Abstract
We researched the lifespan of Drosophila under axenic conditions compared with customary procedure. The experiments revealed that the presence of bacteria during the first week of adult life can enhance lifespan, despite unchanged food intake. Later in life, the presence of bacteria can reduce lifespan. Certain long-lived mutants react in different ways, indicating an interplay between bacteria and longevity-enhancing genes.
Keywords: longevity, aging, axenic flies, antibiotics
Microorganisms have been a major factor in shaping eukaryotic evolution (1). Because the embryonic development of most animals occurs under germ-free conditions, the establishment of a resident fauna is a dynamic process, the initial exposure at birth or hatching determining which bacteria have primary access to the host (2). However, diet and environment during the animal's life can drastically alter its bacterial population (3–5). Even though an animal is exposed to a vast array of bacteria, the complexity of fauna in its gut can vary widely, some harboring a single species, others hosting hundreds (6–10).
To study the interaction between a host and its fauna, axenic culturing techniques and methods for in situ identification of bacteria have been developed (2, 4, 11, 12). Molecular typing by 16S PCR makes it possible to identify species profiles without the need to culture the bacteria (13–15). Axenic techniques have been developed for numerous organisms, including the mouse (16, 17), Drosophila melanogaster (18), Caenorhabditis brigssae (19), and Caenorhabditis elegans (20).
In vertebrates, bacteria can play a beneficial role in development and maintenance of the gut. Zebrafish raised axenically suffer severe gut deterioration, and fail to reach adulthood (21). Mammals use bacteria to extract and process nutrients from the diet (22), to assist in the development and function of the gut and the enteric nervous system (23), and for conditioning of the immune system (24–26). Helicobacter pylori participates in appetite regulation (27, 28); however, on the other hand, it is linked to ulcers and gastric cancer (29).
Bacteria can affect host fitness and longevity. Paramecium, cultured axenically, ages more rapidly, as judged by the accumulation of fragmented DNA (30), and defaunated termites are short-lived (7). For C. elegans and mosquito, axenic culture is deleterious to development but beneficial to lifespan (20, 31–33). In Drosophila, infection by a virulent strain of Wolbachia can shorten lifespan (34), whereas nonvirulent strains can extend lifespan and suppress the Sex-lethal phenotype (35, 36). Bakula (18) observed that axenic cultures of Drosophila develop more slowly but did not test the effect on adult lifespan.
Modulation of the immune system can promote longevity by preventing infection and avoiding autoimmunity. A dramatic, age-dependent up-regulation of immunity-related genes is seen in mice, worms, and flies (37, 38). The expression of these genes in flies is predictive of remaining lifespan (39). Old C. elegans and flies are more susceptible to infection (40) and allow greater bacterial proliferation (41).
The genetic constitution of the host also plays a role. Long-lived insulin-signaling mutations in C. elegans can reduce bacterial load, increase resistance to infection, and up-regulate antibacterial genes (40, 42). In Drosophila, the Thor gene is up-regulated in response to food deprivation or bacterial challenge, putatively by means of an insulin-related signaling mechanism (43–45).
To determine whether the bacteria normally present in Drosophila laboratory cultures affect lifespan, we used axenic cultures and antibiotic treatment and found that for both the Canton-S wild type and the w1118 strain, the presence of bacteria during the first week of adult life enhanced longevity by 30–35%. Conversely, the presence of bacteria in the last stage of life caused a slight decrease. Because signaling by the hormone ecdysone, in initiating metamorphosis, triggers dramatic changes in bacterial titer, the structure of the gut and fat body, and modulation of the immune response (18, 46), we tested the ecdysone receptor mutant EcRv559fs (47), which is long-lived (48). The mutant was little affected by presence or absence of bacteria. In contrast, for another long-lived mutant, DJ817 (37), the beneficial effect of early bacteria was enhanced. Thus, there exist genetic enhancers and suppressors of the bacterial effect on longevity, indicating complex regulation of the interactions involved.
Materials and Methods
Food Preparation. Experiments were performed by using standard Caltech fly food consisting of 5% dextrose, 2.5% sucrose, 1.5% yeast, 17% cornmeal, 0.9% agar, 0.09% propionic acid, and 0.09% phosphoric acid (49). For axenic conditions, plastic bottles containing the food were autoclaved for 30 min, followed by 18 h of irradiation by a radioactive cesium source at a cumulative surface dose of 1.5 megarads (1 rad = 0.01 Gy). Axenic food vials, because of their higher surface-to-volume ratio, only required the irradiation. Antibiotic food contained 500 μg/ml ampicillin, 50 μg/ml tetracycline, and 200 μg/ml rifamycin, prepared by adding 1 ml of a 100× stock of antibiotics in 50% ethanol per 100 ml of liquefied food. Control nonantibiotic food was prepared by adding the same concentration of ethanol alone.
Lifespan Measurements. Lifespan measurements were done by using adult males tested in triplicate at 25–35 flies per vial. Flies were transferred every 3–4 days, and the number of dead flies was recorded. Axenic lifespan experiments and corresponding controls were performed in a sterile hood at 22–23°C by using flies mildly anesthetized with ether on a glass slab. For experiments involving antibiotic and corresponding controls, the flies were collected by using mild CO2 anesthesia and maintained at 25°C. For longevity data, log rank tests were performed with prism 3 software (GraphPad, San Diego) set for survival curve algorithm.
Axenic Fly Cultures. Fly cultures were generated by following the protocol published by Bakula (18). Collections of 12-h embryos were dechorionated for 2 min in 2.7% sodium hypochlorite [2-fold diluted bleach (Kem Tech, St. Ixonia, WI)] and washed twice in 70% ethanol and then twice with sterile, distilled water. These embryos were transferred in a tissue culture hood into axenic food bottles. Axenia of the embryos was confirmed by performing 16S PCR on homogenates of the adult flies and by plating the homogenates on LB agar plates. The flies were transferred every 3–4 days into new, irradiated vials. Vials were also spot checked for contamination by swabbing the food in the axenic vials and plating on LB plates. To generate controls for the axenic flies, 12-hr embryos were collected and washed four times in sterile, distilled water and transferred into axenic food bottles. Where indicated (Fig. 1), bacteria were then introduced at chosen times by addition of a homogenate of nonaxenic flies.
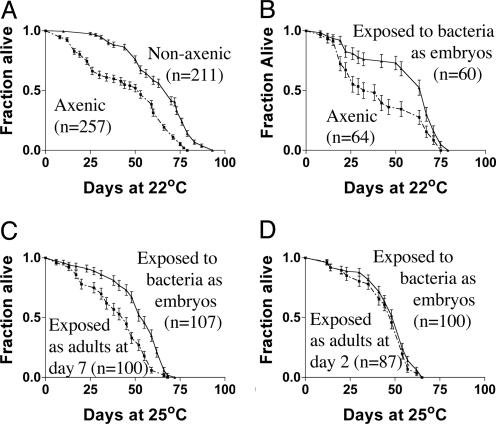
Fig. 1.
Survival curves of axenic flies compared with those of flies maintained under customary, nonaxenic conditions. (A) In the presence of bacteria, mean lifespan was 30% longer (P < 0.0001). (B) Starting with axenic embryos, bacteria were introduced during development by adding a homogenate of nonaxenic flies. Introduction of bacteria at that time produced a 30% increase in longevity (P = 0.002). (C) Starting with axenic embryos, bacteria were introduced either during development or after day 7 of adult life. The latter flies' lives were 25% shorter than those of flies receiving bacteria during development (P < 0.0001). (D) Starting with axenic embryos, bacteria were introduced either during development or after day 2 of adult life. Introduction of bacteria at adult day 2 promoted longevity as well as if introduced during development (P = 0.23). (A and B, 22°C; C and D, 25°C). Error bars represent SEM.
Measurement of Food Intake. We added 70 μCi (1 Ci = 37 GBq) of 32P (α-labeled dCTP; specific activity, 3,000 per μl) to 50 ml of liquefied food. Flies were collected under mild CO2 anesthesia and allowed to recover for 1 day on the same type of food later used for the feeding assay. The flies were then introduced onto the labeled food and allowed to feed for 18 h, at which time they were transferred to scintillation vials and counted.
Identification of Bacterial Species by 16S PCR. A modification of the technique described in ref. 4 was used. Approximately 100 flies were homogenized in 5 ml of sterile saline. The homogenate was filtered through sterile, packed cheesecloth to remove fly parts. DNA was extracted by a modification of a standard alkaline lysis miniprep procedure (Qiagen, Valencia, CA). We incubated 500 μl of homogenate solution plus 500 μl each of solutions P1 and P2 at 70°C for 10 min. N3 buffer was added, followed by extraction with an equal volume of phenol/chloroform. After being vortexed for 1 min and centrifuged at 10,000 × g for 30 min, the supernatant was taken and 3 vol of ethanol was added to precipitate DNA. After being centrifuged and washed with 70% ethanol, the pellet was resuspended in 50 μl of TE buffer (10 mM Tris/1 mM EDTA, pH 8.0), and 2 μl of this was used in a PCR with the universal 16S primers 5′-GGTTACCTTGTTACGACTT (149R) and 5′-AGAGTTTGATCCTGGCTCAG (8F).
Outcrossing of EcRv559fs. Outcrossing was done to minimize any effect of genetic background in the comparison with w1118. Five EcRv559fs/CyO virgin females were collected and crossed to w1118 males. From these progeny, 20 heterozygous virgin females were collected and crossed individually to w1118 males. Progeny were analyzed by PCR by using primers EcR com B1 (AAGATCATATACGCGGGATTCT) and EcR com B2 (TCTGGATCGCTTCGACTAGTT) to identify F1 females harboring the EcRv559fs chromosome, as indicated by the occurrence of two PCR products, 150 bp and 187 bp (wild-type flies produce only a single band of 187 bp). From crosses yielding the diagnostic doublet, 20 virgin F2 females were collected and crossed to w1118 males to generate the F3 outcrossed females. This step was repeated for six cycles of outcrossing. To recover the mutant chromosome, virgin females were then collected and crossed to CyO/Sco males. EcRv559fs was then tested as a heterozygote by crossing virgin EcRv559fs/Cyo females to male w1118.
Results
Under Axenic Conditions, Drosophila Longevity Is Reduced. We eliminated bacteria by a modified version of the axenic culture technique described by Bakula (18), which relies on surface sterilization of the eggs with bleach and 70% ethanol (see Materials and Methods). The food was sterilized by means of irradiation with a cesium source at a cumulative dose of 1.5 megarads (the Centers for Disease Control 99.9% confidence level for sterilization of produce), and sterile techniques were used to handle the cultures. Elimination of bacteria was confirmed by 16S PCR and by plating homogenates of the flies on bacterial culture media (see Materials and Methods). The mean lifespan of axenic flies was ≈30% less than that of untreated flies (Fig. 1 A).
To control for possible deleterious effects of the chemicals used to decontaminate the embryos, the experiment was repeated by making axenic embryos, then reintroducing bacteria by means of a homogenate of untreated flies (see Materials and Methods), simulating the exposure that occurs under ordinary, nonaxenic laboratory conditions. The lifespan of these adults was similar to that of ordinary controls. The result suggests that the diminution of longevity under axenic conditions is due to the elimination of bacteria, not to the method of sterilization (Fig. 1B).
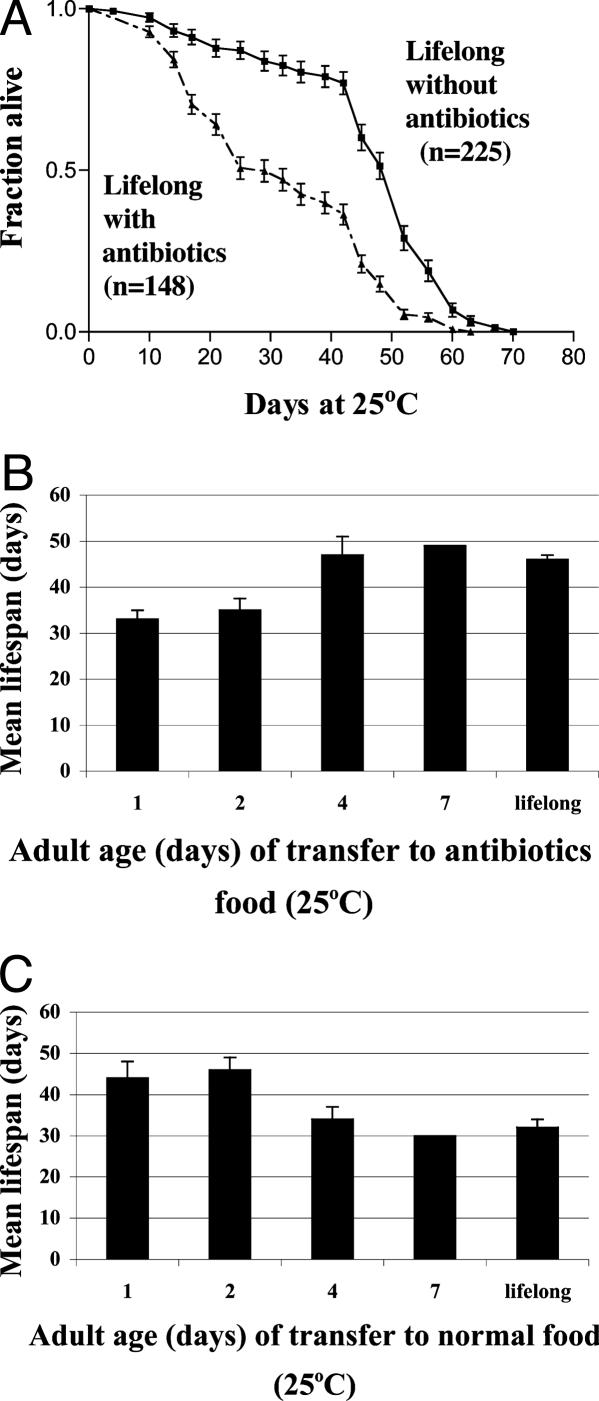
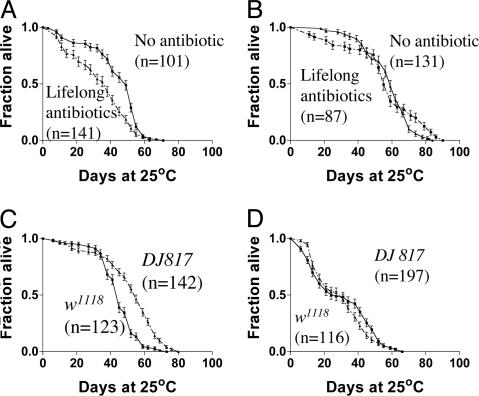
Alternatively, the effect of elimination of bacteria was tested by the addition of antibiotics. w1118 flies were raised for two generations on standard media containing three antibiotics (500 μg/ml ampicillin, 50 μg/ml tetracycline, and 200 μg/ml rifamycin) and maintained on antibiotic-containing food throughout life. These experiments were done in duplicate trials with at least 100 male flies. Elimination of bacteria by this treatment, compared with flies maintained on the same food without antibiotics, resulted in a 35% reduction of mean lifespan, similar to that seen for axenic treatment (Fig. 2A). The same result (data not shown) was obtained with wild-type strain Canton-S.
Fig. 2.
Critical period for extension of longevity by the presence of bacteria by using antibiotics. (A) Lifelong (from embryo onward) antibiotic in the food results in a 35% reduction in longevity (P < 0.0001), consistent with that observed under axenic conditions. (B) Flies developed on normal food. Flies were raised on nonsterile food until adult emergence; they were then transferred, at various times, to antibiotic food. Those transferred to antibiotic food at days 1 and 2 showed a reduction in longevity, compared with flies maintained lifelong on nonsterile food. However, flies not transferred to antibiotic food until days 4 or 7 showed similar lifespans to untreated flies. (C) Flies developed on antibiotic food. Flies were raised on antibiotics from the embryo up to adult emergence and then transferred, at various times, to nonsterile food. Flies transferred at days 1 or 2 showed no difference compared with flies under ordinary conditions. However, flies transferred after days 4 or 7 showed a reduction in longevity comparable to flies maintained lifelong on antibiotics. The bars in B and C correspond to two experiments, each of at least 90 flies (the day 7 experiment was a single trial). Error bars represent SEM.
The First Week of Adult Life Is a Critical Period in Which Bacteria Exert a Positive Effect on Longevity. We then tested the effect of adding bacteria to adult flies that had been raised axenically to adulthood, by shifting them to ordinary (“dirty”) conditions for the remainder of life. For axenic flies collected within 2 days of adult emergence then exposed to bacteria, the full life-extending effect was observed (Fig. 1D). However, for flies kept axenic for 7 days, the subsequent addition of bacteria was ineffective (Fig. 1C). These results indicate the existence of a “window” in early adulthood when the presence of bacteria is important.
To define this critical period, antibiotics were used. Flies were first raised on ordinary food, then, at various times, transferred to antibiotic-containing food and maintained on antibiotics for the remainder of life. It was found that flies transferred to antibiotics after 4–7 days already had the benefit of the early presence of bacteria (Fig. 2B). In a separate experiment, flies were raised either on antibiotics (Fig. 2C) or axenically (Fig. 1 C and D), then transferred to nonsterile food (thereby allowing the reincorporation of bacteria) at various stages. Flies transferred onto nonsterile food after 4–7 days failed to show the benefit, consistent with the conclusion that the first 4–7 days of adult life is a critical period in which the presence of bacteria is important to the extension of lifespan.
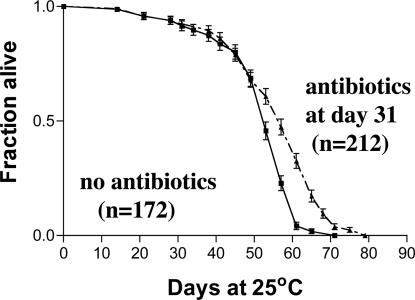
Effect of Exposure to Bacteria Late in Life. A converse, albeit smaller, effect on lifespan was seen when bacteria were present at later age. Separate cohorts of 120–160 flies raised on ordinary food were transferred at various intervals to antibiotics and compared with flies maintained lifelong on ordinary food. Flies receiving antibiotics exclusively during week 2 or week 3 showed no change in lifespan relative to untreated flies. Flies introduced to antibiotic media during week 4 showed an increase in longevity of 9% relative to untreated flies. Flies receiving continued treatment from week 2 showed a 10% increase (Table 1). The results suggest that removing bacteria late in life is beneficial for longevity. Because these experiments were done at 29°C, we checked whether a similar effect occurred at 25°C. Flies were raised under standard conditions until age 31 days, at which point they were either transferred to antibiotic food or maintained on standard medium. Fig. 3A shows that, also at 25°C, late antibiotic addition resulted in an 8% increase in longevity. In C. elegans, removal of bacteria was also shown to increase longevity by suppression of late mortality (31, 32).
Table 1. Effect of exogenous bacteria late in life.
| Period of exposure to antibiotics | No. of flies | Mean lifespan, days | Change in longevity, % | P values |
|---|---|---|---|---|
| No antibiotics | 281 | 23.4 ± 0.3 | — | — |
| Week 2 only | 146 | 23.9 ± 0.6 | +2 | 0.6 |
| Week 3 only | 155 | 22.9 ± 0.5 | -2 | 0.2 |
| Week 4 only | 147 | 25.5 ± 0.4 | +9 | 0.002* |
| From week 2 | 156 | 25.8 ± 0.4 | +10 | 0.001* |
To determine the effect of bacteria on longevity after the first week of adulthood, antibiotics were added for week-long periods and then removed. Each entry represents a single experiment using 120-160 male flies. These experiments were done at 29°C. Addition of antibiotics during week 2 or week 3 had no statistically significant effects on longevity. —, not applicable.
Antibiotics during week 4 produced a 9% increase in mean lifespan, similar to the 10% change in longevity observed when antibiotics were provided from week 2 onward.
Fig. 3.
Presence of bacteria late in life is deleterious. Flies were raised and maintained under ordinary (nonsterile) conditions to age 31 days and then either transferred to antibiotic food or maintained on standard food. This late addition of antibiotics resulted in an 8% increase in mean lifespan (P < 0.0001). These results confirm at 25°C the results shown in Table 1, which were from experiments done at 29°C. Error bars represent SEM.
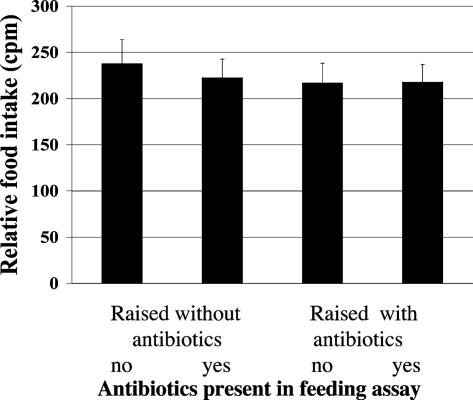
Antibiotics Do Not Significantly Affect Food Intake. Because dietary restriction can dramatically alter longevity, it is necessary to control for this factor. In mammals, gastric bacteria can affect the activity of the appetite-regulating factors ghrelin, gastrin, and leptin (27, 28). Drosophila feeding can be negatively affected by sodium chloride or positively affected by trehalose (50, 51). Food intake was therefore measured by means of addition of the radioactive tracer 32P α-labeled dCTP to the food (see Materials and Methods). Four conditions were used: (i) flies raised from embryo to adult without antibiotics, then tested for feeding without antibiotics; (ii) flies raised without antibiotics, then tested with antibiotics; (iii) flies raised with antibiotics, then tested without antibiotics; (iv) and flies raised and tested with antibiotics.
Male flies 7 days of age were tested by allowing them to feed for 18 h on food containing 32P in groups of 20 flies per vial; the flies were then assayed by scintillation counting. No significant differences in food consumption were observed (Fig. 4). Similar results were obtained for 3-day-old flies (data not shown). Thus, neither the removal of bacteria nor the addition of antibiotics significantly affected food intake in this assay.
Fig. 4.
Feeding rate is unchanged by the antibiotics used. Food intake was determined by using 32P-labeled food under four different conditions. Flies were raised from embryos to adulthood on either antibiotic-containing or standard food, and then, at adult age (7 days), they were transferred either to antibiotic or standard food and allowed to feed for 18 h. Food intake, as determined by scintillation counting, showed no significant differences under the four regimes. Bars represent the results of two trials with 20 flies each. Error bars represent experimental range.
Role of Genetic Makeup of the Host. The above experiments were done on w1118 flies. To test whether the genotype of the host could modulate the response, we tested several mutants. EcRv559fs, an allele of the ecdysone receptor that is long-lived as a heterozygote (48), showed a different response to bacteria. Lifelong antibiotic treatment, which reduced the lifespan of w1118 (Fig. 5A), did not affect EcRv559fs mutant lifespan (Fig. 5B). To minimize any potential effects of genetic background in the mutant strain, the line was outcrossed six times into w1118.
Fig. 5.
Host genotype can enhance or suppress the effect of bacteria on lifespan. (A) w1118 flies. For w1118 flies, the presence of antibiotic throughout lifespan decreases longevity (P < 0.001). (B) EcRv559fs flies. Similar treatment does not affect the long-lived heterozygous EcRv559fs strain (P = 0.2). (C and D) No antibiotics versus with antibiotics. The enhancer trap strain DJ817 is 30% longer lived than w1118 in the presence of bacteria (P < 0.001) (C), but removal of bacteria with antibiotics (D) eliminates the difference in their lifespans (P = 0.3).
A panel of long-lived mutants was similarly tested. One of them, the DJ817 mutant (37), was 30% longer-lived than w1118 when bacteria were present (Fig. 5C), whereas eradicating the bacteria abolished the difference in longevity (Fig. 5D). These varied results with different mutant strains indicate that it should be possible to perform a genetic dissection of the mechanisms of interaction between bacteria and host.
Bacterial Species Occurring in Fly Cultures. Because it is known that many Drosophila stocks in common use harbor Wolbachia, a bacterium known to influence longevity (34, 35), we tested all of the strains used by PCR, and all were negative. To determine which bacteria were present in w1118 grown under ordinary conditions, a 16S PCR library was made by using homogenates of adults washed in 70% ethanol before homogenization (see Materials and Methods). Twenty clones from this library were chosen and analyzed by restriction digest, yielding five different fragment patterns. Ten clones, representing all five classes, were sequenced and compared with sequences in the Ribosomal Database Project II (52). The comparison resulted in the identification of four groups of bacteria, with similarities in sequence to Lactobacillus, Gluconobacter, Enterobacter, and Anaerococcus. The homogenates used for 16S PCR were also plated on various agar media [nutrient broth, LB, potato dextrose agar, and glycerol yeast extract (53)]. Colonies were picked from the various plates and characterized by PCR, which indicated the presence of Staphylococcus and Enterobacter. Much of the bacterial population within the fly is presumably anaerobic and refractory to culture by ordinary methods. This finding may be analogous to the one in humans, where most resident bacteria are refractory to cultivation, even by the most sophisticated culture methods (2).
Discussion
The Early Effect of Bacteria on Drosophila Lifespan. It is known that the bacterial titer in Drosophila increases throughout development, up to pupariation, when it rapidly drops (18). This drop occurs after a pulse of ecdysone that results in a dramatic induction of immune genes (46). During metamorphosis, the gut is remodeled, and, shortly after adult emergence, the larval fat body deteriorates and is replaced with the adult fat body. The critical period of exposure in which the beneficial effect of bacteria is observed overlaps with the timing of the transition from larval to adult fat. This observation is striking in light of the importance of fat in mediating longevity by means of insulin-related signaling in Drosophila (54–56).
The Late Effect of Bacteria. It is interesting to compare our results with Drosophila with those of C. elegans. In the nematode, bacteria are the principal source of nutrients, and their elimination by means of axenic cultures has been considered a form of dietary restriction, thus extending lifespan (32). Larsen and Clarke (33) have shown that coenzyme Q, synthesized in the bacteria eaten by normal worms, has a negative effect on longevity. Garigan et al. (31) have shown that live bacteria present late in life shorten lifespan by causing constipation. Thus, the longevity changes in C. elegans may be due to a combination of dietary effects and others resulting from overproliferation of the bacteria. We observed a similar late, deleterious effect of bacteria in Drosophila. In a previous study, we identified an immune function gene, the expression of which increases with age, consistent with an attempt to ward off infection (37).
Host–Bacteria Interactions. The existence of genetic enhancement or suppression of the early bacterial effect suggests that genetic analysis can be used to analyze these mechanisms. Two of the modifiers we identified, the EcR gene and the enhancer trap DJ817, which has a P-element insertion in the Olf-186 gene, are both expressed in the gut (data not shown). We have found that the expression of the LacZ reporter in DJ817 drops by nearly two orders of magnitude in the first 10 days of adult life (data not shown). Also, by using temperature-sensitive alleles that alter ecdysone signaling, we have found that reducing ecdysone during the first 10 days is sufficient to promote longevity. The timing of these two effects coincides with the early window of bacterial action.
It will be desirable to use pure cultures of identified bacteria introduced to the fly medium under otherwise axenic conditions. Our preliminary characterization of the bacterial species inhabiting our fly cultures indicates the occurrence of at least five genera. Eventually, it may be possible to identify specific substances produced by the bacteria that are capable of extending longevity.
Drosophila geneticists have paid relatively little attention to the bacterial flora within fly stocks (57), yet such flora may increase phenotypic variation. It is remarkable that a high percentage of laboratory stocks in common use contain Wolbachia, which affects longevity and other processes (34). The stocks we used here were Wolbachia-free, as tested by PCR.
Bacteria can alter the chemical and nutritional properties of food and, in some animals, directly alter food intake and gut function, as reviewed in ref. 2. In C. elegans, alterations in chemosensory function can alter lifespan (58). Likewise, changes in food consumption or gene expression in the digestive tracts of nematodes and flies can increase longevity (59–61). The work of Bakula (18) suggests that bacteria may influence nutrition during Drosophila development; axenic flies developed more slowly and yielded lower number of progeny (18), and we observed similar effects. Nevertheless, axenically developed flies displayed a normal lifespan if bacteria were introduced before day 2 of adult life, indicating that the developmental effects were not responsible for the lifespan phenotype. Although we did not detect a difference in rate of food intake for adults in the presence or absence of bacteria, it is possible that secondary nutritional effects could be at play.
The relations between food composition, resident fauna, and gut function in Drosophila remain largely uncharacterized, and the dramatic effects associated with bacteria deserve further study. Antibiotics are routinely used in human and animal populations, and there is a growing concern about such practices. Model organisms such as Drosophila may help to discern their effects on longevity and general well being.
Acknowledgments
We thank Stephanie Cornelison and John Silverlake for expert technical assistance and Todd Chiche, Jared Leadbetter, Micheline Laurent, and members of the Benzer laboratory for helpful discussions. This work was funded by grants (to S.B.) from the National Science Foundation and the National Institute of Aging. L.S. acknowledges funding from the Natural Sciences Engineering Research Council of Canada and the Institute of Aging of the Canadian Institutes of Health Research.
References
- 1.McFall-Ngai, M. J. (2002) Dev. Biol. 242, 1–14. [DOI] [PubMed] [Google Scholar]
- 2.Xu, J. & Gordon, J. I. (2003) Proc. Natl. Acad. Sci. USA 100, 10452–10459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Falk, P. G., Hooper, L. V., Midtvedt, T. & Gordon, J. I. (1998) Microbiol. Mol. Biol. Rev. 62, 1157–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Favier, C. F., Vaughan, E. E., De Vos, W. M. & Akkermans, A. D. (2002) Appl. Environ. Microbiol. 68, 219–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Broderick, N. A., Raffa, K. F., Goodman, R. M. & Handelsman, J. (2004) Appl. Environ. Microbiol. 70, 293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ciche, T. A. & Ensign, J. C. (2003) Appl. Environ. Microbiol. 69, 1890–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Douglas, A. E. (1989) Biol. Rev. Cambridge Philos. Soc. 64, 349–358. [DOI] [PubMed] [Google Scholar]
- 8.Leadbetter, J. R., Schmidt, T. M., Graber, J. R. & Breznak, J. A. (1999) Science 283, 686–689. [DOI] [PubMed] [Google Scholar]
- 9.Endo, Y. & Nickle, W. R. (1991) J. Helminthol. Soc. Wash. 58, 202–212. [Google Scholar]
- 10.Steinhause, E. (1946) Insect Microbiology: An Account of the Microbes Associated with Insects and Ticks, with Special Reference to the Biologic Relationships Involved (Comstock, Ithaca, NY).
- 11.Schultze, M. & Kondorosi, A. (1998) Annu. Rev. Genet. 32, 33–57. [DOI] [PubMed] [Google Scholar]
- 12.Berg, R. D. (1996) Trends Microbiol. 4, 430–435. [DOI] [PubMed] [Google Scholar]
- 13.Relman, D. A. & Falkow, S. (1992) Infect. Agents Dis. 1, 245–253. [PubMed] [Google Scholar]
- 14.Harmsen, H. J., Raangs, G. C., He, T., Degener, J. E. & Welling, G. W. (2002) Appl. Environ. Microbiol. 68, 2982–2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayashi, H., Sakamoto, M., Kitahara, M. & Benno, Y. (2003) Microbiol. Immunol. 47, 557–570. [DOI] [PubMed] [Google Scholar]
- 16.Reyniers, J. A. (1957) Am. J. Vet. Res. 18, 678–687. [PubMed] [Google Scholar]
- 17.Reyniers, J. A. & Sacksteder, M. R. (1958) Appl. Microbiol. 6, 146–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bakula, M. (1969) J. Invertebr. Pathol. 14, 365–374. [DOI] [PubMed] [Google Scholar]
- 19.Buecher, E. J., Jr., & Hansen, E. L. (1969) Experientia 25, 656. [DOI] [PubMed] [Google Scholar]
- 20.Croll, N. A., Smith, J. M. & Zuckerman, B. M. (1977) Exp. Aging Res. 3, 175–189. [DOI] [PubMed] [Google Scholar]
- 21.Rawls, J. F., Buck, S. S. & Gordon, J. I. (2004) Proc. Natl. Acad. Sci. USA 101, 4596–4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hooper, L. V., Midtvedt, T. & Gordon, J. I. (2002) Annu. Rev. Nutr. 22, 283–307. [DOI] [PubMed] [Google Scholar]
- 23.Husebye, E., Hellstrom, P. M. & Midtvedt, T. (1994) Dig. Dis. Sci. 39, 946–956. [DOI] [PubMed] [Google Scholar]
- 24.Macpherson, A. J., Gatto, D., Sainsbury, E., Harriman, G. R., Hengartner, H. & Zinkernagel, R. M. (2000) Science 288, 2222–2226. [DOI] [PubMed] [Google Scholar]
- 25.Kelly, D., Campbell, J. I., King, T. P., Grant, G., Jansson, E. A., Coutts, A. G., Pettersson, S. & Conway, S. (2004) Nat. Immunol. 5, 104–112. [DOI] [PubMed] [Google Scholar]
- 26.Braun-Fahrlander, C., Riedler, J., Herz, U., Eder, W., Waser, M., Grize, L., Maisch, S., Carr, D., Gerlach, F., Bufe, A., et al. (2002) N. Engl. J. Med. 347, 869–877. [DOI] [PubMed] [Google Scholar]
- 27.Nwokolo, C. U., Freshwater, D. A., O'Hare, P. & Randeva, H. S. (2003) Gut 52, 637–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Konturek, J. W., Konturek, S. J., Kwiecien, N., Bielanski, W., Pawlik, T., Rembiasz, K. & Domschke, W. (2001) Scand. J. Gastroenterol. 36, 1148–1154. [DOI] [PubMed] [Google Scholar]
- 29.Wong, B. C., Lam, S. K., Wong, W. M., Chen, J. S., Zheng, T. T., Feng, R. E., Lai, K. C., Hu, W. H., Yuen, S. T., Leung, S. Y., et al. (2004) J. Am. Med. Assoc. 291, 187–194. [DOI] [PubMed] [Google Scholar]
- 30.Holmes, G. & Holmes, N. (1986) Gerontology 32, 252–260. [DOI] [PubMed] [Google Scholar]
- 31.Garigan, D., Hsu, A. L., Fraser, A. G., Kamath, R. S., Ahringer, J. & Kenyon, C. (2002) Genetics 161, 1101–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Houthoofd, K., Braeckman, B. P., Lenaerts, I., Brys, K., De Vreese, A., Van Eygen, S. & Vanfleteren, J. R. (2002) Exp. Gerontol. 37, 1371–1378. [DOI] [PubMed] [Google Scholar]
- 33.Larsen, P. L. & Clarke, C. F. (2002) Science 295, 120–123. [DOI] [PubMed] [Google Scholar]
- 34.Min, K. T. & Benzer, S. (1997) Proc. Natl. Acad. Sci. USA 94, 10792–10796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fry, A. J. & Rand, D. M. (2002) Int. J. Org. Evol. 56, 1976–1981. [DOI] [PubMed] [Google Scholar]
- 36.Star, D. J. & Cline, T. W. (2002) Nature 418, 76–79. [DOI] [PubMed] [Google Scholar]
- 37.Seroude, L., Brummel, T., Kapahi, P. & Benzer, S. (2002) Aging Cell 1, 47–56. [DOI] [PubMed] [Google Scholar]
- 38.DeVeale, B., Brummel, T. & Seroude, L. (2004) Aging Cell 3, 195–208. [DOI] [PubMed] [Google Scholar]
- 39.Landis, G. N., Abdueva, D., Skvortsov, D., Yang, J., Rabin, B. E., Carrick, J., Tavare, S. & Tower, J. (2004) Proc. Natl. Acad. Sci. USA 101, 7663–7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garsin, D. A., Villanueva, J. M., Begun, J., Kim, D. H., Sifri, C. D., Calderwood, S. B., Ruvkun, G. & Ausubel, F. M. (2003) Science 300, 1921. [DOI] [PubMed] [Google Scholar]
- 41.Kim, Y., Nam, H., Chung, H., Kim, N., Ryu, J., Lee, W., Arking, R. & Yoo, M. (2001) J. Am. Aging Assoc. 24, 187–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fabian, T. J. & Johnson, T. E. (1995) Mech. Ageing Dev. 83, 155–170. [DOI] [PubMed] [Google Scholar]
- 43.Bernal, A. & Kimbrell, D. A. (2000) Proc. Natl. Acad. Sci. USA 97, 6019–6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fingar, D. C., Salama, S., Tsou, C., Harlow, E. & Blenis, J. (2002) Genes Dev. 16, 1472–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zinke, I., Schutz, C. S., Katzenberger, J. D., Bauer, M. & Pankratz, M. J. (2002) EMBO J. 21, 6162–6173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tzou, P., De Gregorio, E. & Lemaitre, B. (2002) Curr. Opin. Microbiol. 5, 102–110. [DOI] [PubMed] [Google Scholar]
- 47.Bender, M., Imam, F. B., Talbot, W. S., Ganetzky, B. & Hogness, D. S. (1997) Cell 91, 777–788. [DOI] [PubMed] [Google Scholar]
- 48.Simon, A. F., Shih, C., Mack, A. & Benzer, S. (2003) Science 299, 1407–1410. [DOI] [PubMed] [Google Scholar]
- 49.Lewis, E. B. (1960) Drosophila Inf. Serv. 34, 118–119. [Google Scholar]
- 50.Rodrigues, V. & Siddiqi, O. (1978) Proc. Indian Acad. Sci. Sect. B 87, 146–160. [Google Scholar]
- 51.Dahanukar, A., Foster, K., van der Goes van Naters, W. M. & Carlson, J. R. (2001) Nat. Neurosci. 12, 1188–1186. [DOI] [PubMed] [Google Scholar]
- 52.Cole, J. R., Chai, B., Marsh, T. L., Farris, R. J., Wang, Q., Kulam, S. A., Chandra, S., McGarrell, D. M., Schmidt, T. M., Garrity, G. M. & Tiedje, J. M. (2003) Nucleic Acids Res. 31, 442–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sambrook, J. & Russell, D. (2001) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 54.Kapahi, P., Zid, B. M., Harper, T., Koslover, D., Sapin, V. & Benzer, S. (2004) Curr. Biol. 14, 885–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hwangbo, D. S., Gersham, B., Tu, M. P., Palmer, M. & Tatar, M. (2004) Nature 429, 562–566. [DOI] [PubMed] [Google Scholar]
- 56.Giannakou, M. E., Goss, M., Junger, M. A., Hafen, E., Leevers, S. J. & Partridge, L. (2004) Science 305, 361. [DOI] [PubMed] [Google Scholar]
- 57.Ashburner, M. (1989) Drosophila: A Laboratory Handbook (Cold Spring Harbor Lab. Press, Plainview, NY).
- 58.Alcedo, J. & Kenyon, C. (2004) Neuron 41, 45–55. [DOI] [PubMed] [Google Scholar]
- 59.Libina, N., Berman, J. R. & Kenyon, C. (2003) Cell 115, 489–502. [DOI] [PubMed] [Google Scholar]
- 60.Clancy, D., Gems, D., Harshman, L., Oldham, S., Stocker, H., Hafen, E., Leevers, S. & Partridge, L. (2001) Science 292, 104–106. [DOI] [PubMed] [Google Scholar]
- 61.Rogina, B., Reenan, R. A., Nilsen, S. P. & Helfand, S. L. (2000) Science 290, 2137–2140. [DOI] [PubMed] [Google Scholar]