Abstract
CCAAT enhancer-binding protein β (C/EBPβ), a basic-leucine zipper transcription factor, is an important effector of signals in physiologic growth and cancer. The identification of direct C/EBPβ targets in vivo has been limited by functional compensation by other C/EBP family proteins and the low stringency of the consensus sequence. Here we use the combined power of expression profiling and high-throughput chromatin immunoprecipitation to identify direct and biologically relevant targets of C/EBPβ. We identified 25 potential C/EBPβ targets, of which 88% of those tested were confirmed as in vivo C/EBPβ-binding sites. Six of these genes also displayed differential expression in C/EBPβ–/– livers. Computational analysis revealed that bona fide C/EBPβ target genes can be distinguished by the presence of binding motifs for specific additional transcription factors in the vicinity of the C/EBPβ site. This approach is generally applicable to the discovery of direct, biologically relevant targets of mammalian transcription factors.
CCAAT enhancer-binding proteins (C/EBPs) constitute a family of basic-leucine zipper (bZIP) transcription factors that are critical for the regulation of numerous biological processes, including differentiation, metabolic homeostasis, proliferation, tumorigenesis, inflammation, and apoptosis (1–9). C/EBP proteins are regulated at multiple levels, including gene transcription, translation, and phosphorylation, in response to a variety of stimuli including hormonal, cytokine and growth factor-signaling pathways (1). C/EBP proteins are able to form hetero- and homodimeric complexes with other C/EBP family members, thereby creating additional diversity in target sequence recognition.
C/EBPβ is an important effector of growth signals in experimental models of physiologic and neoplastic growth, the acutephase response, and metabolic homeostasis (1–11). Livers from C/EBPβ–/– mice exhibit a blunted regenerative response associated with prolonged hypoglycemia and altered expression of several cell-cycle-associated genes (10). In addition, a recent microarray analysis of human tumors has implicated C/EBPβ as a downstream mediator of cyclin D (12). Although these studies provide strong support for the role of C/EBPβ as a regulator of cell growth, at present, neither the mechanism by which C/EBPβ modulates the growth effects of cyclin D1 nor the targets of C/EBPβ in this pathway have been elucidated.
A variety of approaches has been used to identify C/EBPβ-binding sites, including cell culture systems, C/EBPβ–/– mice, and analyses of promoter sequences. However, several obstacles have limited the identification of direct C/EBPβ-dependent transcriptional targets in vivo.AllC/EBP family members with the exception of C/EBPξ possess identical in vitro DNA-binding affinity for C/EBP consensus sequences, suggesting that other C/EBP family members may be able to compensate for the loss of C/EBPβ (13). Second, the application of computational sequence analysis to identify C/EBP promoter sequences has been impeded by the fact that significant variations from the optimal C/EBP-binding sequence are tolerated, limiting the discriminative power of the C/EBP consensus sequence. Furthermore, the ability of C/EBPβ to heterodimerize with other basic-leucine zipper (bZIP) and non-bZIP transcription factors is associated with alterations in transactivation and DNA-binding specificity that may not be predicted based on consensus C/EBP-binding sequences (14–17). Here, we leverage the combined power of expression profiling and high-throughput chromatin immunoprecipitation (ChIP) to characterize direct and biologically relevant targets of C/EBPβ.
Materials and Methods
Animals. The derivation of mice homozygous for the C/EBPβ null mutation has been described (2). Mouse partial hepatectomies were performed as described (10).
ChIP. Mouse liver was minced in cold PBS and passed through a 21-gauge needle. The minced tissue was crosslinked in 1% formaldehyde/PBS for 15 min with constant shaking. Crosslinking was quenched by the addition of glycine to a final concentration of 0.125 M with constant shaking for an additional 5 min. The tissue was rinsed in PBS and homogenized with a Dounce homogenizer. The cells were pelleted by centrifugation at 13,000 × g for 5 min, resuspended in cell lysis buffer (5 mM Pipes, pH 8.0/85 mM KCl/0.5% Nonidet P-40/10 μM aprotinin/10 μM leupeptin/1mM PMSF), and incubated on ice for 15 min. Nuclei were pelleted by centrifugation at 13,000 × g for 5 min, resuspended in nuclear lysis buffer (50 mM Tris·Cl, pH 8.1/10 mM EDTA/1% SDS/10 μM aprotinin/10 μM leupeptin/1 mM PMSF), and incubated on ice for 10 min. The lysate was divided into 500-μl aliquots and sonicated by using the Sonic Dismembrator Model 100 (Fisher Scientific) with a microtip probe set to a power output of 4–6 W for three cycles of 20 s each. Insoluble debris was removed by centrifugation and the supernatant was collected and flash frozen in liquid nitrogen. Quantitative PCR was performed on the input DNA fractions to ensure that equal amounts of chromatin DNA were used in all immunoprecipitations. Immunoprecipitations were performed as described (18). Enrichment of the target gene phosphoenolpyruvate carboxykinase (PEPCK) relative to the 28S rRNA gene was calculated as follows: enrichment = 2^[(28SChIP – PEPCKChIP) – (28SInput – PEPCKInput)]. Potential C/EBPβ-binding sites in candidate target genes were identified by searching the region represented on the mouse promoter microarray plus 500 bp on either end, by using the transcriptional element search system (TESS) program (19). Primer sequences are available on request.
Mouse Promoter Microarray. The mouse promoter microarray was constructed by intersecting the Refseq gene collection with genes expressed in fetal and adult pancreas and liver. The first nucleotide of the Golden Path BLAT alignment of the Refseq sequence onto the mouse genome was taken as the transcriptional start site (TSS). primer3 was used to design PCR primers amplifying ≈1 kb of genomic promoter sequence in the region of (–2,000, +200) the TSS, including the TSS when possible (20). These primers were used to generate PCR products from CD1 mouse genomic DNA. All the PCR products were purified by using the Qiaquick PCR kit (Qiagen, Valencia. CA), eluted with deionized sterile water, diluted with an equal volume of DMSO (Sigma), and printed on poly-l-lysine-coated slides with a Biorobotics Microgrid II (Genomic Solutions, Ann Arbor, MI).
Genome-Wide Location Analysis. Amplification and labeling of immunoprecipitated DNA were performed as described (21) with modifications as noted below. Purified DNA obtained from ChIP was treated with T4 DNA polymerase to generate blunt ends and was then ligated to the annealed linker and amplified by PCR. After amplification, ≈1 μg of amplified ChIP DNA was labeled for each hybridization. The PCR products were purified by using the QIA-quick PCR purification kit (Qiagen). Labeling was performed with Ready-To-Go labeling beads (Amersham Pharmacia) according to the manufacturer's instructions. Each 50-μl reaction included 5 μl of Cy3- or Cy5-dCTP (Amersham Pharmacia). Each hybridization comprised a nonimmune IgG ChIP labeled with Cy3 and a C/EBPβ-specific ChIP labeled with Cy5. The labeling reactions for each slide were pooled and purified by using the Minelute PCR purification kit (Qiagen). Hybridization, washing, and scanning were performed as described below. Genes with a difference in intensity between wild-type and C/EBPβ–/– not exceeding the background intensity were excluded. All slides were normalized globally to the same average intensity. Genes having a ratio of wild-type/C/EBPβ–/– signal ≥1.2 were included in the combined location and expression microarray analyses.
Microarray Analysis of Gene Expression. Liver RNA was isolated from four wild-type and four C/EBPβ–/– mice at 0, 2, and 16 h after partial hepatectomy. Twenty micrograms of RNA for each sample was prepared by guanidine isothiocyanate and cesium chloride ultracentrifugation. In addition, 200 μg of RNA containing a pool of both wild-type and C/EBPβ–/– RNA was used as a common control in each hybridization. All samples were analyzed by using a Bioanalyzer Nano 6000 (Agilent Technologies, Palo Alto, CA) to determine the integrity and the concentration of the samples. RNA was labeled with amino-allyl dUTP and d(T)21 to prime reverse transcription. The fluorescent label was coupled to the cDNA and the cDNA was hybridized to pancchip v. 5.0 (22, 23). The median intensities of each spot were measured by an Agilent scanner with the genepix software. These data were normalized by using print tip lowess normalization, and the ratio of specific intensity to common control intensity was used in all further comparisons. For the combined location and expression microarray analyses, we included genes with altered expression (>50% change in C/EBPβ–/– mutant liver versus wild type) in quiescent liver and genes with abnormal induction after hepatectomy. The latter were defined as those genes showing a 50% change in expression level at either 2 or 16 h after partial hepatectomy in the wild-type mice but a <25% change at the corresponding time point in the C/EBPβ–/– mice.
Quantitative Real-Time PCR. PCR reaction mixes were assembled by using the Brilliant SYBR Green QPCR Master Mix (Stratagene), 10 μM primers, and the included reference dye at a 1:200 dilution according to manufacturer's instructions. Reactions were performed with the SYBR Green (with dissociation curve) program on the Mx4000 Multiplex Quantitative PCR System (Stratagene). Cycling parameters were 95°C for 10 min and then 40 cycles of 95°C (30 s), 58°C or 60°C (1 min), and 72°C (30 s) followed by a melting-curve analysis. All reactions were performed with two to six biological replicates and three technical replicates with reference dye normalization. The median cycle threshold value was used for analysis. Primer sequences are available on request.
DNA-Binding Site Detection. All 674 position-sensitive scoring matrices (PSSM) in transfac v7.3 were scored against a training set of the 15 ChIP-confirmed C/EBP target genes and 138 negative control genes. The negative control genes were selected on the basis of a lack of binding to the mouse promoter microarray in our study and a lack of dependence on C/EBPβ for mRNA expression based on our pancchip 5.0 microarray experiment. Sites that scored a log-likelihood ratio score of at least 5.0 were considered. This threshold was adjusted upward if needed to ensure that no more than three sites were predicted on average for each promoter. For each PSSM, we created a receiver operating characteristics (ROC) graph by counting the fraction of genes in each group that contained a site with a score at least as high as a threshold that ranged between the minimum and maximum observed scores. We then computed the area under the ROC graph (AUC), which can range from 0 to 1; a higher number indicates that the PSSM has a stronger preference for the C/EBPβ-binding amplicons. PSSMs were ranked by their AUC score, and those that are not expressed in the adult mouse liver (based on the gene expression database GXD) were excluded. We retained the top three PSSMs (M00624/DBP_Q6, M00258/ISRE_01, and M00763/PPAR_DR1_01) and the C/EBP PSSM (M00109, CEBPB_02), and computed ROC graphs for all pairs and sets of three or four PSSMs. In all cases, we allowed the PSSMs to occur in any order and orientation. We computed the ROC graph by varying the individual score thresholds and the maximum allowed distance from the 5′ most to the 3′ most PSSM in the group. We noted the scoring and distance thresholds for the point on the ROC curve that yielded the largest difference of positives between the 15 C/EBPβ target genes (true positives, TP) and the negative control set. We then counted the number of the 2,122 genes represented on the promoter chip that contained a PSSM set that passed these thresholds (chip positives, CP). Using the hypergeometric distribution, we computed the P value of seeing at least TP matches when drawing 15 samples from a population of 2,122 that contains CP-marked instances. We compensated for multiple testing effects by multiplying each P value by the number of groups (with repetition) of two, three, or four PSSMs that include at least one of the eight PSSMs for C/EBP.
Results
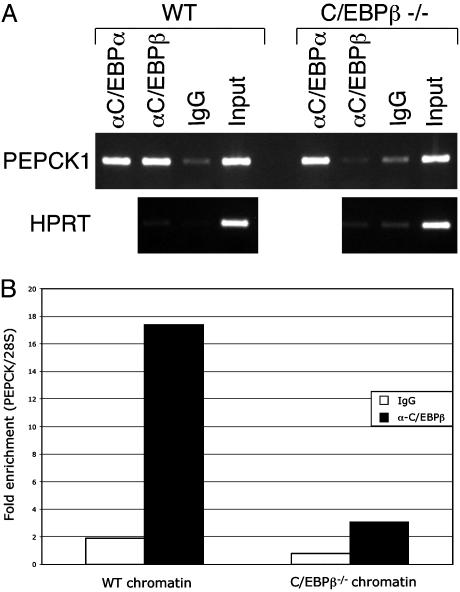
To identify target genes regulated by C/EBPβ in normal liver and during the process of liver regeneration, we performed an orthogonal analysis by combining genome-wide location analysis and mRNA expression profiling. A prerequisite for the identification of these target genes is the detection C/EBPβ DNA-binding activity in mouse liver, as cultured cells cannot reproduce the state of hepatocytes responding to a growth stimulus. We confirmed the binding of C/EBPβ to its well characterized binding site in the PEPCK gene by using chromatin prepared from adult mouse liver and immunoprecipitated with an antibody specific for C/EBPβ (Fig. 1A). To quantify the degree of C/EBPβ target sequence enrichment, we performed real-time PCR of the immunoprecipitated material (Fig. 1B). The C/EBPβ antibody provided a 17.4-fold enrichment of PEPCK DNA from wild-type liver chromatin, whereas minimal enrichment was obtained when a nonimmune IgG was used. To further confirm the specificity of the C/EBPβ antibody, we performed ChIP using chromatin obtained from C/EBPβ–/– liver chromatin. The 3.1-fold enrichment of PEPCK DNA observed (Fig. 1) indicates that the antibody used has a slight degree of nonspecific binding activity, either to other C/EBP family members or to other factors. For this reason, use of the mutant chromatin as a control throughout the location analysis is crucial (see below), because this results in enhanced specificity for bona fide C/EBPβ-bound DNA sequences.
Fig. 1.
ChIP of C/EBPβ bound to the PEPCK gene in mouse liver. Crosslinked chromatin prepared from wild-type or C/EBPβ–/– mouse liver was immunoprecipitated with anti-C/EBPα, anti-C/EBPβ, or nonimmune IgG as indicated. (A) Input chromatin and precipitated material were amplified with primers specific for PEPCK or the negative control gene HPRT. (B) Quantitative realtime PCR analysis of ChIP performed by using C/EBPβ or nonimmune antibody. Enrichment of the target gene PEPCK was calculated by using the 28S rRNA gene as a nonspecific control.
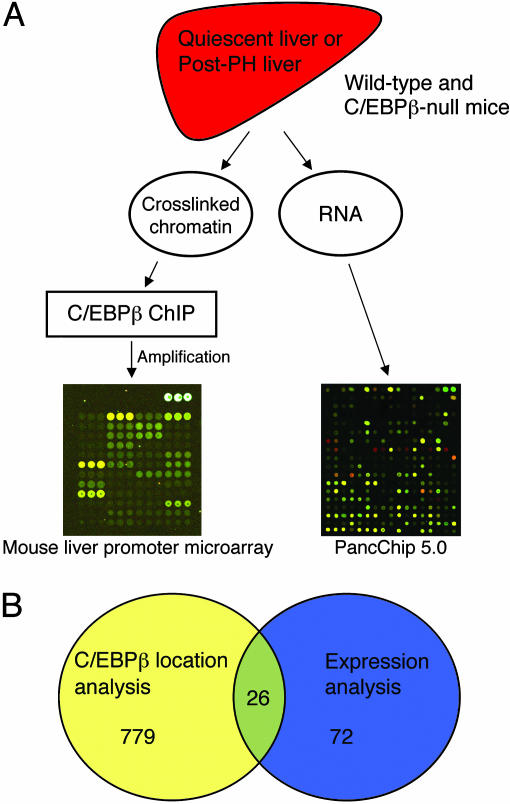
Orthogonal Analysis of C/EBPβ Target Genes. We proceeded to identify genes that are both bound by C/EBPβ and depend on C/EBPβ for proper induction during liver regeneration (Fig. 2A). Toward this end, we manufactured a mouse promoter microarray by PCR amplification of ≈1 kb of genomic sequence in the immediate vicinity of the promoters of 2,122 genes expressed in the liver. Using this microarray, we performed location analysis with chromatin obtained from quiescent adult mouse liver and chromatin obtained 2 h after partial hepatectomy. Livers from mice homozygous for a null mutation of the C/EBPβ gene were used as a negative control throughout for antibody specificity. Purified DNA obtained from ChIP by using the C/EBPβ antibody was amplified by ligation-mediated PCR, fluorescently labeled, and hybridized to the mouse promoter microarray. Genes with at least 20% greater signal intensity in the wild-type versus C/EBPβ–/– samples were considered positive. This low stringency was chosen to maximize the sensitivity of the location analysis; specificity was enhanced by applying the additional screen provided by expression profiling (Fig. 2B). Furthermore, we have noted that amplification, labeling, and microarray hybridization result in a compression of enrichments when compared with quantitative PCR of unamplified ChIP samples (P.P.L., J.R.F., and K.H.K., unpublished results). For the expression profiling, partial hepatectomies were performed on four wild-type and four C/EBPβ–/– mutant mice, and liver RNA was extracted at 0, 2, and 16 h after partial hepatectomy. This RNA was reverse-transcribed, fluorescently labeled, and hybridized to the pancchip 5.0 cDNA microarray (22). (Complete results of the pancchip 5.0 and mouse promoter microarray hybridizations are listed in Tables 3 and 4, which are published as supporting information on the PNAS web site.) We focused our analysis on genes that require C/EBPβ for proper expression in quiescent liver (i.e., those having a >50% change in C/EBPβ–/– mutant liver versus wild-type) and genes with abnormal induction after hepatectomy. The latter were defined as those genes showing a 50% change in expression level at either 2 or 16 h after partial hepatectomy in the wild-type mice but a <25% change at the corresponding time point in the C/EBPβ–/– mice. Direct C/EBPβ target genes in quiescent and regenerating liver identified by this combined screening method are listed in Table 1. Three of these [Aldh1a1 (24), Pepck (25, 26), and Saa1 (27–29)] have been reported to be targets of C/EBPβ, providing support for the utility of our orthogonal analysis.
Fig. 2.
Combining location and expression microarray analysis. (A) Schematic representation of the experimental approach. Partial hepatectomies were performed on wild-type and C/EBPβ–/– mice. RNA and crosslinked chromatin were prepared at 0, 2, and 16 (RNA only) h after partial hepatectomy. ChIP was performed by using an antibody specific for C/EBPβ, and the resulting material was amplified by ligation-mediated PCR (see Materials and Methods), fluorescently labeled, and hybridized to the mouse promoter microarray. The RNA was reverse-transcribed and the cDNA was labeled and hybridized to pancchip 5.0.(B) Candidate genes directly regulated by C/EBPβ after partial hepatectomy were defined as those within the intersection of the set of genes identified by location analysis with those genes with altered expression in quiescent or post-partial hepatectomy (Post-PH) liver (see Materials and Methods).
Table 1. Candidate direct C/EBPβ target genes in quiescent and/or regenerating liver.
| Expression analysis
|
Location analysis
|
||||
|---|---|---|---|---|---|
| Gene name | Function | Quiescent liver | Post-PH, 2 h, 16 h | Quiescent liver | Post-PH, 2 h |
| Aldh 1a1 (aldehyde dehydrogenase 1A1) | Retinoic acid synthesis, xenobiotic transformation | -2.5 | -2.3, -2.1 | 8.5 | 1.1 |
| Anxa5 (Anx5, annexin V) | Unknown | 1.5 | 1.9, -1.1 | 6.7 | 1.0 |
| Bcap37 (B cell receptor-associated protein 37) | Signal transduction | 1.4 | -1.1, 1.4 | 1.3 | 1.2 |
| Car3 (carbonic anhydrase 3) | CO2/bicarbonate conversion | -2.5 | -3.1, -3.9 | 2.1 | 1.9 |
| Cln8 (mnd, motor neuron degeneration) | Ceramide/lipid synthesis | 1.1 | 1.1, 1.3 | 0.8 | 2.0 |
| Clic4 (chloride intracellular channel 4, mitochondrial) | Unknown | 1.4 | 1.4, 1.0 | 1.5 | 0.9 |
| Chd1 | Chromatin modification or RNA splicing? | 1.0 | 1.6, 1.1 | 1.5 | 1.6 |
| Csrp1 (CRP1) | Unknown | 1.5 | 1.4, 1.4 | 1.6 | 0.6 |
| Dnm2 (dynamin 2) | Cytoskeletal organization | 3.3 | 4.9, 1.8 | 1.3 | 1.3 |
| Es1 (Ee-1, Es-1, Es-4, Es-N) | Carboxylesterase | 2.1 | 1.9, 1.6 | ND | 1.5 |
| Fad2 (fatty acid coenzyme A ligase, long chain 2) | Fatty acid metabolism | -2.5 | -2.1, 1.6 | 1.5 | 1.3 |
| Fkbp11 | FKBP-type peptidyl-prolyl-cis-trans-isomerase | 1.2 | 1.1, 1.7 | 1.8 | 1.0 |
| Foxa3 (Hnf3g, Tcf-3g) | Transcription factor | 1.7 | 1.2, 1.9 | 0.7 | 1.2 |
| Gstt1 (Gstt1-1) | Glutathione-S-transferase | -1.3 | 1.4, -1.6 | 0.7 | 1.3 |
| Grb2 (growth factor receptor bound protein 2) | SH2/SH3 adaptor | -1.4 | 1.0, -2.6 | 1.8 | ND |
| Krt1-18 (keratin complex 1, acidic, gene 18) | Structural protein | 6.2 | 10.7, 1.8 | 1.2 | 1.1 |
| Pck1 (PEPCK, Pck-1) | Gluconeogenesis | 1.4 | 1.4, 2.1 | ND | 2.8 |
| Arhb (Arh6, RhoB) | Ras homologue, multiple functions | 1.5 | 1.1, 1.5 | 1.6 | 1.2 |
| RIKEN cDNA 2310008M10 | Unknown | 1.2 | -1.1, 1.5 | 1.2 | 1.0 |
| RIKEN cDNA 2010203J19 | Unknown | 1.4 | 1.2, 1.3 | 1.8 | 1.1 |
| RIKEN cDNA 2900019C14 | Unknown | 1.1 | 1.1, 1.9 | 1.4 | 1.4 |
| RIKEN cDNA 3930402F23 | Unknown | 1.8 | 1.6, 1.2 | 1.7 | 1.1 |
| Rnf19 | Centrosomal protein | 1.2 | -1.1, 1.3 | 1.3 | 0.9 |
| S 100a 10 (annexin II, p11, Cal11) | Unknown | 1.7 | 2.0, 1.1 | 1.2 | 3.0 |
| Saa1 (serum amyloid A1) | High-density apolipoproteins | 92.7 | 24.9, 101.9 | 1.4 | 1.2 |
The expression analysis in quiescent liver is given as the fold expression change in wild-type versus C/EBPβ-/- liver. The post-partial hepatectomy (post-PH) results are given as the ratio of induction (i.e., expression at 2 or 16 h after partial hepatectomy divided by expression in quiescent liver) in wild-type versus C/EBPβ-/- liver. The location analysis is given as the intensity ratio of wild-type chromatin versus C/EBPβ-/- chromatin obtained from quiescent liver or 2 h after partial hepatectomy. ND, no data.
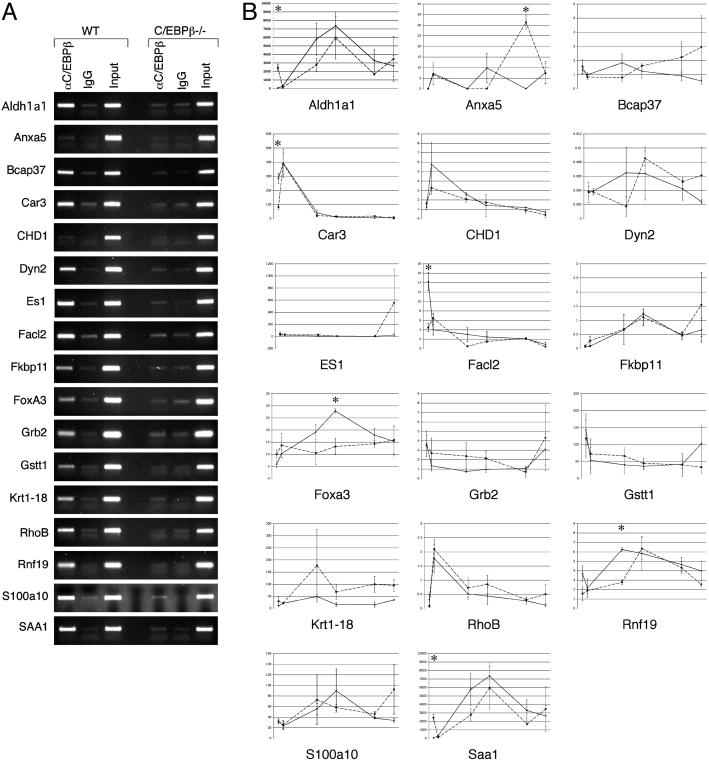
Confirmation of C/EBPβ Target Genes. Seventeen target genes were selected for further confirmation. To ascertain whether the selected gene promoters are directly bound by C/EBPβ, we performed conventional ChIP (Fig. 3A). Fifteen (88.2%) of the genes were specifically enriched in the ChIPs from wild type but not the C/EBPβ–/– mouse liver chromatin. This finding is likely to be an underestimate, because some of the negative genes may by bound by C/EBPβ at a location too distant from the ChIP PCR amplicon to be detected by this method. Using quantitative reverse transcription/real-time PCR, we found that the selected genes were induced or repressed in a variety of patterns in quiescent liver or after partial hepatectomy in wild-type mice (Fig. 3B). The pattern and/or timing of expression of six of these genes (35%) is significantly altered in C/EBPβ–/– mice relative to wild-type mice. Several of the genes that we have shown to be bound by C/EBPβ are nevertheless expressed at normal levels in the livers of C/EBPβ–/– mice, illustrating the redundancy present in complex regulatory networks. We conclude that the combination of location analysis and expression profiling makes it possible to identify genes that are not only directly bound by C/EBPβ, but also particularly those for which this binding is functionally relevant in quiescent and/or regenerating liver.
Fig. 3.
Confirmation of C/EBPβ targets identified by orthogonal analysis. (A) Conventional ChIP of C/EBPβ target genes. Chromatin was prepared from quiescent wild-type or C/EBPβ–/– mutant mouse livers and immunoprecipitated by using C/EBPβ or nonimmune antibody. The immunoprecipitates and input chromatin were subjected to PCR by using primers specific for the indicated genes. Typical results from experiments performed in duplicate are presented. (B) Expression of C/EBPβ targets in quiescent and regenerating liver. Quantitative real-time PCR was performed by using reverse-transcribed RNA obtained from wild-type and C/EBPβ–/– mutant mouse livers before and at 2, 16, 24, 40, and 48 h after partial hepatectomy. The expression of each gene is presented as a ratio to the expression of TATA binding protein. Error bars indicate standard error of the mean. *, P ≤ 0.05.
Identification of Potential C/EBPβ-Binding Partners. Random oligonucleotide-binding selection has identified the consensus C/EBP family DNA-binding sequence as ATTCGGCAAT (13). This and closely related sites appear at a high frequency in mammalian genomic sequence; indeed, an analysis of C/EBPβ binding in vitro to random fragments of genomic DNA implied that there are 3 × 107 C/EBPβ sites in the rat genome (30). It is likely that only a subset of these sites are actually bound in vivo, and we hypothesize that this specificity arises in part from the binding of other transcription factors to nearby sites. The binding of such factors may promote the binding of C/EBPβ through direct interactions with C/EBPβ or by relieving repressive chromatin structure. Our discovery of 15 genes bound in vivo by C/EBPβ provided an opportunity to identify some of these sites. Using all 674 transcription factor-binding sites in the TRANSFAC database, we performed a computational search for sites that are found more frequently in the set of C/EBPβ-bound genes (n = 15) than in genes that were neither bound by C/EBPβ in our location analysis nor dependent on C/EBPβ for expression (n = 138). The highest ranking sites were the IFN-stimulated response element, the peroxisome proliferator-activated receptor direct-repeat 1 site (PPAR-DR1), and the albumin D-site-binding protein consensus site (Table 2). Remarkably, higher-order combinations of a C/EBPβ site and these three sites were present at a significantly increased frequency in the C/EBPβ-bound genes relative to the total sequences present on the mouse promoter microarray (80% versus 4–12%). These results suggest that in vivo, the binding of C/EBPβ to target genes is determined by the binding of multiple additional tissue-specific factors.
Table 2. Transcription factor-binding sites associated with C/EBPβ sites bound in vivo.
| Binding sites | Maximum total length, bp | Frequency in C/EBP targets (n = 15), % | Frequency in microarray set (n = 2,122), % | P value (corrected) |
|---|---|---|---|---|
| C/EBPβ, DBP | 342 | 80 | 16 | 4.0 × 10-4 |
| C/EBPβ, ISRE | 608 | 80 | 14 | 7.7 × 10-5 |
| C/EBPβ, PPAR-DR1 | 736 | 73 | 19 | 0.04 |
| C/EBPβ, DBP, ISRE | 790 | 80 | 6.4 | 1.9 × 10-6 |
| C/EBPβ, DBP, PPAR-DR1 | 963 | 80 | 12 | 3.5 × 10-3 |
| C/EBPβ, ISRE, PPAR-DR1 | 663 | 80 | 9.3 | 1.9 × 10-4 |
| C/EBPβ, DBP, ISRE, PPAR-DR1 | 768 | 80 | 4.2 | 2.7 × 10-6 |
Potential C/EBPβ DNA-binding partners were evaluated as described in Materials and Methods. For each combination, the maximum distance between the outermost sites is given, and the frequency of positive genes in the 15 ChIP-positive targets and in the entire set of genes present on the mouse promoter microarray. The sites may be in any order or orientation. The P value is corrected for multiple testing. The LOD scores for each site are as follows: C/EBPβ, 8.9; D-site-binding protein (DBP), 9.2; ISRE, 9.0; PPAR-DR1, 9.0 for the pair with C/EBPβ and 8.1 for the higher order combinations. The transfac v. 7.3 position-sensitive scoring matrices used are as follows: M00109/C/EBPB_01, M00624/DBP_Q6, M00258/ISRE_01, and M00763/PPAR_DR1_01.
Discussion
The direct C/EBPβ targets identified in our study segregate into two categories: those that depend on C/EBPβ in quiescent liver but are normally induced after partial hepatectomy, and those that depend on C/EBPβ only after partial hepatectomy. Two of the genes in the former category (Car3 and Saa1) are involved in the prevention of oxidative damage (31, 32), and C/EBPβ has been implicated in this process as well (33–35). The C/EBPβ-dependent induction of Car3 and Saa1 after partial hepatectomy reinforces the concept that gene regulation is a dynamic process in which different transcription factors may regulate a single gene, depending on the state of the cell.
The second class of C/EBPβ targets includes Foxa3 and Rnf19. The identification of Foxa3 as a direct C/EBPβ target in the regenerating liver is of particular interest because Foxa3 is required for maintenance of glucose homeostasis during a prolonged fast (36). It is conceivable that decreased activation of Foxa3 may contribute to the prolonged hypoglycemia observed in C/EBPβ–/– mice after partial hepatectomy. Another C/EBPβ target identified by our analysis is Rnf19, a ubiquitin ligase localized to the centrosomes of somatic cells (37, 38). Its centrosomal localization and ubiquitin ligase activity suggest that Rnf19 may function in the microtubule organizing center (MTOC). Ubiquitin-ligase-mediated degradation of centrosome-associated proteins contained in the MTOC is required for centrosome duplication, a necessary component of the G1/S phase transition (39). Expression of Rnf19 was significantly reduced in C/EBPβ–/– livers 16 h after partial hepatectomy, corresponding to mid-G1 phase of the hepatocyte cell cycle. This reduced expression could result in increased levels of proteins inhibitory to G1/S phase progression and might contribute to the decrease in DNA synthesis observed in C/EBPβ–/– livers after partial hepatectomy.
The high frequency of C/EBPβ-binding sites predicted by in vitro and computational analysis (13, 30) suggests that additional features must distinguish bona fide C/EBPβ target genes. This added specificity may be achieved through a requirement that other factors be bound in the region surrounding the C/EBPβ site. These other factors may interact directly with C/EBPβ and cooperate in DNA binding. In the former case, looping of the DNA may allow C/EBPβ to simultaneously interact with its own DNA-binding site and with the other DNA-bound transcription factor; this is analogous to the mechanism whereby a transcriptional activator bound to a distant enhancer element brings the RNA polymerase II complex to the target gene promoter. The frequent occurrence of ISREs in proximity to C/EBPβ sites identified in this study lends support to this model, as C/EBPβ and the IFN-response factor 1 (IRF-1) have been shown to physically interact and jointly regulate the IL-18-binding protein promoter (40). Our studies suggest that IRF-1 and C/EBPβ may cooperate in a similar manner to activate liver-specific genes.
The binding of C/EBPβ to its consensus sequence may also be facilitated in vivo by local changes in chromatin structure that are induced by other bound transcription factors, as proposed to occur at the albumin enhancer during embryonic liver development after the binding of Foxa3 and GATA-4 (41). In this light, the high frequency of PPAR-DR1 sites near C/EBPβ sites is particularly suggestive, because members of the PPAR family have been shown to interact with the SWI/SNF chromatin remodeling complex (42). The joint presence of C/EBPβ and PPAR sites is also consistent with the cooperative nature of gene regulation by these two factors in fat metabolism in liver and adipogenesis (43, 44). The coincidence of D-site-binding protein and C/EBPβ sites is also consistent with previous findings of synergy between these two factors in activating the factor IX gene (45). Finally, we were surprised to find a high frequency of combinations of a C/EBPβ site with two or three other transcription factor-binding sites. This finding suggests that several mechanisms may be used simultaneously to achieve tissue-specific binding of C/EBPβ to target genes.
In conclusion, we have leveraged the combined power of expression profiling and high-throughput ChIP to identify direct and biologically relevant targets of C/EBPβ. The combination of screening approaches identified 26 potential C/EBPβ targets. Of these, 17 were tested and confirmed as in vivo C/EBPβ-binding sites by conventional ChIP and six of these were shown to be physiological targets by their altered expression in C/EBPβ–/– mice. We expect that this technique should be generally applicable to the discovery of biologically relevant target genes of other transcription factors.
Supplementary Material
Acknowledgments
We thank Liping Zhang for help with quantitative PCR and Amy Weinmann of the Peggy Farnham laboratory for technical assistance. This work was supported by National Institute of Diabetes and Digestive and Kidney Diseases Grants DK056669 (to L.E.G.), DK49210 (to K.H.K.), and DK56947 (to K.H.K.). J.R.F. was supported by National Institute of General Medical Sciences Grant 5T32GM008638.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: C/EBP, CCAAT enhancer-binding protein; ChIP, chromatin immunoprecipitation; PSSM, position-sensitive scoring matrices; ROC, receiver operating characteristics; PEPCK, phosphoenolpyruvate carboxykinase; PPAR, peroxisome proliferator-activated receptor.
References
- 1.Ramji, D. P. & Foka, P. (2002) Biochem. J. 365, 561–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Poli, V. (1998) J. Biol. Chem. 273, 29279–29282. [DOI] [PubMed] [Google Scholar]
- 3.Croniger, C., Leahy, P., Reshef, L. & Hanson, R. (1998) J. Biol. Chem. 273, 31629–31632. [DOI] [PubMed] [Google Scholar]
- 4.Rask, K., Thorn, M., Ponten, F., Kraaz, W., Sundfeldt, K., Hedin, L. & Enerback, S. (2000) Int. J. Cancer 86, 337–343. [DOI] [PubMed] [Google Scholar]
- 5.Sundfeldt, K., Ivarsson, K., Carlsson, M., Enerback, S., Janson, P. O., Brannstrom, M. & Hedin, L. (1999) Br. J. Cancer 79, 1240–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zahnow, C. A., Younes, P., Laurcirica, R. & Rosen, J. M. (1997) J. Natl. Cancer Inst. 89, 1887–1891. [DOI] [PubMed] [Google Scholar]
- 7.Zhu, S., Yoon, K., Sterneck, E., Johnson, P. F. & Smart, R. C. (2002) Proc. Natl. Acad. Sci. USA 99, 207–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buck, M., Poli, V., van der Geer, P., Chojkier, M. & Hunter, T. (1999) Mol. Cell 4, 1087–1092. [DOI] [PubMed] [Google Scholar]
- 9.Bundy, L. M. & Sealy, L. (2003) Oncogene 22, 869–883. [DOI] [PubMed] [Google Scholar]
- 10.Greenbaum, L. E., Li, W., Cressman, D., Peng, Y., Ciliberto, G., Poli, V. & Taub, R. (1998) J. Clin. Invest. 102, 996–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buck, M. & Chojkier, M. (2003) Hepatology 37, 731–738. [DOI] [PubMed] [Google Scholar]
- 12.Lamb, J., Ramaswamy, S., Ford, H. L., Contreras, B., Martinez, R. V., Kittrell, F. S., Zahnow, C. A., Patterson, N., Golub, T. R. & Ewen, M. E. (2003) Cell 114, 323–334. [DOI] [PubMed] [Google Scholar]
- 13.Osada, S., Yamamoto, H., Nishihara, T. & Imagwa, M. (1996) J. Biol. Chem. 271, 3891–3896. [DOI] [PubMed] [Google Scholar]
- 14.LeClair, K. P., Blanar, M. A. & Sharp, P. A. (1992) Proc. Natl. Acad. Sci. USA 89, 8145–8149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vallejo, M., Ron, D., Miller, C. P. & Habener, J. F. (1993) Proc. Natl. Acad. Sci. USA 90, 4679–4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsu, W., Kerppola, T. K., Chen, P.-L., Curran, T. & Chen-Kiang, S. (1994) Mol. Cell. Biol. 14, 268–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lekstrom-Himes, J. & Xanthopoulos, K. G. (1998) J. Biol. Chem. 273, 28545–28548. [DOI] [PubMed] [Google Scholar]
- 18.Wells, J., Boyd, K. E., Fry, C. J., Bartley, S. M. & Farnham, P. J. (2000) Mol. Cell. Biol. 20, 5797–5807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schug, J. & Overton, G. C. (1997) Proc. Int. Conf. Intell. Syst. Mol. Biol. 5, 268–271. [PubMed] [Google Scholar]
- 20.Rozen, S. & Skaletsky, H. (2000) Methods Mol. Biol. 132, 365–386. [DOI] [PubMed] [Google Scholar]
- 21.Li, Z., Van Calcar, S., Qu, C., Cavenee, W. K., Zhang, M. Q. & Ren, B. (2003) Proc. Natl. Acad. Sci. USA 100, 8164–8169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaestner, K. H., Lee, C. S., Scearce, L. M., Brestelli, J. E., Arsenlis, A., Le, P. P., Lantz, K. A., Crabtree, J., Pizarro, A., Mazzarelli, J., et al. (2003) Diabetes 52, 1604–1610. [DOI] [PubMed] [Google Scholar]
- 23.Scearce, L. M., Brestelli, J. E., McWeeney, S. K., Lee, C. S., Mazzarelli, J., Pinney, D. F., Pizarro, A., Stoeckert, C. J., Jr., Clifton, S. W., Permutt, M. A., et al. (2002) Diabetes 51, 1997–2004. [DOI] [PubMed] [Google Scholar]
- 24.Elizondo, G., Corchero, J., Sterneck, E. & Gonzalez, F. J. (2000) J. Biol. Chem. 275, 39747–39753. [DOI] [PubMed] [Google Scholar]
- 25.Croniger, C., Trus, M., Lysek-Stupp, K., Cohen, H., Liu, Y., Darlington, G. J., Poli, V., Hanson, R. W. & Reshef, L. (1997) J. Biol. Chem. 272, 26306–26312. [DOI] [PubMed] [Google Scholar]
- 26.Mohn, K. L., Laz, T. M., Melby, A. E. & Taub, R. (1990) J. Biol. Chem. 265, 21914–21921. [PubMed] [Google Scholar]
- 27.Li, X. X. & Liao, W. S. (1991) J. Biol. Chem. 266, 15192–15201. [PubMed] [Google Scholar]
- 28.Ray, A., Hannink, M. & Ray, B. K. (1995) J. Biol. Chem. 270, 7365–7374. [DOI] [PubMed] [Google Scholar]
- 29.Ray, A. & Ray, B. K. (1994) Mol. Cell. Biol. 14, 4324–4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Falvey, E., Marcacci, L. & Schibler, U. (1996) Biol. Chem. 377, 797–809. [PubMed] [Google Scholar]
- 31.Liao, F., Lusis, A. J., Berliner, J. A., Fogelman, A. M., Kindy, M., de Beer, M. C. & de Beer, F. C. (1994) Arterioscler. Thromb. 14, 1475–1479. [DOI] [PubMed] [Google Scholar]
- 32.Ikeda, M., Ishii, Y., Kato, H., Akazawa, D., Hatsumura, M., Ishida, T., Matsusue, K., Yamada, H. & Oguri, K. (2000) Arch. Biochem. Biophys. 380, 159–164. [DOI] [PubMed] [Google Scholar]
- 33.Fawcett, T. W., Martindale, J. L., Guyton, K. Z., Hai, T. & Holbrook, N. J. (1999) Biochem. J. 339, Pt. 1, 135–141. [PMC free article] [PubMed] [Google Scholar]
- 34.Seo, S. J., Kim, H. T., Cho, G., Rho, H. M. & Jung, G. (1996) Gene 178, 177–185. [DOI] [PubMed] [Google Scholar]
- 35.Seo, S. J., Kang, S. S., Cho, G., Rho, H. M. & Jung, G. (1997) Gene 203, 11–15. [DOI] [PubMed] [Google Scholar]
- 36.Shen, W., Scearce, L. M., Brestelli, J. E., Sund, N. J. & Kaestner, K. H. (2001) J. Biol. Chem. 276, 42812–42817. [DOI] [PubMed] [Google Scholar]
- 37.Parraga, M. & del Mazo, J. (2000) Mech. Dev. 90, 95–101. [DOI] [PubMed] [Google Scholar]
- 38.Niwa, J., Ishigaki, S., Doyu, M., Suzuki, T., Tanaka, K. & Sobue, G. (2001) Biochem. Biophys. Res. Commun. 281, 706–713. [DOI] [PubMed] [Google Scholar]
- 39.Hansen, D. V., Hsu, J. Y., Kaiser, B. K., Jackson, P. K. & Eldridge, A. G. (2002) Oncogene 21, 6209–6221. [DOI] [PubMed] [Google Scholar]
- 40.Hurgin, V., Novick, D. & Rubinstein, M. (2002) Proc. Natl. Acad. Sci. USA 99, 16957–16962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cirillo, L. A., Lin, F. R., Cuesta, I., Friedman, D., Jarnik, M. & Zaret, K. S. (2002) Mol. Cell 9, 279–289. [DOI] [PubMed] [Google Scholar]
- 42.Debril, M. B., Gelman, L., Fayard, E., Annicotte, J. S., Rocchi, S. & Auwerx, J. (2004) J. Biol. Chem. 279, 16677–16686. [DOI] [PubMed] [Google Scholar]
- 43.Grimaldi, P. A. (2001) Curr. Opin. Clin. Nutr. Metab. Care 4, 433–437. [DOI] [PubMed] [Google Scholar]
- 44.Grimaldi, P. A. (2001) Prog. Lipid Res. 40, 269–281. [DOI] [PubMed] [Google Scholar]
- 45.Picketts, D. J., Lillicrap, D. P. & Mueller, C. R. (1993) Nat. Genet. 3, 175–179. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.