of product to two-component regulators. Science. 1993;262:539–544. doi: 10.1126/science.8211181. [DOI] [PubMed] [Google Scholar]
- 21.Kakimoto T. CKI1, a histidine kinase homolog implicated in cytokinin signal transduction. Science. 1996;274:982–985. doi: 10.1126/science.274.5289.982. [DOI] [PubMed] [Google Scholar]
- 22.Imamura A, et al. Response regulators implicated in His-to-Asp phosphotransfer signaling in Arabidopsis. Proc. Natl Acad. Sci. USA. 1998;95:2691–2696. doi: 10.1073/pnas.95.5.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inoue T, et al. Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature. 2001;409:1060–1063. doi: 10.1038/35059117. [DOI] [PubMed] [Google Scholar]
- 24.To JP, et al. Cytokinin regulates type-A Arabidopsis response regulator activity and protein stability via two-component phosphorelay. Plant Cell. 2007;19:3901–3914. doi: 10.1105/tpc.107.052662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tan X, et al. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature. 2007;446:640–645. doi: 10.1038/nature05731. [DOI] [PubMed] [Google Scholar]
- 26.Shimada A, et al. Structural basis for gibberellin recognition by its receptor GID1. Nature. 2008;456:520–523. doi: 10.1038/nature07546. [DOI] [PubMed] [Google Scholar]
- 27.Nishimura N, et al. Structural mechanism of abscisic acid binding and signaling by dimeric PYR1. Science. 2009;326:1373–1379. doi: 10.1126/science.1181829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Santiago J, et al. The abscisic acid receptor PYR1 in complex with abscisic acid. Nature. 2009;462:665–668. doi: 10.1038/nature08591. [DOI] [PubMed] [Google Scholar]
- 29.Miyazono K, et al. Structural basis of abscisic acid signalling. Nature. 2009;462:609–614. doi: 10.1038/nature08583. [DOI] [PubMed] [Google Scholar]
- 30.Sheard LB, et al. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature. 2010;468:400–405. doi: 10.1038/nature09430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hothorn M, et al. Structural basis of steroid hormone perception by the receptor kinase BRI1. Nature. 2011;474:467–471. doi: 10.1038/nature10153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.She J, et al. Structural insight into brassinosteroid perception by BRI1. Nature. 2011;474:472–476. doi: 10.1038/nature10178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun Y, et al. Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex. Science. 2013;342:624–628. doi: 10.1126/science.1243825. [DOI] [PubMed] [Google Scholar]
- 34.Brunoud G, et al. A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature. 2012;482:103–106. doi: 10.1038/nature10791. [DOI] [PubMed] [Google Scholar]
- 35.Liao CY, et al. Reporters for sensitive and quantitative measurement of auxin response. Nature Methods. 2015;12:207–210. doi: 10.1038/nmeth.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Larrieu A, et al. A fluorescent hormone biosensor reveals the dynamics of jasmonate signalling in plants. Nature Commun. 2015;6:6043. doi: 10.1038/ncomms7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones AM, et al. Abscisic acid dynamics in roots detected with genetically encoded FRET sensors. Elife. 2014;3:e01741. doi: 10.7554/eLife.01741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Waadt R, et al. FRET-based reporters for the direct visualization of abscisic acid concentration changes and distribution in Arabidopsis. Elife. 2014;3:e01739. doi: 10.7554/eLife.01739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park SY, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peterson FC, et al. Structural basis for selective activation of ABA receptors. Nature Struct. Mol. Biol. 2010;17:1109–1113. doi: 10.1038/nsmb.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park SY, et al. Agrochemical control of plant water use using engineered abscisic acid receptors. Nature. 2015;23:545–548. doi: 10.1038/nature14123. [DOI] [PubMed] [Google Scholar]
- 42.Thornton JW. Resurrecting ancient genes: experimental analysis of extinct molecules. Nature Rev. Genet. 2004;5:366–375. doi: 10.1038/nrg1324. [DOI] [PubMed] [Google Scholar]
- 43.Harms MJ, Thornton JW. Analyzing protein structure and function using ancestral gene reconstruction. Curr. Opin. Struct. Biol. 2010;20:360–366. doi: 10.1016/j.sbi.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nelson DC, et al. F-box protein MAX2 has dual roles in karrikin and strigolactone signaling in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA. 2011;108:8897–8902. doi: 10.1073/pnas.1100987108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Waters MT, et al. Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis. Development. 2012;139:1285–1295. doi: 10.1242/dev.074567. [DOI] [PubMed] [Google Scholar]
- 46.Delaux PM, et al. Origin of strigolactones in the green lineage. New Phytol. 2012;195:857–871. doi: 10.1111/j.1469-8137.2012.04209.x. [DOI] [PubMed] [Google Scholar]
- 47.Toth R, van der Hoorn RA. Emerging principles in plant chemical genetics. Trends Plant Sci. 2010;15:81–88. doi: 10.1016/j.tplants.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 48.Blackwell HE, Zhao Y. Chemical genetic approaches to plant biology. Plant Physiol. 2003;133:448–455. doi: 10.1104/pp.103.031138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hayashi K, et al. Small-molecule agonists and antagonists of F-box protein–substrate interactions in auxin perception and signaling. Proc. Natl Acad. Sci. USA. 2008;105:5632–5637. doi: 10.1073/pnas.0711146105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takeuchi J, et al. Designed abscisic acid analogs as antagonists of PYL-PP2C receptor interactions. Nature Chem. Biol. 2014;10:477–482. doi: 10.1038/nchembio.1524. [DOI] [PubMed] [Google Scholar]
- 51.Shani E, et al. Gibberellins accumulate in the elongating endodermal cells of Arabidopsis root. Proc. Natl Acad. Sci. USA. 2013;110:4834–4839. doi: 10.1073/pnas.1300436110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Irani NG, et al. Fluorescent castasterone reveals BRI1 signaling from the plasma membrane. Nature Chem. Biol. 2012;8:583–589. doi: 10.1038/nchembio.958. [DOI] [PubMed] [Google Scholar]
- 53.Tsuda E, et al. Alkoxy-auxins are selective inhibitors of auxin transport mediated by PIN, ABCB, and AUX1 transporters. J. Biol. Chem. 2011;286:2354–2364. doi: 10.1074/jbc.M110.171165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hayashi K, et al. Auxin transport sites are visualized in planta using fluorescent auxin analogs. Proc. Natl Acad. Sci. USA. 2014;111:11557–11562. doi: 10.1073/pnas.1408960111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rasmussen A, et al. A fluorescent alternative to the synthetic strigolactone GR24. Mol. Plant. 2013;6:100–112. doi: 10.1093/mp/sss110. [DOI] [PubMed] [Google Scholar]
- 56.Tsuchiya Y, et al. strigolactone receptors in Striga hermonthica with fluorescence. Science. 2015;349:846–848. doi: 10.1126/science.aab3831. [DOI] [PubMed] [Google Scholar]
- 57.Schaumberg KA, et al. Quantitative characterization of genetic parts and circuits for plant synthetic biology. Nature Methods. 2016;13:94–100. doi: 10.1038/nmeth.3659. [DOI] [PubMed] [Google Scholar]
- 58.Ishizaki K, Nishihama R, Yamato KT, Kohchi T. Molecular genetic tools and techniques for Marchantia polymorpha research. Plant Cell Physiol. 2015 doi: 10.1093/pcp/pcv097. http://dx.doi.org/10.1093/pcp/pcv097. [DOI] [PubMed]
- 59.Vernoux T, et al. The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol. Syst. Biol. 2011;7:508. doi: 10.1038/msb.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Prigge MJ, Bezanilla M. Evolutionary crossroads in developmental biology: Physcomitrella patens. Development. 2010;137:3535–3543. doi: 10.1242/dev.049023. [DOI] [PubMed] [Google Scholar]
- 61.Sun B, Xu Y, Ng KH, Ito T. A timing mechanism for stem cell maintenance and differentiation in the Arabidopsis floral meristem. Genes Dev. 2009;23:1791–1804. doi: 10.1101/gad.1800409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sun B, et al. Timing mechanism dependent on cell division is invoked by Polycomb eviction in plant stem cells. Science. 2014;343:1248559. doi: 10.1126/science.1248559. [DOI] [PubMed] [Google Scholar]
- 63.Wu M-F, et al. Auxin-regulated chromatin switch directs acquisition of flower primordium founder fate. eLife. 2015;4:e09269. doi: 10.7554/eLife.09269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hashimoto T. Microtubules in plants. Arabidopsis Book. 2015;13:e0179. doi: 10.1199/tab.0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Meagher RB, Fechheimer M. The Arabidopsis cytoskeletal genome. Arabidopsis Book. 2003;2:e0096. doi: 10.1199/tab.0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Smith LG, Oppenheimer DG. Spatial control of cell expansion by the plant cytoskeleton. Annu. Rev. Cell Dev. Biol. 2005;21:271–295. doi: 10.1146/annurev.cellbio.21.122303.114901. [DOI] [PubMed] [Google Scholar]
- 67.Gutierrez R, Lindeboom JJ, Paredez AR, Emons AM, Ehrhardt DW. Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nature Cell Biol. 2009;11:797–806. doi: 10.1038/ncb1886. [DOI] [PubMed] [Google Scholar]
- 68.Li S, Bashline L, Lei L, Gu Y. Cellulose synthesis and its regulation. Arabidopsis Book. 2014;12:e0169. doi: 10.1199/tab.0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Morimatsu M, et al. The molecular structure of the fastest myosin from green algae, Chara. Biochem. Biophys. Res. Commun. 2000;270:147–152. doi: 10.1006/bbrc.2000.2391. [DOI] [PubMed] [Google Scholar]
- 70.Tominaga M, et al. Cytoplasmic streaming velocity as a plant size determinant. Dev. Cell. 2013;27:345–352. doi: 10.1016/j.devcel.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 71.Shcherbakova DM, Shemetov AA, Kaberniuk AA, Verkhusha VV. Natural photoreceptors as a source of fluorescent proteins, biosensors, and optogenetic tools. Annu. Rev. Biochem. 2015;84:519–550. doi: 10.1146/annurev-biochem-060614-034411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sorokina O, et al. A switchable light-input, light-output system modelled and constructed in yeast. J. Biol. Eng. 2009;3:15. doi: 10.1186/1754-1611-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kennedy MJ, et al. Rapid blue-light-mediated induction of protein interactions in living cells. Nature Methods. 2010;7:973–975. doi: 10.1038/nmeth.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shimizu-Sato S, Huq E, Tepperman JM, Quail PH. A light-switchable gene promoter system. Nature Biotechnol. 2002;20:1041–1044. doi: 10.1038/nbt734. [DOI] [PubMed] [Google Scholar]
- 75.Tyszkiewicz AB, Muir TW. Activation of protein splicing with light in yeast. Nature Methods. 2008;5:303–305. doi: 10.1038/nmeth.1189. [DOI] [PubMed] [Google Scholar]
- 76.Wong S, Mosabbir AA, Truong K. An engineered split intein for photoactivated protein trans-splicing. PLoS ONE. 2015;10:e0135965. doi: 10.1371/journal.pone.0135965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Beyer HM, et al. Red light-regulated reversible nuclear localization of proteins in mammalian cells and zebrafish. ACS Synth. Biol. 2015;4:951–958. doi: 10.1021/acssynbio.5b00004. [DOI] [PubMed] [Google Scholar]
- 78.Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nature Methods. 2009;6:917–922. doi: 10.1038/nmeth.1401. [DOI] [PubMed] [Google Scholar]
- 79.Havens KA, et al. A synthetic approach reveals extensive tunability of auxin signaling. Plant Physiol. 2012;160:135–142. doi: 10.1104/pp.112.202184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang L, Ward JD, Cheng Z, Dernburg AF. The auxin-inducible degradation (AID) system enables versatile conditional protein depletion in C. elegans. Development. 2015;142:4374–4384. doi: 10.1242/dev.129635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liang FS, Ho WQ, Crabtree GR. Engineering the ABA plant stress pathway for regulation of induced proximity. Sci. Signal. 2011;4:rs2. doi: 10.1126/scisignal.2001449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Khakhar A, Bolten NJ, Nemhauser J, Klavins E. Cell–cell communication in yeast using auxin biosynthesis and auxin responsive CRISPR transcription factors. ACS Synth. Biol. 2015 doi: 10.1021/acssynbio.5b00064. http://dx.doi.org/10.1021/acssynbio.5b00064. [DOI] [PubMed]
- 83.Pierre-Jerome E, Jang SS, Havens KA, Nemhauser JL, Klavins E. Recapitulation of the forward nuclear auxin response pathway in yeast. Proc. Natl Acad. Sci. USA. 2014;111:9407–9412. doi: 10.1073/pnas.1324147111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Guseman JM, et al. Auxin-induced degradation dynamics set the pace for lateral root development. Development. 2015;142:905–909. doi: 10.1242/dev.117234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ort DR, et al. Redesigning photosynthesis to sustainably meet global food and bioenergy demand. Proc. Natl Acad. Sci. USA. 2015;112:8529–8536. doi: 10.1073/pnas.1424031112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jiang W, et al. Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 2013;41:e188. doi: 10.1093/nar/gkt780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Baltes NJ, Gil-Humanes J, Cermak T, Atkins PA, Voytas DF. DNA replicons for plant genome engineering. Plant Cell. 2014;26:151–163. doi: 10.1105/tpc.113.119792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wu HY, et al. AGROBEST: an efficient Agrobacterium-mediated transient expression method for versatile gene function analyses in Arabidopsis seedlings. Plant Methods. 2014;10:19. doi: 10.1186/1746-4811-10-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fahlgren N, Gehan MA, Baxter I. Lights, camera, action: high-throughput plant phenotyping is ready for a close-up. Curr. Opin. Plant Biol. 2015;24C:93–99. doi: 10.1016/j.pbi.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 90.Yordanov B, et al. A computational method for automated characterization of genetic components. ACS Synth. Biol. 2014;3:578–588. doi: 10.1021/sb400152n. [DOI] [PubMed] [Google Scholar]
- 91.Jang SS, Oishi KT, Egbert RG, Klavins E. Specification and simulation of synthetic multicelled behaviors. ACS Synth. Biol. 2012;1:365–374. doi: 10.1021/sb300034m. [DOI] [PubMed] [Google Scholar]
- 92.Fernandez-Castane A, Feher T, Carbonell P, Pauthenier C, Faulon JL. Computer-aided design for metabolic engineering. J. Biotechnol. 2014;192:302–313. doi: 10.1016/j.jbiotec.2014.03.029. [DOI] [PubMed] [Google Scholar]
- 93.Oberortner E, Densmore D. Web-based software tool for constraint-based design specification of synthetic biological systems. ACS Synth. Biol. 2015;4:757–760. doi: 10.1021/sb500352b. [DOI] [PubMed] [Google Scholar]
- 94.Stevens JT, Myers CJ. Dynamic modeling of cellular populations within iBioSim. ACS Synth. Biol. 2013;2:223–229. doi: 10.1021/sb300082b. [DOI] [PubMed] [Google Scholar]