Abstract
PF20 was first identified in Chlamydomonas rheinhardtii as an essential component of the axoneme central apparatus. We discovered that the mouse Pf20 gene encodes two major transcripts (2.5 and 1.4 kb), which are expressed in different patterns during spermatogenesis, yielding proteins of 71 and 35 kDa, respectively. Both proteins contain contiguous WD repeats in their C termini. The meiotically expressed 71-kDa protein is incorporated into the central apparatus, whereas the 35-kDa protein, which accumulates in postmeiotic male germ cells, is abundant in the nucleus. We disrupted the Pf20 gene domains that encode the C-terminal WD repeats in embryonic stem cells. Highly chimeric mice carrying the mutant Pf20 allele had impaired spermatogenesis with a significant loss of germ cells at the round spermatid stage, in association with disorganization of sperm axoneme structure. The mutated Pf20 allele was never transmitted, indicating that Pf20 haploinsufficiency caused the defects in spermatogenesis. The 35-kDa PF20 protein was shown to bind to meiosis-expressed gene 1 (MEIG1), a chromosome/chromatin-binding protein initially expressed during meiosis but retained in the germ cell nucleus throughout later stages of spermatogenesis. Our findings reveal an essential role for Pf20 in mouse spermatogenesis, sustaining postmeiotic germ cell viability. The different patterns of expression of the two PF20 proteins suggest the possibility that the Pf20 gene has multiple functions during spermatogenesis.
Axonemes, critical components of cilia and flagella, have a structure consisting of nine outer doublet microtubules encircling a central pair of microtubules, which has remained virtually unchanged during evolution (1–3). Defective assembly or function of axonemes results in immotile cilia, which causes defects in lateralization, respiratory disease, hydrocephalus, and infertility (4–7). The best-studied mutations in mammals that cause immotile cilia are in genes encoding dynein arm proteins, which generate the force that drives axonemal motility (8–10). Less is known about other axonemal proteins, especially those of the central pair. However, studies of Chlamydomonas rheinhardtii revealed important roles for the central apparatus. Mutant Chlamydomonas strains lacking functional PF16, PF6, and PF20 have paralyzed flagella, and the C1 microtubule, the projections connecting the C1 microtubule and the entire central pair, respectively, are missing in isolated axonemes (11–13).
To understand the function of the central apparatus in mammals, we cloned the human and mouse orthologues of Chlamydomonas, PF16 (SPAG6) (14, 15) and PF20 (16). SPAG6-deficient mice are hydrocephalic, and males surviving to maturity are infertile as a result of a marked sperm motility defect associated with disorganization of flagellar structures, including loss of the central pair microtubules and disorganization of the outer dense fibers and fibrous sheath (6). These observations suggested an important role for central apparatus proteins in mammalian axoneme function.
In the present study, we examined the function of the mouse Pf20 gene. We discovered that this gene encodes two different proteins, which are expressed in different patterns and have different subcellular localizations in male germ cells. The PF20 proteins contain contiguous WD repeats in their C termini, suggesting that these molecules interact with other proteins, a notion substantiated in our previous work, which demonstrated that PF20 binds to SPAG6 (16). Targeted disruption of Pf20 sequences encoding the C terminus of both proteins resulted in severe impairment of spermatogenesis and disorganization of sperm flagellar structure. Our findings suggest that the products of the Pf20 gene have multiple functions during spermatogenesis that include survival of developing germ cells and maintenance of the integrity of the sperm axoneme and associated structures.
Materials and Methods
Generation of Anti-PF20 C-Terminal Antibody. A cDNA encoding the C terminus (amino acids 330–639) of mouse PF20 was cloned into the pET28a vector. The fusion protein was purified and the rabbit polyclonal antibody generated (16).
Yeast Two-Hybrid Screen. A mouse testis cDNA library was screened by using the C terminus of PF20 as the bait (16). The interaction of 35-kDa PF20 with proteins was confirmed in yeast with the Matchmaker Lex A system (Clontech). Briefly, the full-length 35-kDa PF20 was cloned into the pLexA vector and the target proteins into pB42AD vector. The two plasmids were transformed into yeast (EGY48) containing p8op-lacZ plasmid, and the yeasts were grown on SD plates lacking His, Trp, and URA, with or without galactose and raffinose as inducers.
Expression Vector Constructs. cDNAs containing the 71- and 35-kDa coding sequences and the N terminus of the 71-kDa protein (amino acids 1–329) from the mouse PF20 cDNA were generated by RT-PCR. After sequencing, the cDNAs were cloned into EcoRI/BamHI sites of the pEGFP-N2 vector, creating the three pEGFP-N2/PF20 plasmids. Meiosis-expressed gene 1 (MEIG1) fusions with both GFP and DsRed tags were generated. PF20 (35 kDa) was also cloned into pcDNA3 for coimmunoprecipitation experiments.
Cell Culture and Transient Transfection. Chinese hamster ovary (CHO), COS-1, Madin–Darby canine kidney, and NIH 3T3 cells were cultured in two-well chamber slides with DMEM containing FBS at 37°C. The cells were transfected with the pEGFP-N2/PF20, or 35-kDa PF20/pEGFP-N2 was transfected into CHO cells together with MEIG1–DsRed. Forty-eight hours after transfection, the cells were visualized by confocal microscopy.
Coimmunoprecipitation. CHO cells were transfected with 35-kDa PF20/pcDNA3 and MEIG1/pEGFP-N2, and 48 h later, the cells were harvested into immunoprecipitation buffer (150 mM NaCl/50 mM Tris·HCl, pH 8.0/5 mM EDTA/1% Triton X-100/1 mM PMSF/proteinase inhibitor mixture), and the lysates were passed through a 20-gauge needle. After centrifugation at 11,600 × g for 5 min, the supernatants were precleared with protein A beads at 4°C for 30 min. The supernatants were then incubated with C- or N-terminal (negative control) anti-PF20 polyclonal antibodies at 4°C for 2 h, and protein A beads were added with a further incubation at 4°C overnight. The beads were washed with immunoprecipitation buffer three or four times, and 1× protein loading buffer was then added to the beads, which were boiled at 100°Cfor 10 min; the samples were then processed for Western blotting with monoclonal anti-GFP antibody.
Targeted Disruption of Pf20. The Pf20 targeting vector, which contained exon 11 replaced by a neo selection cassette vector, was linearized with ClaI and electroporated into 129Sv embryonic stem (ES) cells. ES cells clones surviving G418 selection were evaluated for homologous recombination by Southern analysis. ES cells were injected into C57BL/6J blastocysts, and chimeric mice produced were mated to C57BL/6J females. Mice were genotyped by PCR with tail DNA. Two sets of primers were used in the PCRs. One set of primers corresponded to the neo (sense: 5′GACTCCAGGAGATAGGAAGA-3′) and Pf20 genes (antisense: 5′-GAACTTCGGAATAGGAACTTC-3′) separately. The other set corresponded to the deleted region of the Pf20 gene (sense: 5′-GCATTCACAGCTGTAAACTG-3′ and antisense: 5′-CAACAATACCATTGTGCATG-3′).
Sperm Genotyping. Sperm preparation and primers used to distinguish the C57BL/6J and 129/Sv alleles were as described (17).
Northern Blot Analysis. Northern blots containing total testicular RNA (30 μg per lane) were probed with a full-length mouse PF20 cDNA. Blots were reprobed with mouse SPAG6 and AKAP4 cDNAs (16).
Western Blot Analysis. An equal amount of protein (50 μg per lane) was subjected to Western analysis by using antibodies against the N- and C-terminal domains of PF20, mouse SPAG6, or AKAP4, as described (16).
Southern Blot Analysis. Fifteen micrograms of DNA from ES cells was digested with XbaI and separated by 0.8% agarose gel. The DNA was transferred to nylon membranes and probed with a Pf20 gene-specific probe downstream of the homologous recombination region.
Histology, Immunochemistry, Terminal Deoxynucleotidyltransferase-Mediated dUTP Nick End Labeling (TUNEL) Staining, and Transmission Electron Microscopy. Cauda epididymal sperm, testes, and bronchi were prepared for light and electron microscopy and immunochemistry by using standard methods (6). Immunochemistry was performed on testes with the N- and C-terminal antibodies by using immunoperoxidase methods. TUNEL staining was performed with the DeadEnd Colorimetric TUNEL System (Promega). Biotin was incorporated with the Vector ABC kit (Vector Laboratories) to increase the signal.
Results
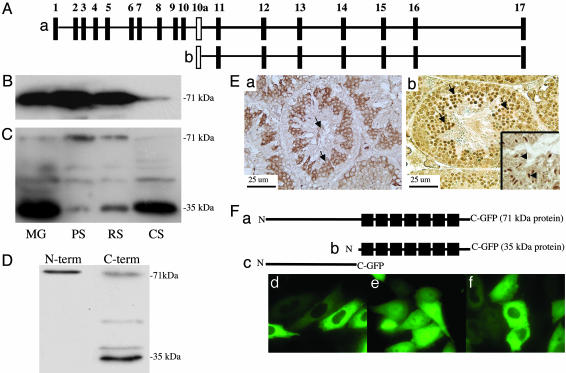
The Pf20 Gene Encodes Two Major Transcripts Expressed in Different Patterns During Spermatogenesis, Yielding Two Proteins with Different Subcellular Localizations. The mouse Pf20 gene encodes a WD repeat protein highly expressed in testis and localized to the central apparatus of the sperm flagella (16). Two transcripts were detected of 2.5 and 1.4 kb derived from a single gene consisting of 17 exons. The 2.5-kb transcript includes exons 1–17, whereas the 1.4-kb transcript consists of exons 11–17, which encompass 7 WD repeats (Fig. 1A). The protein encoded by the 2.5-kb transcript is 71 kDa, whereas the 1.4-kb transcript encodes a 35-kDa protein. Analysis of PF20 transcript levels during spermatogenesis revealed that the 2.5-kb transcript is expressed at a relatively constant level from the pachytene spermatocyte to the condensing spermatid stage, whereas the 1.4-kb transcript appears at the round spermatid stage to the condensing spermatid stage (16). This transcript was detected only in testis in a screen of various mouse tissues by RT-PCR (data not shown). The 71-kDa protein is detectable at similar levels in isolated pachytene spermatocytes and round spermatids but reduced in isolated condensing spermatids that lose their flagella during isolation (Fig. 1 B and C). The 35-kDa protein appeared in round spermatids, consistent with the results of Northern blotting, and was prominent in condensing spermatids, indicating a different pattern of expression and subcellular location compared with the 71-kDa PF20 protein. Epididymal sperm contain both the 71-kDa protein as well as the 35-kDa PF20 (Fig. 1D).
Fig. 1.
Deduced structure of the Pf20 gene and analysis of the two PF20 proteins expressed during spermatogenesis. (A) Map of the Pf20 gene. (a) Exons encoding the 2.5-kb transcript. (b) Exons encoding the 1.4-kb transcript. (B) Western blot analysis of germ cell extracts (MG, mixed germ cells; PS, pachytene spermatocytes; RS, round spermatids; CS, condensing spermatids) with anti-PF20 antibody to the N-terminal domain. (C) Western blot analysis of germ cell extracts with anti-PF20 antibody to the C-terminal domain. The C-terminal antibody weakly reacts with a protein of 50 kDa, which may be a fragment of the larger PF20 protein. (D) Western analysis of epididymal sperm with anti-N- and anti-C-terminal antibodies showing the presence of both PF20 isoforms. The 38-kDa PF20 band detected in this blot likely represents a phosphorylated form of 35-kDa PF20. (E) Immunochemistry of normal testis with anti-PF20 antibodies to the N (a) and C(b) termini. The N-terminal antibody stains the cytoplasm of maturing germ cells and the flagella of sperm. The C terminus antibody similarly stains the cytoplasm but shows much more intense staining of the nuclei of round (arrows) and condensing (Inset, arrows) spermatids. (F) Map of GFP fusion proteins (a–c) and CHO cells transfected with plasmids expressing the 71-(d) and 35-(e) kDa and N-terminal (f) GFP fusion proteins showing cytoplasmic localization of the 71-kDa and N-terminal fusion proteins and cytoplasmic and nuclear localization of the 35-kDa fusion protein.
To investigate the in vivo localization of the two proteins, immunochemistry was performed on testes with the antibody against the N terminus of PF20, which can recognize only the 71-kDa protein, and the antibody against the C terminus, which should recognize both proteins. The N-terminal antibody gave cytoplasmic staining in pachytene spermatocytes, round and condensing spermatids, and flagellar staining in sperm, whereas the C-terminal antibody gave some cytoplasmic staining in germ cells and staining of the sperm tails, but a stronger nuclear stain in round and condensing spermatids (Fig. 1E). These observations demonstrate that the two PF20 proteins indeed have different cellular localizations.
Plasmids expressing the 71- and 35-kDa PF20/GFP fusion proteins, respectively, were transfected into CHO cells. The 71-kDa-GFP protein was detected only in the cytoplasm, whereas the 35-kDa-GFP protein was detected both in the cytoplasm and nucleus, with the nucleus showing a more intense signal (Fig. 1F). The PF20 N terminus-GFP fusion protein was localized to the cytoplasm. Similar results were obtained when the GFP fusion protein plasmids were introduced into COS-1, Madin–Darby canine kidney, and NIH 3T3 cells (data not shown). These findings are consistent with the in vivo patterns of localization of the 71- and 35-kDa PF20 proteins and suggest that the PF20 N terminus restricts the protein from entering the nucleus.
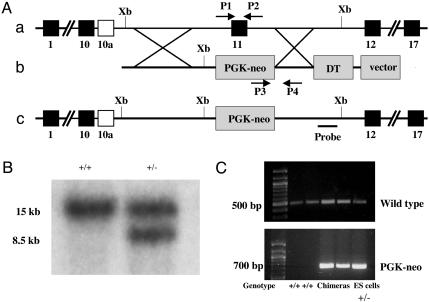
Targeted Disruption of the Pf20 Gene. We disrupted the Pf20 gene in murine ES cells by replacing exon 11 with the phosphoglycerate kinase-neo cassette (Fig. 2A). This prevents expression of the 35-kDa protein and the WD repeats of the 71-kDa protein. Fig. 2A illustrates the targeting strategy. Three independently targeted ES clones were identified by Southern blotting (Fig. 2B). We obtained a total of 23 male chimeric mice from the three different ES cell clones carrying a mutant Pf20 allele (clone 1, 12; clone 2, 7; clone 3, 4) (Fig. 2C). All were viable and showed no gross abnormalities. Male chimeric mice (>6 weeks of age) were mated with WT female C57BL/6J mice for up to 5–7 months. Four males fathered only black progeny, and six fathered both black and agouti offspring (a total of 104 black offspring). The other chimeric mice were infertile. None of the agouti progeny (a total of 204 mice fathered by 6 different chimeric males) carried the mutant allele.
Fig. 2.
Targeted disruption of the mouse Pf20 gene. (A) Schematic representation of the strategy used to disrupt Pf20. (a) Partial genomic structure of the Pf20 gene. (b) Structure of the targeting vector. (c) Structure of the mutated allele. Xb, XbaI; PGK, phosphoglycerate kinase; DT, diphtheria toxin; P1/P2, primers for amplifying the WT allele; P3/P4, primers for amplifying the mutant allele. (B) Southern blotting analysis of targeted ES cell clones. An external probe gave rise to a single 15-kb band in WT genomic DNA digested with XbaI and an 8.5-kb band in the mutant allele. (C) Genotyping by PCR. The WT allele yielded a 500-bp amplicon. A 700-bp amplicon representing the phosphoglycerate kinase-neo cassette was detectable only when a targeted allele was present.
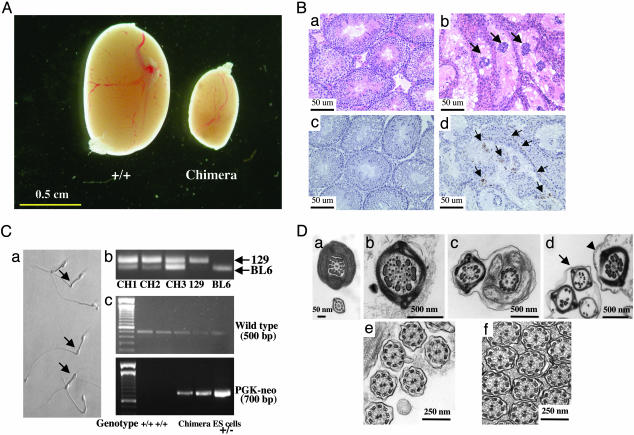
The testes of the chimeric mice killed between 7 weeks and 8 months of age were significantly smaller than those of WT 129/Sv/C57BL/6J mice [Fig. 3A; WT testis (mg/gm, means ± SEM) 7.74 ± 0.30, n = 10; chimeric testis, 1.94 ± 0.38, n = 10]. The same phenotype was observed in chimeric mice generated from each of the three independent ES cell clones. Light microscopy revealed tubules in chimeric testis tubules, with a central collection of degenerating cells and others with no evident germ cells and vacuolated Sertoli cells, interspersed with a few tubules showing apparently normal spermatogenesis (Fig. 3B). In tubules with degenerating germ cells, spermatogenesis was arrested at the round spermatid stage. TUNEL staining demonstrated increased apoptosis, primarily at the round spermatid stage (Fig. 3B).
Fig. 3.
Phenotype of Pf20 chimeric mice. (A) Representative testes from WT (+/+) and chimeric mice. (B) Histological (a and b) and TUNEL (c and d) analyses of testes from WT (a and c) and chimeric (b and d) mice. Arrows in b show clusters of degenerating germ cells in the center of seminiferous tubules. Arrows in d (brown-staining cells) indicate apoptotic cells. (C) Chimeric mice contain sperm from 129/Sv and C57BL/6J germ cells. (a) Epididymal sperm from a chimeric mouse. The arrows indicate sperm with bent tails. (b) PCR of sperm DNA to detect the129/Sv and C57BL/6J genomes (CH, chimeric). (c) PCR of sperm DNA to distinguish WT and mutant alleles. (D) Ultrastructure of sperm (a–d) and bronchial cilia (e and f) from WT (a and e) and chimeric mice (b–d and f). The arrows indicate the sperm with disorganized axonemes from the chimeric mice. The arrowhead points to a normal-appearing axoneme.
The number of sperm recovered from the cauda epididymis of chimeric mice was markedly reduced compared with WT males [epididymal sperm count × 106 per mouse: WT 22.20 ± 1.70 (n = 10); chimeric, 3.19 ± 0.85 (n = 10)]. Moreover, many of the epididymal sperm from chimeric mice had bent tails and displayed a quaking motion (Fig. 3C). Immunostaining revealed that the sperm with bent tails consistently stained weakly for PF20 with the C-terminal antibody compared with apparently normal sperm, indicating that they were derived from ES cells with the targeted allele. Sperm motility averaged 23.3 ± 4.4% for chimeric mice compared with 70 ± 2.8% in WT males. To confirm that epididymal sperm collected from the chimeric mice were derived in part from the 129/Sv ES cells and contained the mutant allele, we performed PCR on sperm DNA by using primers that distinguish 129/Sv DNA from C57BL/6J DNA based on a polymorphism (17) and also with the same primers used in the PCR genotyping reaction. 129/Sv genomic DNA was consistently detected in the epididymal sperm with variable amounts of C57BL/6J DNA (Fig. 3C), demonstrating the coexistence of germ cells derived from both the injected ES cells and the host germ line. Sperm from WT mice contained only the WT Pf20 allele, whereas sperm from chimeric mice displayed the WT and phosphoglycerate kinase-neo bands (Fig. 3C), establishing the presence of a mixture of sperm containing the WT and mutant Pf20 alleles.
Normal sperm ultrastructure was observed in the WT epididymal sperm, but sperm with disorganized axonemes were observed in the chimeric mice (Fig. 3D). Ninety-eight percent of the transverse images (480 axonemes) of sperm from two WT mice had a normal 9 + 2 structure, but 27% and 31% transverse sections of sperm (214 axonemes) from two chimeric mice lacked the central pair of microtubules, and 14% and 16% displayed marked disorganization of the outer doublet microtubules and outer dense fibers (Fig. 3D). These findings, in conjunction with the genotyping observation of epididymal sperm described above, indicate that germ cells derived from ES cells with the targeted Pf20 allele can complete maturation but suffer from the consequences of deficiency of PF20 proteins. Interestingly, transmission electron microscopic studies of testis revealed a much smaller percentage of abnormal axonemes (91.4% normal; missing central pair, 6.4%; missing outer doublets, 3.2%), suggesting a maturation-dependent change in axonemal structure. Transmission electron microscopy of bronchiolar cilia revealed no ultrastructural defects in samples from three chimeric mice with a marked spermatogenesis defect, indicating that the axonemal abnormalities may be restricted to sperm flagella (Fig. 3D).
The Leydig cells of the chimeric testis were evidently normal, because serum testosterone levels were similar in WT and chimeric mice as were seminal vesicle weights [weights mg/gm, WT: 9.13 ± 0.75 (n = 10); chimeric: 8.47 ± 0.82 (n = 10)], reflecting normal androgenization. Serum follicle-stimulating hormone and luteinizing hormone levels were also similar in WT and chimeric male mice (Table 1).
Table 1. Serum hormone levels in mature male WT and chimeric mice.
| Group | Testosterone, ng/dl | FSH, ng/ml | LH, ng/ml |
|---|---|---|---|
| WT | 22.85 ± 5.29 | 36.61 ± 2.45 | 0.23 ± 0.05 |
| Chimeric | 19.50 ± 3.72 | 39.58 ± 2.02 | 0.21 ± 0.03 |
Blood was taken from 5- to 7-month-old male WT and chimeric mice, and serum hormone levels were analyzed. n = 10 for total testosterone and FSH (each group); n = 9 for LH (each group). FSH, follicle-stimulating hormone; LH, luteinizing hormone.
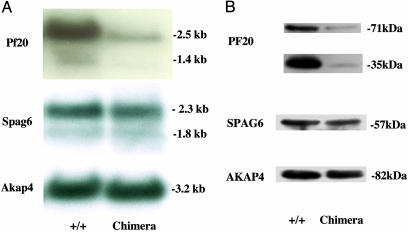
Northern blot analysis demonstrated reduced levels of the 2.5- and 1.4-kb PF20 transcripts relative to the mRNAs for SPAG6, which encodes another central apparatus protein, and AKAP4, which encodes the major protein of the fibrous sheath (Fig. 4A) in chimeric mice, which gave agouti offspring. Western blotting revealed that both the 71- and 35-kDa proteins were reduced in these mice, relative to SPAG6 and AKAP4 (Fig. 4B). Notably, the reduction in the 35-kDa protein, which accumulates postmeiotically, was more dramatic than the 71-kDa protein. We did not detect aberrant PF20 transcripts or a truncated protein using the N-terminal antibody in testis of chimeric mice, arguing against the expression of a N-terminal fragment derived from the mutant allele, which could act as a dominant negative.
Fig. 4.
Analysis of testicular mRNA and protein expression in WT and chimeric mice. (A) PF20, SPAG6, and AKAP4 mRNA expression in WT and chimeric mice. (B) PF20, SPAG6, and AKAP4 protein expression in WT and chimeric mice.
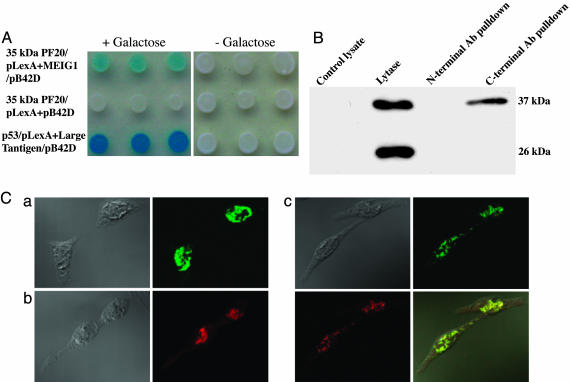
PF20 (35-kDa) Protein Binds to MEIG1 in the Nucleus. The spermatogenic abnormalities in the chimeric mice were unexpected and suggested a role for PF20 beyond a function in the axoneme, possibly regulating postmeiotic events in the nucleus where the 35-kDa PF20 is localized. To explore possible functions of nuclear PF20, we conducted a yeast two-hybrid screen to identify interacting partners. This screen revealed that MEIG1, a small protein, was a potential partner. This interaction was confirmed in a subsequent yeast protein interaction assay and by coimmunoprecipitation of 35-kDa PF20 and MEIG1 from cotransfected cell lysates (Fig. 5 A and B). In CHO cells cotransfected with MEIG1/DsRed-N1, and 35-kDa PF20/pEGFP-N2, the proteins were colocalized in the nucleus (Fig. 5C).
Fig. 5.
Interaction of 35-kDa PF20 with MEIG1. (A) Yeast two-hybrid assay with (+Galactose) or without (–Galactose) inducer shows specific binding of 35-kDa PF20 to MEIG1. p53–large T antigen interaction is displayed as a positive control. PF20 (35 kDa) and empty pB42D serve as negative control. (B) Coimmunoprecipitation of MEIG1-GFP and 35-kDa PF20 from lysates of nontransfected CHO cells (control) and cotransfected CHO cells with N- or C-terminal anti-PF20 antibodies. The immunoprecipitated material was subjected to Western blotting with anti-GFP antibody. The lysate of cotransfected cells contains the MEIG1-GFP (37 kDa) and free GFP (26 kDa), but only MEIG1-GFP is detected in the pull-down with the C-terminal antibody. (C) Colocalization of 35-kDa PF20 and MEIG1 in CHO cells. CHO cells were transfected with 35-kDa PF20/pEGFP-N2 (a), MEIG1/DsRed-N1 (b), or both plasmids (c). Phase contrast and fluorescence images are shown, and in c, a merged image (Lower Right) shows colocalization of PF20-GFP and MEIG1-DsRed.
Discussion
Motility is a prerequisite for mammalian sperm to fertilize eggs in vivo (18–20), and an intact 9 + 2 structure of the axoneme is essential for normal sperm motility. Mutations in genes encoding proteins of the axoneme dynein arms are known to cause immotile cilia syndromes. A wealth of evidence from the study of Chlamydomonas and more limited work in the mouse suggest that the central apparatus is also essential for sperm flagellar function. The present study explored the function of the murine orthologue of the Chlamydomonas PF20 gene, which when mutated causes flagellar paralysis and loss of the central pair of microtubules in isolated axonemes. The Chlamydomonas PF20 gene encodes a 67-kDa protein localized in the bridges connecting the two central microtubules (12). The mouse Pf20 gene encodes two major transcripts, regulated by two different promoters (Z.Z. and J.F.S., unpublished observations), which translate into 71- and 35-kDa proteins, respectively. These proteins accumulate during spermatogenesis in different patterns, paralleling the changes in abundance of their respective transcripts. We previously localized the meiotically expressed larger PF20 protein to the central apparatus of the axoneme by using an antibody recognizing the protein's N terminus, consistent with the known location of the Chlamydomonas PF20 protein (16). Unexpectedly, the smaller protein, which contains the contiguous C-terminal WD repeats, appears to be primarily targeted to the nucleus, despite the absence of a consensus nuclear localization signal.
To investigate the function of the two PF20 proteins, we generated chimeric mice carrying a mutant Pf20 allele that disrupts the C-terminal WD repeats. Most of the male chimeric mice were infertile, and the mutant allele was never transmitted to the offspring, probably because of the low sperm count and the disorganization of axonemal structures in the sperm, which affected motility. The phenotype of impaired spermatogenesis was surprising and suggested a role for PF20 beyond its functions in the axoneme. The finding that spermatogenesis was impaired in chimeric testis at the round spermatid stage, a time of assembly of the sperm flagellum, and increased expression of the 35-kDa PF20 protein suggest that the relative absence of this smaller protein in postmeiotic germ cells affects progression of spermatogenesis and germ cell survival, as evidenced by increased apoptotic germ cell death. The absence of germ cells from some tubules may reflect a secondary response to death of the round spermatids.
The phenotype of the PF20 mutation in Chlamydomonas, where the flagella are paralyzed and the central apparatus is missing from isolated axonemes (12), predicted an abnormality in axonemal structure in mutant mice. Indeed, the ultrastructural disorganization seen in transverse sections of flagella of epididymal sperm from the chimeric mice could be viewed as being consistent with a key role for PF20 in stability of the axoneme. However, axonemal defects per se are unlikely to be the cause of the loss of germ cells at the round spermatid stage, because this has not been observed in SPAG6-deficient mice, which have sperm with ultrastructural abnormalities related to those seen in the chimeric mice (16), or for that matter in most males with primary ciliary dyskinesia who have sperm motility defects. The nuclear localization of the 35-kDa PF20 protein also suggests that the impaired process of spermatogenesis is not merely the consequence of abnormalities in the axoneme but rather a reflection of a more global abnormality in gene expression at the round spermatid stage due to lack of the nuclear 35-kDa PF20.
Although the exact function of the nuclear 35-kDa PF20 protein in spermatogenesis remains to be elucidated, we have identified at least one interacting partner, MEIG1, a chromosome/chromatin-binding protein initially expressed at meiosis but retained in the nucleus through later stages of spermatogenesis (21–23). MEIG1 is thought to play a role in meiosis in female and male germ cells, but its presence in postmeiotic male germ cells in conjunction with the current findings raises the possibility that it participates in regulation of chromosome structure and/or gene expression after meiosis is completed, a function that is presumably impaired in the absence of 35-kDa PF20.
Conclusion
Defective spermatogenesis in chimeric mice is evidently due to haploinsufficiency of Pf20. Our findings suggest that the products of the Pf20 gene have multiple functions during spermatogenesis, including survival of postmeiotic germ cells and possibly maintenance of the integrity of the sperm axoneme and associated structures.
Acknowledgments
We thank Yang Luo for assistance in photographing light microscopic histology preparations and Jean Richa and Peifu He (Transgenic Core, University of Pennsylvania Medical Center, Philadelphia) for help with injection of the ES cells. This work was supported by National Institutes of Health Grants R01 HD37416 and P01 HD06724. Transmission electron microscopy was performed in the Imaging Core of the University of Pennsylvania's Diabetes Center (DK19525). Follicle-stimulating hormone and luteinizing hormone levels were measured at the University of Virginia Center for Research in Reproduction Ligand Assay and Analysis Core (U54-HD28934).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CHO, Chinese hamster ovary; ES, embryonic stem; TUNEL, terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling; MEIG1, meiosis-expressed gene 1.
References
- 1.Smith, E. F. & Lefebvre, P. A. (1997) Cell Motil. Cytoskeleton 38, 1–8. [DOI] [PubMed] [Google Scholar]
- 2.Porter, M. E. & Sale, W. S. (2000) J. Cell Biol. 151, F37–F42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dutcher, S. K. (1995) Trends Genet. 11, 398–404. [DOI] [PubMed] [Google Scholar]
- 4.Nonaka, S., Tanaka, Y., Okada, Y., Takeda, S., Harada, A., Kanai, Y., Kido, M. & Hirokawa, N. (1998) Cell 195, 829–837. [DOI] [PubMed] [Google Scholar]
- 5.Kobayashi, Y., Watanabe, M., Okada, Y., Sawa, H., Takai, H., Nakanishi, M., Kawase, Y., Suzuki, H., Nagashima, K., Ikeda, K. & Motoyama, N. (2002) Mol. Cell. Biol. 22, 2769–2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sapiro, R., Kostestkii, I., Olds-Clarke, P., Gerton, G. L., Radice, G. L. & Strauss, J. F., III (2002) Mol. Cell. Biol. 22, 6298–6305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.El Zein, L., Omran, H. & Bouvagnet, P. (2003) Trends Genet. 19, 162–167. [DOI] [PubMed] [Google Scholar]
- 8.Kamiya, R.. (2002) Int. Rev. Cytol. 219, 115–155. [DOI] [PubMed] [Google Scholar]
- 9.Mencarelli, C., Lupetti, P., Rosetto, M., Mercati, D., Heuser, J. E. & Dallai, R. (2001) Cell Motil. Cytoskeleton 50, 129–146. [DOI] [PubMed] [Google Scholar]
- 10.Casey, D. M., Inaba, K., Pazour, G. J., Takada, S., Wakabayashi, K., Wilkerson, C. G., Kamiya, R. & Witman, G. B. (2003) Mol. Biol. Cell 14, 3650–3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith, E. F. & Lefebvre, P. A. (1996) J. Cell Biol. 132, 359–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith, E. F. & Lefebvre, P. A. (1997) Mol. Biol. Cell 8, 455–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rupp, G., O'Toole, E. & Porter, M. E. (2001) Mol. Biol. Cell 12, 739–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sapiro, R., Tarantino, L. M., Velazquez, F., Kiriakidou, M., Hecht, N. B., Bucan, M. & Strauss, J. F., III (2000) Biol. Reprod. 62, 511–518. [DOI] [PubMed] [Google Scholar]
- 15.Neilson, L. I., Schneider, P. A., Van Deerlin, P. G., Kiriakidou, M., Driscoll, D. A., Pellegrini, M. C., Millinder, S., Yamamoto, K. K., French, C. K. & Strauss, J. F., III (1999) Genomics 60, 272–280. [DOI] [PubMed] [Google Scholar]
- 16.Zhang, Z., Sapiro, R., Kapfhamer, D., Bucan, M., Bray, J., Chennathukuzhi, V., McNamara, P., Curtis, A., Zhang, M., Blanchette-Mackie, E. J., et al. (2002) Mol. Cell. Biol. 22, 7993–8004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cho, C., Willis, W. D., Goulding, E. H., Jung-Ha, H., Choi, Y. C., Hecht, N. B. & Eddy, E. M. (2001) Nat. Genet. 28, 82–86. [DOI] [PubMed] [Google Scholar]
- 18.Inaba, K. (2003) Zool. Sci. 20, 1043–1056. [DOI] [PubMed] [Google Scholar]
- 19.Turner, R. M.. (2003) J. Androl. 24, 790–803. [DOI] [PubMed] [Google Scholar]
- 20.Cosson, J., Huitorel, P. & Gagnon, C. (2003) Cell Motil. Cytoskeleton 54, 56–63. [DOI] [PubMed] [Google Scholar]
- 21.Don, J. & Wolgemuth, D. J. (1992) Cell Growth Differ. 3, 495–505. [PubMed] [Google Scholar]
- 22.Don, J., Winer, M. A. & Wolgemuth, D. J. (1994) Mol. Reprod. Dev. 38, 16–23. [DOI] [PubMed] [Google Scholar]
- 23.Steiner, R., Ever, L. & Don, J. (1999) Dev. Biol. 216, 635–645. [DOI] [PubMed] [Google Scholar]