Abstract
Recent advances in aging research have uncovered genes and genetic pathways that influence lifespan in such diverse organisms as yeast, nematodes, flies, and mice. The discovery of genes and drugs that affect lifespan has been delayed by the absence of a phenotype other than survivorship, which depends on the measurement of age at death of individuals in a population. The use of survivorship to identify genetic and pharmacological interventions that prolong life is time-consuming and requires a large number of homogeneous animals. Here, we report the development of an assay in Drosophila melanogaster using the expression of molecular biomarkers that accelerates the ability to evaluate potential lifespan-altering interventions. Coupling the expression of an age-dependent molecular biomarker to a lethal toxin reduces the time needed to perform lifespan studies by 80%. The assay recapitulates the effect of the three best known environmental life-span-extending interventions in the fly: ambient temperature, reproductive status, and calorie reduction. Single gene mutations known to extend lifespan in the fly such as Indy and rpd3 also extend lifespan in this assay. We used this assay as a screen to identify drugs that extend lifespan in flies. Lipoic acid and resveratrol were identified as being beneficial in our assay and shown to extend lifespan under normal laboratory conditions. We propose that this assay can be used to screen pharmacological as well as genetic interventions more rapidly for positive effects on lifespan.
Keywords: aging, biomarker, lifespan extension, drug screen, resveratrol
Changes in reproductive and physical activity in insects and ambient temperature in poikilotherms have been shown to extend lifespan, but the only intervention to increase lifespan robustly in various species is calorie restriction (1, 2). Research into the causes of aging is confounded by the fact that aging is a multifaceted process that lacks a clear phenotype other than age at the time of death itself. The most commonly noted feature is lifespan, determined by survivorship curves that measure the age at death of individuals in a population. Furthermore, the aging process is influenced by various physiological systems that are not easy to separate experimentally (3). For example, both calorie restriction and virginity lead to an extension of lifespan, but caloric restriction also reduces fertility (4), thereby indicating a significant extent of cross talk between those different biological systems.
Advances in recent years have begun to greatly expand our understanding of the aging process and even have begun to shed light on the genes that mediate known lifespan-extending interventions. In the nematode Caenorhabditis elegans, >100 genes have been identified that extend lifespan. In the fruit fly, however, only a handful of genes have been shown to prolong life (2), and even a smaller number have been identified in mammals. Most of the genes identified so far in Drosophila melanogaster appear to affect the metabolic status of the organism by altering nutrient uptake (5), modulating gene expression through chromatin silencing (6) or altering insulin signaling (7, 8). Other physiological systems in which lifespan-extending genes have been identified involve response pathways to intracellular stressors like oxidative damage to macromolecules (9–11) or heat-shock proteins (12, 13). These advances, however, were achieved at a fairly slow pace. The need for survivorship analysis of a sufficiently large and genetically homogenous population to detect lifespan-changing interventions places a “speed limit” on discovery. Although this handicap is relatively unobtrusive in lower organisms such as yeast or nematodes, it becomes more of a problem in more complex model organisms with longer lifespans, such as mice or even D. melanogaster. Therefore, even the relatively short lifespan of the fruit fly (80 days) makes comprehensive investigation of aging difficult.
An alternate approach to assessing interventions that alter lifespan and aging is to use biomarkers that can track physiological age or predict lifespan expectancy relatively early in the life of the organism. Various physiological markers, such as reproduction, mobility, and behavioral tests, have been proposed as means of tracking physiological age in organisms as diverse as nematodes, insects, and mammals (14). Molecular markers such as the accumulation of oxidized macromolecules have also received attention as biomarkers of aging (15).
Recently, the realization that the regulation of gene expression remains a dynamic, well regulated process throughout adult life has led to the suggestion that this sensitive physiological measure may serve as a biomarker. In Drosophila it has been shown that the expression of a number of genes changes in a dynamic, yet characteristic pattern through adult life (16–22). For some of these genes, their temporal pattern of expression is altered in conjunction with interventions that alter lifespan. For example, when flies were raised under conditions that extend lifespan, the temporal pattern of gene expression was delayed. Conversely, the temporal pattern of expression was accelerated when flies were raised under lifespan-shortening conditions. The temporal pattern of expression for such genes may thus serve as a biomarker for tracking physiological age or predicting lifespan. Because the normal trajectory of expression of these genes changes early in adult life, when a lifespan-altering manipulation is imposed on the flies, it has been suggested that the temporal pattern of gene expression may be useful for identifying interventions that alter lifespan. For example, an alteration extending lifespan should correspondingly slow the timing of gene expression for these biomarkers. Identification of interventions that extend lifespan can be achieved by looking for a delay in biomarker profile. To identify such a delay in biomarker profile, a suitable reporter needs to be coupled to the gene-expression driver and read out visually or by means of a form of selection. Reporters such as lacZ, luciferase, and GFP can be used as a method for visual selection. However, the selection process can be expedited if, instead of having to visually score a change in biomarker profile, a lethal toxin is used as a reporter. In this case, expression of the toxin will kill all flies reaching a lethal toxin-level threshold at the normal or unaltered chronological age. Flies with delayed biomarker profile will reach that threshold later and, thus, can be readily identified as “survivors.”
Here, we report the generation of such a system by using an age-dependent biomarker coupled to the expression of the tetanus toxin light chain. This system measures changes in biomarker trajectory by killing off flies with normal trajectory earlier than flies with delayed trajectory. It recapitulates the lifespan-extending effect of lower ambient temperature, dietary restriction, and reduced fecundity. Furthermore, single gene mutations that have been demonstrated to extend lifespan also extend the survival times of flies in this rapid assay. We then used this system to rapidly screen for lifespan-extending drugs. Our results demonstrate that this system can be used as a screening tool in pharmacological and genetic assays for lifespan-extending interventions.
Materials and Methods
Fly Strains and Culture. All flies were kept in a humidified, temperature-controlled incubator with 12:12 h on/off light cycle at 25°C in vials containing standard cornmeal medium with the addition of a few grains of yeast. The tetanus toxin light chain expressing UAS-TNT-E strain was obtained from C. O'Kane (23), the diphtheria toxin-carrying strain UAS-DTI was obtained from R. Davis (24), and the age-dependent DJ enhancer trap lines were obtained from L. Seroude (22). Freshly isogenized Indy206 and Indy302 lines, their genetic control line 1085 (5), as well as the hypomorph rpd3P-UTR and the deficiency rpd3Def24 lines and their genetic control line rpd3P-1.8 (6) were used in the genetic experiments.
To test lethality effects of different driver/toxin combinations, each of the biomarker lines DJ634, DJ651, DJ656, and DJ694 was crossed individually to either UAS-TNT-E or UAS-DTI. For DJ651-driven tetanus toxin (DTT) survival assays, DJ651 flies were crossed to UAS-TNT-E flies. A fly line homozygous for both UAS-TNT-E and DJ651 was generated by standard breeding procedures. These double-DTT flies were then crossed to the lifespan-extending mutants.
DTT Survival-Time Analysis. For survival time analysis of DTT flies, newly eclosed flies were collected and housed at a density of 20–30 males and 20–30 females each per vial. At least 100 males and 100 females were tested for each treatment. Flies were collected without anesthesia because DTT flies exhibited high mortality under light CO2 anesthesia or even after collection on ice. Virgin males and females and their controls were collected individually without anesthesia. For studies involving ambient-temperature changes, after development at 25°C, adult flies from the same cohort were collected upon eclosion, divided randomly into two groups, and kept in a humidified 12:12 h on/off light-cycle incubator at 18°C or 25°C. For studies using calorie-restricted food, flies were kept on cornmeal food containing only 50% of the ingredients of standard cornmeal food and without addition of yeast. For drug treatments, drugs were added during the preparation of the standard cornmeal food after it had cooled down to 55°C. A few grains of yeast were added to the vials, except in the case of lipoic acid. In all survival-time studies, flies were passed to new vials every other day, and the number of dead flies was counted daily. To test lifespan-extending genetic interventions in the DTT assay, double-DTT males were crossed to female virgins of the experimental lines. Offspring were collected and assayed as described.
Lifespan Analysis. Canton-S flies were collected under light anesthesia and housed at a density of 25 males and 25 females each per vial without added yeast. At least 10 vials were used per treatment (total of 250 males and 250 female flies per lifespan). Flies were passed every other day, and the number of dead flies was recorded.
Drugs and Pharmaceuticals. All drugs were obtained from either Sigma or Calbiochem. Lipoic acid and resveratrol were obtained from Sigma and dissolved in 100% ethanol. Stock solutions of at least 1,000× were then used to prepare food containing the drugs. Up to 10% additional ethanol was tested as a solvent control and showed no effect in either the DTT or the lifespan assay.
Statistical Analysis. Log-rank tests of survivorship curves were performed by using prism (GraphPad, San Diego).
Results
Generation of a System Exhibiting Age-Dependent Toxicity. We aimed to develop a rapid screening tool that causes age-dependent lethality during the first few weeks of adult life to identify interventions that change lifespan. To do this, we used a series of age-dependent GAL4 enhancer-trap lines as biomarkers (22) and the tetanus toxin light chain and diphtheria toxin under control of UAS sequences as lethal reporters. The tetanus and diphtheria toxins have been reported to be able to kill D. melanogaster when expressed in minute amounts (23, 24). Although many of the GAL4 enhancer-trap lines show a down-regulation of GAL4 expression with age, several of them demonstrate rapid up-regulation after eclosion.
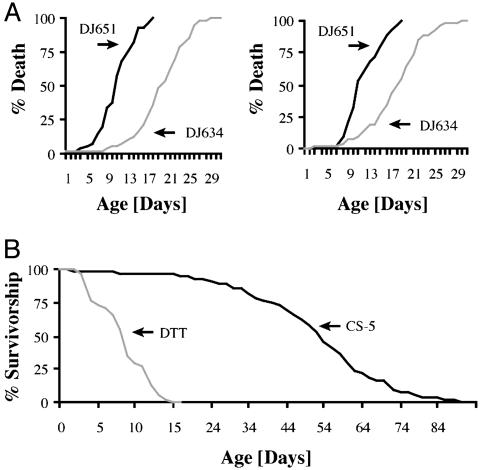
Four different GAL4 age-dependent biomarker driver lines specifically selected for an increased expression in GAL4 during the first few weeks of life were combined with either UAS-DTI (diphtheria) or UAS-TNT-E (tetanus), and offspring were assessed for age-dependent lethality. Despite high expression levels, as evidenced by a UAS-lacZ reporter (ref. 22 and data not shown), all four crosses with UAS-DTI failed to demonstrate any obvious lethality. In contrast, when coupled with UAS-TNT-E, two of the four lines showed age-dependent lethality, a third line showed no lethality, and the fourth line caused lethality during development. Of the two biomarker lines (DJ651 and DJ634) that showed age-dependent lethality with UAS-TNT-E, DJ651 exhibits the earlier adult lethality, with an average survival of 8 days and a maximal survival of 15 days. The other biomarker demonstrating age-dependent lethality (DJ634) had an 18-day average and 30-day maximal survivorship (Fig. 1A). The shorter survival times of the DJ651-driven flies expressing tetanus toxin led us to use this driver for most of our subsequent experiments. The lifespan of this combination is 20% of the lifespan of the normal wt Canton-S flies (Fig. 1B). The combination of DJ651 driver and UAS-TNT-E flies was termed the DTT system.
Fig. 1.
Death curves of two different biomarkers of aging coupled to tetanus toxin expression. Flies were kept at a density of 25 flies per sex and vial on lightly yeasted standard cornmeal food and passed every other day. Dead flies were counted daily. (A) DJ651-driven toxin expression leads to average survival times of 8 days for both males and females. Toxin expression is delayed in DJ634-driven flies, and therefore, average survival times increased to 18 days. (B) Compared with survivorship curves of Canton-S flies, DTT females live 80% shorter.
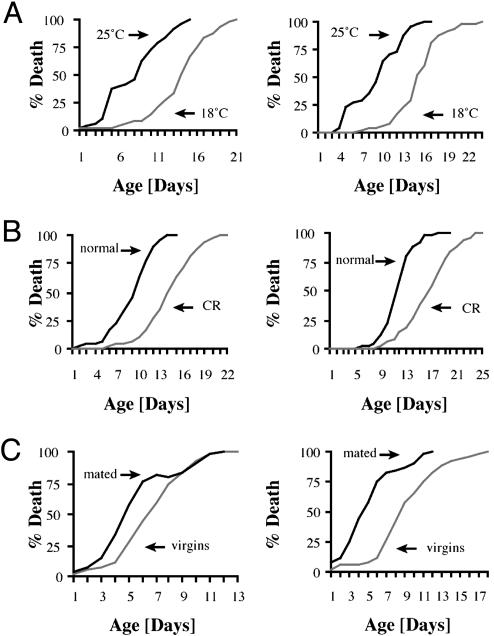
The DTT System Recapitulates the Effects of Known Environmental Interventions That Extend Lifespan. To determine whether the DTT system could be used to detect lifespan-extending interventions, we tested whether environmental interventions known to extend lifespan in normal flies also led to prolonged survival of DTT flies. The three environmental interventions that are best known to extend lifespan in flies are lowering ambient temperature, restricting reproduction, and reducing calories in the food (2).
The effect of decreasing ambient temperature on DTT survival. As shown in Fig. 2A, lowering the temperature to 18°C increased average survival times 74% for males and 63% for females as compared with DTT flies living at 25°C. This increase in survival is similar to, but slightly less than, what is seen when normal flies are cultured at 18°C versus 25°C (2).
Fig. 2.
Environmental interactions that increase lifespan also extend survival times of DTT flies. Both male (Left) and female (Right) flies live longer in the DTT assay when subjected to environmental interventions that are known to extend lifespan of D. melanogaster. Lower ambient temperature (A), reduced caloric intake (B), and virginity (C) extend survival times up to 74%.
The effect of decreasing calorie availability on DTT survival. Next, we tested whether dietary restriction would increase survival of the DTT flies. When DTT flies were grown on food containing 50% less calories than normal food, males lived 41% longer and females lived 43% longer (Fig. 2B). The percentage of lifespan extension seen with the DTT flies on low-calorie food is similar in magnitude to the extension seen for normal adults when placed on low-calorie food (6).
The effect of decreasing reproduction on DTT survival. We next investigated whether reproductive status influenced the survival of DTT flies. A decrease in reproduction, particularly in females, is known to extend lifespan significantly. For example, virgin females can live up to 100% longer than fully mated females, whereas virgin males live only 10–20% longer (25). By using the DTT system, we found that virgin females lived 74% longer than mated females, whereas virgin males lived 22% longer than mated males (Fig. 2C).
Thus, in all three cases, environmental manipulations that are known to extend the lifespan of normal adult flies cause the extension of survival times of DTT flies. Interestingly, the lifespan extension seen with the DTT flies also shows a similar magnitude as with normal flies. Furthermore, when mortality curves are compared, the effect on the slope and y-intercept of the mortality curve is similar in the DTT flies and wt (data not shown). Lower ambient temperature and calorie restriction primarily change the slope of the mortality curve in the DTT flies and Canton-S, whereas virginity typically changes the y-intercept (2).
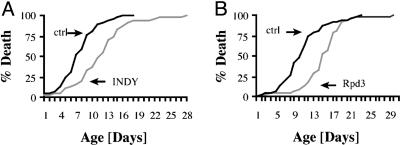
Lifespan-Extending Mutations Increase Survival Times of DTT Flies. In addition to environmental interventions, we also investigated whether genetic interventions that are known to extend lifespan in D. melanogaster could change the DJ651 biomarker trajectory and, therefore, the survival times of DTT flies. We examined two different lifespan-extending mutations, Indy and rpd3, both of which extend lifespan as heterozygotes (5, 6). A DTT line, homozygous for both the biomarker DJ651 GAL4 driver and UAS-TNT-E was created (double DTT) and then tested with Indy and rpd3.
The effect of the long-lived Indy mutant on DTT survival. The double-DTT line was crossed to flies carrying one of two different long-lived Indy alleles, Indy206 or Indy302. For controls, genetically matched fly lines that did not carry the lifespan-extending mutation were used (5). As shown in Fig. 3A, male Indy206/DTT flies lived 50% longer than control. Similar results were seen with the Indy302/DTT flies, although unexpectedly only a small increase in lifespan was seen with the Indy206/DTT or Indy302/DTT female flies (data not shown).
Fig. 3.
Long-lived mutants extend survival times of DTT flies. Homozygous DTT flies were crossed to the indicated stocks, and death curves of offspring were generated. (A) When crossed to freshly isogenized Indy mutants, males lived 50% longer than genetically matched control. (B) Male offspring of a cross to the hypomorph rpd3P-UTR mutant lived significantly longer than genetically matched control. Males lived 40% longer.
The effect of the long-lived rpd3 mutant on DTT survival. The double-DTT line was then crossed to flies carrying one of two different long-lived rpd3 alleles, rpd3P-UTR or rpd3def24. An additional control was the rpd3P-1.8 allele, a P-element insertion in the rpd3 locus, which is in the same genetic background as rpd3P-UTR and has an effect only on gene expression in the eye and has no effect on lifespan (6). Both the rpd3P-UTR and rpd3def24 alleles showed a significant increase in survival times for males (40%) and, as expected, only the rpd3P-UTR allele extended lifespan significantly in females (45%) (Fig. 3B, rpd3def24, and data not shown).
These studies demonstrate that the DTT system is capable of detecting genetic changes that extend lifespan. Together with the data described above on environmental interventions that extend lifespan, it suggests that the DTT system can be used to rapidly ascertain whether a given environmental, pharmaceutical, or genetic intervention might lead to an extended lifespan in normal flies.
The DTT System Can Be Used to More Rapidly Screen for Lifespan-Extending Drugs. Having demonstrated the ability of the DTT system to respond appropriately to several environmental and genetic lifespan-extending manipulations, we investigated the use of the DTT system for detecting pharmaceutical interventions that extend lifespan. Currently, most hypotheses about the aging process and lifespan involve damage to macromolecules and/or delayed clearance and repair of these lesions. Therefore, we tested several drugs that are assumed to modify these processes. Furthermore, we also tested drugs that are thought to influence the endoplasmic reticulum stress response, modulate Sir2 activity, or inhibit apoptosis. Quite often, it was necessary to test more than one concentration of a given drug because high doses of drugs were often associated with very early mortality in the DTT system as well as in normal flies. Because the rapid assay can make a predication about the efficacy of a treatment within 2 weeks, screening time is reduced substantially. This feature allowed us to screen >20 different drugs at different concentrations in only a few months. Some of the drugs that we tested are given in Table 1.
Table 1. Drugs tested in the DTT assay.
| Drug | Survival-time extension |
|---|---|
| Lipoic acid | |
| 0.5% | No |
| 0.05% | No |
| 0.005% | Yes |
| 0.001% | Yes |
| Geldanamycin | |
| 1 μM | No |
| 200 nM | No |
| 100 nM | No |
| Catechin | |
| 80 μM | No |
| EGCG | |
| 50 μM | No |
| Caffeine | |
| 0.04% | No |
| 0.004% | No |
| Tocopherol | |
| 1 mM | No |
| 100 μM | No |
| 4-PBA | |
| 5 mM | No |
| 2.5 mM | No |
| Resveratrol | |
| 500 μM | No |
| 200 μM | Yes |
| 100 μM | Yes |
| 50 μM | No |
| Sirtinol | |
| 100 μM | No |
| zVAD-fmk | |
| 10 μM | No |
| 5 μM | No |
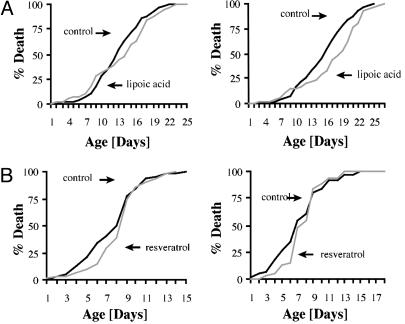
One class of drugs that we tested were drugs thought to effect the accumulation of oxidative damage. We observed that low doses of the antioxidant lipoic acid led to an increase in survival time of DTT flies. Females raised on 0.005% lipoic acid lived 12% longer, whereas males lived 4% longer (Fig. 4A). This effect was lost when higher doses were used (data not shown). Other antioxidants, such as tocopherol or catechin, did not demonstrate any beneficial effect at the tested concentrations (data not shown).
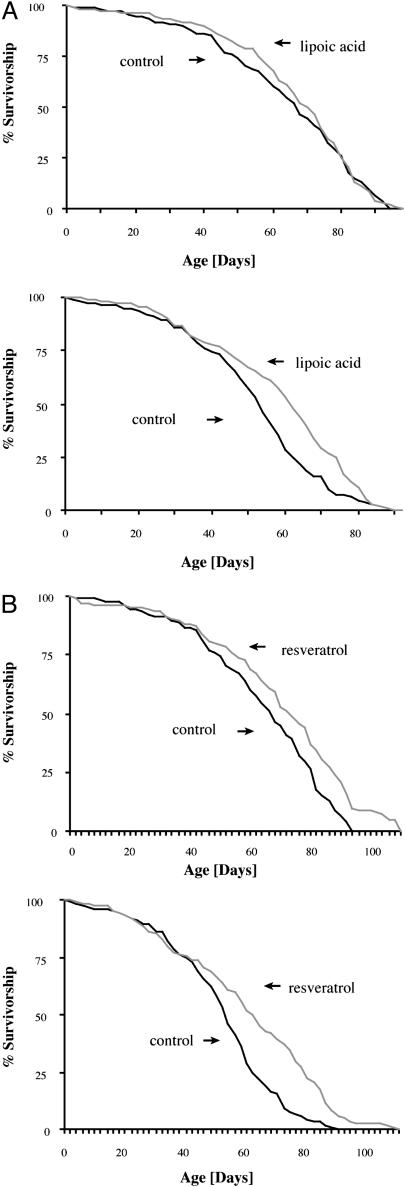
Fig. 4.
The DTT system can be used as a drug-discovery tool. (A) Culturing DTT flies on food containing 0.005% lipoic acid increases the average survival times of males (Left) by 4% and of females (Right) by 12%. No yeast was added because the yeast/lipoic acid combination at any concentration was toxic. (B) Food containing 200 μM polyphenol resveratrol increased the average survival times of males (Left) by 9% and of females (Right) by 8%.
Another drug that we tested that had a positive effect was resveratrol, a component of red wine, which recently has been shown to extend mother-cell lifespan in yeast (26). The mechanisms of this effect are unclear because resveratrol has been shown to function both as an antioxidant and as an activator of the histone deacetylase Sir2 (27). When DTT flies were raised on food containing 200 μM resveratrol, males showed a 9% increase in average survival time and females showed an 8% increase (Fig. 4B) over untreated controls.
Drug-Induced Survival-Time Increases in the DTT System Are Reflected in Lifespan Increases of Normal Flies. The survival-time increases that we observed in our drug treatments were smaller than those seen with the environmental or genetic interventions. Therefore, we investigated whether treatment of normal flies with these drugs would lead to lifespan extension in a normal lifespan assay. When Canton-S flies were cultured on food containing 0.005% lipoic acid from the day of eclosion, there was a high rate of mortality during the first 10 days of adult life and a shortening in lifespan (data not shown). Examination of the survivorship and mortality curves, however, showed that after the first 10 days, there was a dramatic decrease in the rate of mortality. This observation prompted us to repeat the experiments, this time by starting the administration of lipoic acid on day 10 after eclosion. By using this regimen, female flies on lipoic acid lived 12% longer and males lived 4% longer than controls (Fig. 5A).
Fig. 5.
Drugs extending survival times of DTT flies also extend lifespan of Canton-S flies. Canton-S flies were kept on food without yeast according to the conditions established in Fig. 4, and survivorship curves were generated. (A) The 0.005% lipoic acid increased average and median lifespan of males (Upper) by 4% and 6% (P = 0.5498), respectively, and average, median, and maximum lifespan of females (Lower) by 12%, 15%, and 5% (P = 0.0025), respectively, which were the same as increases seen in DTT flies. (B) Average, median, and maximum lifespan extension by 16%, 13%, and 15%, respectively, in females (Lower)(P < 0.0001) and 10%, 10%, and 18%, respectively, in males (Upper)(P < 0.0001) is observed when flies are fed 200 μM resveratrol.
Finally, we tested whether resveratrol would increase the lifespan of Canton-S flies. Canton-S flies cultured on food containing 200 μM resveratrol showed an increase in average lifespan of 17% for females and 10% for males and an increase in maximum lifespan of 22% and 20% for females and males, respectively (Fig. 5B).
Discussion
The recent renaissance in aging research has largely been spurred by the discovery of a plethora of single gene mutations that extend lifespan in the model system C. elegans. In addition to stimulating great interest in the molecular mechanisms of aging, these studies have provided evidence for conservation of physiological systems associated with longevity determination across distant species barriers. Although the information from C. elegans is of great value and has provided insight into those pathways regulating lifespan, there remains a need for identifying both genetic and pharmacological interventions that regulate longevity in more complex organisms such as flies and mammals.
Unfortunately, in higher organisms, research into the molecular mechanisms of lifespan determination and aging has been slowed by practical problems associated with performing survivorship analyses, such as length of time and number of animals. Because of the stochastic nature of the survivorship assay, the necessity of having a large enough homogeneous cohort of synchronously aged animals makes using survivorship studies as a general screen for lifespan-extending interventions problematic. Survivorship assays could be used for screening drugs, for which a large cohort of normal flies could be generated, but genetic screens requiring a cohort of genetically identical (F2) individuals is impractical. The handful of pharmacologic and genetic interventions that have been identified to extend lifespan in D. melanogaster indicate that the dearth of information in comparison with the nematode is likely due to these logistical and procedural problems and is not of a fundamental nature (5, 28, 29). Methods accelerating the screening of genetic and pharmacological agents for lifespan-extending affects are needed.
Here, we report the development of an assay that shortens the time it takes to identify potential life-extending interventions in D. melanogaster. The DTT system shortens the time that it takes to select or enrich for lifespan-extending manipulations by combining an age-dependent GAL4 driver (biomarker) that tracks lifespan and a threshold-dependent lethal toxin that selects for lifespan extension. Thus, it is possible to select against a population or individual whose biomarker trajectory is unchanged from normal and to select for a population or individual on a slower biomarker trajectory. This system shortens screening time by ≈80%, from several months to 2 weeks, thereby greatly facilitating drug or gene discovery. Different age-dependent biomarker drivers have different survival times in this system, yet we found that they behave similarly with respect to lifespan-extending interventions (driver DJ634; data not shown). The DTT system faithfully reproduces the effects of known environmental changes (such as ambient temperature, dietary restriction, and reproductive status) and genetic mutations (such as rpd3 and Indy) on lifespan, as indicated by the prolonged survival times of DTT flies that were subjected to these lifespan-extending interventions. Importantly, the percentage of extension in lifespan seen in the DTT assay is very similar to that seen in normal lifespan studies.
Chemical or genetic interventions that slow the biomarker trajectory and allow the flies to live longer in the DTT assay are candidates for lifespan extension in normal flies. By using different biomarkers associated with genes from different physiological systems, an intervention found to change the trajectory for a particular biomarker could be confirmed as a likely lifespan-extending alteration by using a second biomarker, before having to perform a standard survival analysis.
The DTT system can be used to search for drugs that extend lifespan. In the initial drug screens presented in this article, we were able to identify two drugs that led to average lifespan extension of up to 16% from only 20 different drugs tested. Lipoic acid has previously been shown to improve memory function in old rats, where it also acts to prevent age associated mitochondrial decay and reduces oxidative damage. This effect was even more pronounced when carnitine was added (30–32). We initially found that addition of carnitine proved to be toxic to the DTT flies (data not shown), but more careful analysis is needed to determine whether lower carnitine doses may lead to beneficial effects. At the doses that we used, lipoic acid modestly extended lifespan of female D. melanogaster. Further studies need to be done to clarify the effect of lipoic acid on males. These data confirm and expand the data on lipoic acid treatment in rats, demonstrating that this compound bestows beneficial effects on health or lifespan across species.
The other drug that tested positive for increased survival time in our initial drug screen was resveratrol. Resveratrol has been shown to extend the reproductive lifespan of the budding yeast Saccharomyces cerevisiae (26). Although it is known to exert pleiotropic effects (27), the lifespan-extending effect of resveratrol is thought to be mediated by means of the activation of the histone deacetylase Sir2 (26). Sir2 has been implicated in lifespan extension of yeast, nematodes, and fruit flies (6, 33–35), and it has been proposed to mediate some of the effects of calorie restriction (36). Consistent with the importance of Sir2 activity in lifespan determination, our data showed that treatment of DTT flies with the Sir2 inhibitor sirtinol leads to shortening of lifespan (Table 1 and data not shown).
Although drugs are critical tools for identifying aging pathways, another method is to identify the genes involved in lifespan extension directly. Traditionally, the most powerful and systematic approach with which to investigate the genes involved in complex biological phenomena like aging or developmental processes has been to perform large-scale mutageneses. As noted above, the logistical problems of screening a mutagenesis by using survivorship are daunting. In addition to serving as a rapid tool for drug discovery, the DTT system would appear to be particularly useful as a means of enriching for potential lifespan-extending genetic alterations. The similarity of the percentages of lifespan increases seen with the DTT assay and with normal survivorship curves when lifespan was increased through environmental, long-lived genetic mutations or drugs is striking. The fact that an increase in lifespan was in no case seen in the DTT assay and not in the normal survivorship assay is of additional benefit. This function, coupled with the ability of the DTT assay to identify alterations that could increase lifespan by as little as 10%, suggests that the DTT screen is a sensitive assay with a low false-positive rate. A low false-positive rate is particularly critical when screening for genes that extend lifespan because the normal survivorship studies that are necessary for confirming any potential lifespan-extending mutations are very time-consuming. Therefore, in addition to serving as a tool for drug discovery, the DTT system should be valuable in a standard mutagenesis screen by providing a rapid method for enriching for potential lifespan-extending alterations.
Acknowledgments
We thank Robert Reenan (University of Connecticut Health Center) for suggesting the use of toxins as a selection system; C. O'Kane (University of Cambridge, Cambridge, United Kingdom), R. Davis (Baylor College of Medicine, Houston), and B. Rogina (University of Connecticut Health Center) for fly stocks; and, particularly, Laurent Seroude (Queen's University, Kingston, ON, Canada) for generously sharing his GAL4 P-element lines. We also thank Suzanne Kowalski and Dianna Schwarz for technical assistance. This work was supported by National Institute on Aging Grant AG16667, as well as grants from the Donaghue Foundation and the Ellison Medical Foundation (to S.L.H.). S.L.H. is an Ellison Medical Research Foundation Senior Investigator and a member of the Scientific Advisory Board of Elixir Pharmaceuticals, Inc. (Cambridge, MA).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: DTT, DJ651-driven tetanus toxin.
References
- 1.Masoro, E. J. (2003) Sci. Aging Knowledge Environ. 8, RE2. [DOI] [PubMed] [Google Scholar]
- 2.Helfand, S. L. & Rogina, B. (2003) Adv. Genet. 49, 67–109. [DOI] [PubMed] [Google Scholar]
- 3.Guarente, L. & Kenyon, C. (2000) Nature 408, 255–262. [DOI] [PubMed] [Google Scholar]
- 4.Kirkwood, T. B. & Austad, S. N. (2000) Nature 408, 233–238. [DOI] [PubMed] [Google Scholar]
- 5.Rogina, B., Reenan, R. A., Nilsen, S. P. & Helfand, S. L. (2000) Science 290, 2137–2140. [DOI] [PubMed] [Google Scholar]
- 6.Rogina, B., Helfand, S. L. & Frankel, S. (2002) Science 298, 1745. [DOI] [PubMed] [Google Scholar]
- 7.Clancy, D. J., Gems, D., Harshman, L. G., Oldham, S., Stocker, H., Hafen, E., Leevers, S. J. & Partridge, L. (2001) Science 292, 104–106. [DOI] [PubMed] [Google Scholar]
- 8.Tatar, M., Kopelman, A., Epstein, D., Tu, M. P., Yin, C. M. & Garofalo, R. S. (2001) Science 292, 107–110. [DOI] [PubMed] [Google Scholar]
- 9.Orr, W. C. & Sohal, R. S. (1994) Science 263, 1128–1130. [DOI] [PubMed] [Google Scholar]
- 10.Sun, J. & Tower, J. (1999) Mol. Cell. Biol. 19, 216–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun, J., Folk, D., Bradley, T. J. & Tower, J. (2002) Genetics 161, 661–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tatar, M., Khazaeli, A. A. & Curtsinger, J. W. (1997) Nature 390, 30. [DOI] [PubMed] [Google Scholar]
- 13.Morrow, G., Samson, M., Michaud, S. & Tanguay, R. M. (2004) FASEB. J. 18, 598–599. [DOI] [PubMed] [Google Scholar]
- 14.Johnson, T. E., Conley, W. L. & Keller, M. L. (1988) Exp. Gerontol. 23, 281–295. [DOI] [PubMed] [Google Scholar]
- 15.Rao, G., Xia, E., Nadakavukaren, M. J. & Richardson, A. (1990) J. Nutr. 120, 602–609. [DOI] [PubMed] [Google Scholar]
- 16.Helfand, S. L., Blake, K. J., Rogina, B., Stracks, M. D., Centurion, A. & Naprta, B. (1995) Genetics 140, 549–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rogina, B. & Helfand, S. L. (1995) Genetics 141, 1043–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rogina, B., Benzer, S. & Helfand, S. L. (1997) Proc. Natl. Acad. Sci. USA 94, 6303–6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rogina, B. & Helfand, S. L. (1997) Mech. Dev. 63, 89–97. [DOI] [PubMed] [Google Scholar]
- 20.Rogina, B., Vaupel, J. W., Partridge, L. & Helfand, S. L. (1998) Curr. Biol. 8, 475–478. [DOI] [PubMed] [Google Scholar]
- 21.Pletcher, S. D., Macdonald, S. J., Marguerie, R., Certa, U., Stearns, S. C., Goldstein, D. B. & Partridge, L. (2002) Curr. Biol. 12, 712–723. [DOI] [PubMed] [Google Scholar]
- 22.Seroude, L., Brummel, T., Kapahi, P. & Benzer, S. (2002) Aging Cell 1, 47–56. [DOI] [PubMed] [Google Scholar]
- 23.Sweeney, S. T., Broadie, K., Keane, J., Niemann, H. & O'Kane, C. J. (1995) Neuron 14, 341–351. [DOI] [PubMed] [Google Scholar]
- 24.Roman, G., Endo, K., Zong, L. & Davis, R. L. (2001) Proc. Natl. Acad. Sci. USA 98, 12602–12607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Partridge, L. & Prowse, N. (1997) J. Insect Physiol. 43, 501–512. [DOI] [PubMed] [Google Scholar]
- 26.Howitz, K. T., Bitterman, K. J., Cohen, H. Y., Lamming, D. W., Lavu, S., Wood, J. G., Zipkin, R. E., Chung, P., Kisielewski, A., Zhang, L. L., et al. (2003) Nature 425, 191–196. [DOI] [PubMed] [Google Scholar]
- 27.Pervaiz, S. (2003) FASEB. J. 17, 1975–1985. [DOI] [PubMed] [Google Scholar]
- 28.Kang, H. L., Benzer, S. & Min, K. T. (2002) Proc. Natl. Acad. Sci. USA 99, 838–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin, Y. J., Seroude, L. & Benzer, S. (1998) Science 282, 943–946. [DOI] [PubMed] [Google Scholar]
- 30.Liu, J., Head, E., Gharib, A. M., Yuan, W., Ingersoll, R. T., Hagen, T. M., Cotman, C. W. & Ames, B. N. (2002) Proc. Natl. Acad. Sci. USA 99, 2356–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hagen, T. M., Liu, J., Lykkesfeldt, J., Wehr, C. M., Ingersoll, R. T., Vinarsky, V., Bartholomew, J. C. & Ames, B. N. (2002) Proc. Natl. Acad. Sci. USA 99, 1870–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hagen, T. M., Ingersoll, R. T., Lykkesfeldt, J., Liu, J., Wehr, C. M., Vinarsky, V., Bartholomew, J. C. & Ames, A. B. (1999) FASEB. J. 13, 411–418. [DOI] [PubMed] [Google Scholar]
- 33.Kaeberlein, M., McVey, M. & Guarente, L. (1999) Genes Dev. 13, 2570–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim, S., Benguria, A., Lai, C. Y. & Jazwinski, S. M. (1999) Mol. Biol. Cell 10, 3125–3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tissenbaum, H. A. & Guarente, L. (2001) Nature 410, 227–230. [DOI] [PubMed] [Google Scholar]
- 36.Lin, S. J., Defossez, P. A. & Guarente, L. (2000) Science 289, 2126–2128. [DOI] [PubMed] [Google Scholar]