Abstract
The activation-induced cytidine deaminase (AID) is required for somatic hypermutation (SHM) and class-switch recombination of Ig genes. It has been shown that in vitro, AID protein deaminates C in single-stranded DNA or the coding-strand DNA that is being transcribed but not in double-stranded DNA. However, in vivo, both DNA strands are mutated equally during SHM. We show that AID efficiently deaminates C on both DNA strands of a supercoiled plasmid, acting preferentially on SHM hotspot motifs. However, this DNA is not targeted by AID when it is relaxed after treatment with topoisomerase I, and thus, supercoiling plays a crucial role for AID targeting to this DNA. Most of the mutations are in negatively supercoiled regions, suggesting a mechanism of AID targeting in vivo. During transcription the DNA sequences upstream of the elongating RNA polymerase are negatively supercoiled, and this transient change in DNA topology may allow AID to access both DNA strands.
A major advance in the study of somatic hypermutation (SHM) and class-switch recombination (CSR) has been the discovery of the activation-induced cytidine deaminase (AID) (1–3), which appears to be the long-sought SHM mutator factor and inducer of CSR (4). Current in vitro data show that AID is a DNA-specific cytidine deaminase that preferentially removes the amino group of cytidine in single-stranded DNA and in the nontranscribed strand when transcription is active (5–11). These findings are consistent with previous experiments in which SHM is linked to transcription (12, 13). However, in vivo, both DNA strands are equally mutated (14). Furthermore, in ung–/– mice, where almost all of the C or G mutations are transitions due to unrepaired AID lesions, equal targeting of both strands is confirmed (15). Single-stranded DNA would also occur in vivo as a potential AID target during DNA replication, which would explain why both DNA strands are targeted during SHM and CSR. However, experiments with a cell line that undergoes SHM in culture support the conclusion that AID can act during the G1 phase of the cell cycle, and therefore, is not restricted to the S phase (16) (S. Gasior and U.S., unpublished data). A nonexclusive third possibility is that DNA topology (e.g., supercoiling) creates an AID-accessible conformation. In vivo, Ig genes are associated in nucleosomes with histones and other chromatin proteins. These associations, as well as the process of transcription, affect the topology of DNA. To test the possibility that supercoiled DNA as it exists in vivo may be a target for AID, we carried out cytidine deamination assays in vitro with AID purified from insect cells (5). The supercoiled target DNA was an Escherichia coli plasmid that had been manipulated to allow bacterial resistance to carbenicillin only when the initiator AUG of an ampicillin resistance (Ampr) gene was created by AID deamination of an ACG triplet.
Materials and Methods
Plasmid Construction. The kanamycin-resistance (Kanr) gene was inserted into the SacII site in pBluescript KS(II), and the plasmid was transformed into DH5α E. coli. A colony growing in both kanamycin and carbenicillin was selected to further amplify the plasmid. To mutate the start codon in the Ampr gene, PCR was performed by using a pair of primers in which the 5′ primer has a mutation from ATG to ACG in the start codon (5′-TGCTTCAATAATATTGAAAAAGGAAGAGTACGTATATTC/CTGCAATGATACCGCGAGACC). The PCR product and the plasmid were cut with BsaI/SspI, and the products were then inserted into the plasmid. The mutation was confirmed by sequence analysis. To remove several potential start codons in the Ampr gene-promoter region, the SspI fragment was deleted (≈420 bp). The deletion of the original promoter did not affect Ampr gene expression, and the new promoter driving the Ampr gene has not been defined. There are two potential start codons located at positions 196–201 in the Ampr gene, which were changed by using the QuikChange site-directed mutagenesis kit (Stratagene), and the primers were as follows: 5′-CTTTAAAAGTGCTAATTATTGGAAAACG/CGTTTTCCAATAATTAGCACTTTTAAAG. Clones that grew in the kanamycin condition but not in the carbenicillin condition were confirmed by sequence analysis. The final version of the modified plasmid is called pKM2.
AID and Other Enzyme Assays. The baculovirus GST-AID plasmid vector was a gift from M. Goodman (University of Southern California, Los Angeles). The GST-AID protein was expressed in Sf9 insect cells, and the protein was purified by using glutathione–Sepharose 4B (Pharmacia). The purified protein was dialyzed twice against 20 mM Tris, pH 7.5/10 mM NaCl/0.1 mM DTT/20% glycerol (5). AID treatment was performed as described by Bransteitter et al. (5), with modifications. Briefly, 100 fmol of pKM2 was treated with 450 ng of AID at 37°C for 2 h in 10 mM Tris·HCl, pH 8.0/1 mM EDTA/1 mM DTT in the presence of 1 μg of RNase A. The AID-treated plasmid was purified by using phenol/chloroform/isoamyl alcohol (25:24:1) and then transformed into BW504, a uracil–DNA glycosylase-deficient E. coli strain (a gift from A. Bhagwat, Wayne State University, Detroit). Selection was done in 50 μg/ml kanamycin or 100 μg/ml carbenicillin. We also treated the plasmid with AID in the absence of RNase A, and no Ampr colonies were observed (data not shown), which is consistent with the findings of Goodman and colleagues (5) and Lieber and colleagues (7) that GST-AID protein isolated from insect cells requires RNase for activity.
To determine the effect of supercoiling on AID deamination, we treated pKM2 with topoisomerase I (Topo I) (Sigma) at 37°C for 1.5 h in 50 mM Tris·HCl, pH 7.5/100 mM NaCl/2.5 mM MgCl2/0.1 mM EDTA. The treated plasmid was purified by using the Qiaquick gel-extraction kit (Qiagen, Valencia, CA) before it was treated further with AID.
Sequence Analysis. Plasmid DNAs were prepared from carbenicillin- or kanamycin-resistant bacterial colonies. Each plasmid DNA was sequenced with eight different primer pairs, which resulted in overlapping sequences so that errors, and even ambiguities, are avoided. Automated sequencing was done with the 3730XL apparatus (Applied Biosystems), and sequence analysis was performed by using sequencher 4.1.
Results
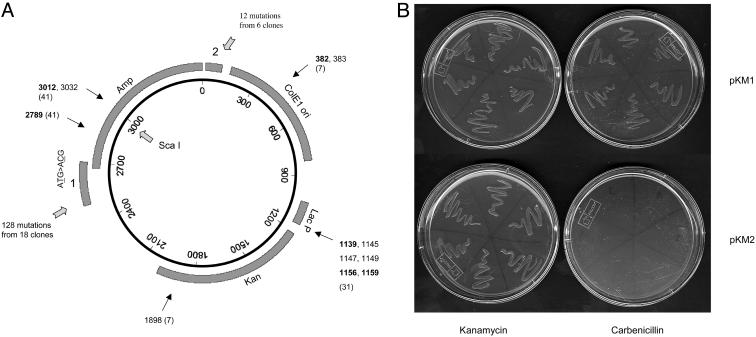
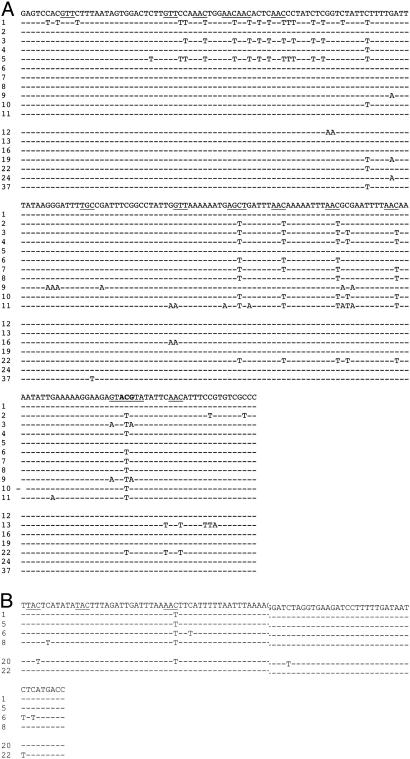
AID Can Target Double-Stranded DNA in Vitro. To test the possibility that supercoiled DNA may be a target for AID, we used a supercoiled circular DNA E. coli plasmid as a substrate for AID deamination in vitro. The plasmid pKM2 contains an Ampr β-lactamase gene whose initiator ATG codon was changed to ACG (Fig. 1A). The plasmid should confer bacterial resistance to ampicillin or carbenicillin if AID activity has reverted the ACG to ATG. The plasmid also contains a Kanr gene that was used to estimate the transformation efficiency and reversion frequency of ACG to ATG in the Ampr gene. The modified plasmid, pKM2, does not encode any β-lactamase protein, and bacteria carrying the plasmid cannot survive in carbenicillin (Fig. 1B). Control and AID-treated plasmids were transformed into BW504, a uracil–DNA glycosylase-deficient E. coli strain (17). The transformants were grown on either kanamycin or carbenicillin plates. Healthy colonies growing in the carbenicillin plates indicated that the Ampr gene start codon was reestablished in the AID-treated pKM2. We expected that, if AID treatment led to mutations, the number of carbenicillin-resistant colonies would be low because pKM2 DNA contains over 1,800 cytidines and only one mutation (ACG→ATG) would restore Ampr gene expression. If AID deamination is random, the frequency of ACG→ATG would be ≈5.6 × 10–4. However, the frequency of recovering Ampr colonies was ≈3.3 × 10–3 (colonies in carbenicillin per colonies in kanamycin) (Table 1), which is well above the expected frequency of 5.6 × 10–4, suggesting that mutation at the target ACG is not random and that, at a minimum, 1 of ≈300 input DNA molecules was a substrate for AID. We sequenced the complete 3.5-kb plasmid DNA of 11 independent Ampr clones, and we found that most clones carried the reverted start codon (ACG→ATG) in the Ampr gene (Fig. 2A). There are two carbenicillin resistant clones without a conventional start codon (clones 1 and 5), suggesting that undefined start codons may exist in these plasmids. Multiple mutations in each sequenced clone were found upstream and at the 5′ end of the Ampr gene, extending over ≈210 bp, including the ACG triplet (Fig. 2 A). Of 117 mutations observed in Ampr clones, 106 were located in this hypermutable region 1 (Fig. 1 A, region 1). DNA mutations from Kanr clones showed the same pattern, in which 22 of 35 mutations reside in the hypermutable region 1 (Fig. 2 A, clones 12–37). Mutations also accumulated 3′ of the Ampr gene in hypermutable region 2 (Figs. 2B and 1 A, region 2). About 50% of mutations are located in the SHM hotspot motif WRC/GYW (where W = A/T, R = purine, and Y = pyrimidine), which is roughly the same proportion as found in SHM in vivo (18). Interestingly, the mutations in the hypermutable region 1 form three clusters in sequences enriched for hotspots, and several clones have similar mutations (Fig. 2 A).
Fig. 1.
pKM2 is an effective substrate for AID. (A) The pKM2 map. The start codon in the Ampr gene was mutated to ACG. Hypermutable regions 1 and 2 are indicated, together with the numbers of mutations and mutated clones in these regions. Black arrows indicate other mutation sites, and the numbers give the mutation positions. The numbers shown in bold are in the WRC/GYW motif. Clone numbers are given in parentheses. (B) Bacteria carrying pKM2 do not grow on carbenicillin plates. pKM2 is identical with pKM1, except that in pKM2, the Ampr gene start codon has been changed from ATG to ACG.
Table 1. Frequency of recovering colonies from five independent experiments.
| Experiment | Kanamycin colonies* | Carbenicillin colonies† | Carbenicillin/kanamycin |
|---|---|---|---|
| 1 | 645 | 18 | 0.0040 |
| 2 | 847 | 33 | 0.0056 |
| 3 | 1,680 | 37 | 0.0031 |
| 4 | 1,105 | 20 | 0.0026 |
| 5 | 1,164 | 16 | 0.0020 |
Two different batches of GST-AID protein were used to perform experiments 1-3 and 4 and 5. Plasmid DNA from each AID or control reaction was purified and transformed into BW504 E. coli.
One plate.
Seven plates.
Fig. 2.
AID targets hypermutable regions 1 and 2. The top lines in A and B are unmutated sequences. (A) Mutations in hypermutable region 1. Clones 1–11 grew under carbenicillin selection and clones 12–37 grew under kanamycin selection. The underlined triplets are the WRC/GYW motif, and the start codon is shown in bold. (B) Mutations in hypermutable region 2. Clones 1, 5, 6, and 8 are from carbenicillin selection, and clones 20 and 22 are from kanamycin.
In pKM2, both DNA Strands Are Equally Accessible to AID Activity. In general, each bacterial colony contained the following two types of plasmids: one type that was derived from the top strand, and another type that was derived from the bottom strand of an original double-stranded DNA molecule. Although most mutations in carbenicillin-selected clones were on the nontranscribed (top) strand (containing the ACG→ATG mutation) selected for Ampr gene expression, we also observed mutations in the other (bottom) strand and in both strands in single carbenicillin-selected colonies, suggesting that AID targets both DNA strands. To determine whether AID attacks both strands equally, we further sequenced 40 Kanr clones that are not selected for mutations in the hypermutable region 1. The eight mutated clones that we obtained (20%) showed no strand bias for AID targeting (i.e., five of the clones had G→A mutations, and five of the clones had C→T mutations) (Fig. 2). Additional G→A and C→T mutations were seen also in two other clones outside of the hypermutable regions (clones 41 and 31, respectively; Fig. 1 A). Thus, in double-stranded DNA, AID can target either strand. When mutations were seen in both strands in single bacterial colonies, they were always close to each other (Fig. 2 A; clones 3, 9, 11, 13, and 19; C→TorG→A are deaminations that arose on the top or bottom strands, respectively). In several cases, consecutive bases on the top and bottom strand were mutated (Fig. 2 A; clones 3, 9, 11, and 13). Thus, AID either functions on both DNA strands at the same time or it acts repeatedly on either strand. If the latter were true, mutations in the top strand would not necessarily be located at the same cluster as mutations in the bottom strand. However, when mutations occur on both strands, they are always in the same cluster, suggesting that AID may be able to jump from one strand to the other strand or that it may exist as a complex of multiple AID proteins, as has been suggested (6, 8, 19). Several restriction sites in the hypermutable region 1 are completely digestible (data not shown), indicating that the region is a double-stranded, and not single-stranded, bubble, suggesting that AID indeed acts on double-stranded DNA.
AID Appears to Act Processively. Most of the sequences have multiple C deaminations, with some of them having as many as 14 consecutive mutated Cs; for example, sequence 1 (Fig. 2 A) has all 14 Cs in a stretch of 39 nt changed to T. Of these Cs, only four are in WRC hotspots. Because these consecutive C-deaminations and only ≈20% of the analyzed total plasmids show any C-deamination (Table 2), it is possible that AID acts processively, as proposed (9). Alternatively, an initial C→U deamination by an AID complex that then leaves the DNA may have created a more favorable substrate conformation for further deaminations of nearby Cs by an independent AID complex.
Table 2. Mutations occur only in the supercoiled plasmid.
| Plasmid type | Sequenced clones | Mutated clones | Clone-mutation frequency | Sequenced bases | Mutated bases | Base-mutation frequency |
|---|---|---|---|---|---|---|
| Supercoiled | 40* | 8 | 0.2 | 141,160 | 35 | 2.5 × 10-4 |
| Relaxed | 40† | 0 | 0 | 141,160 | 0 | 0 |
E. coli transformed with the AID treated pKM2 were grown in kanamycin. We picked 40 colonies from supercoiled- or relaxed-plasmid transformations.
Colonies from experiments with supercoiled DNA, given in Table 1.
Colonies were picked for sequencing from two plates from two separate experiments containing 1,121 and ≈500 kanamycin-resistant colonies, respectively. No colonies were observed in these two experiments with the AID-treated relaxed plasmid in 14 carbenicillin-containing plates.
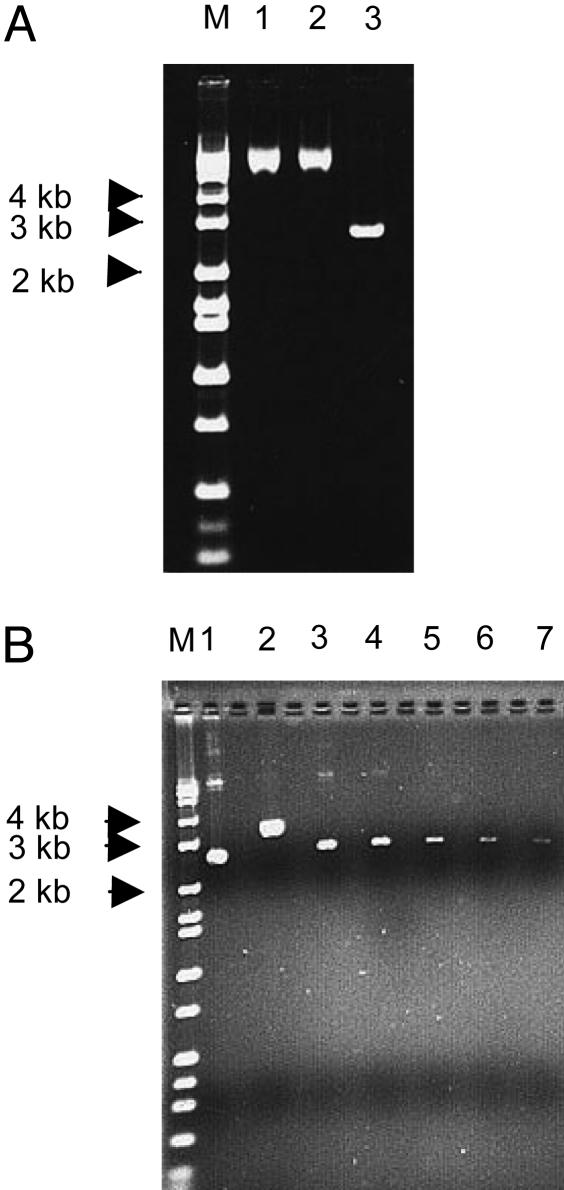
The Double-Stranded DNA Has to Be Supercoiled to Be Deaminated by AID. To determine whether pKM2 DNA is an AID substrate because it is supercoiled, we asked whether a relaxed circular plasmid is an AID target by treating the pKM2 plasmid with Topo I (Fig. 3A). AID treatment of the relaxed plasmid did not result in any Ampr colonies, whereas the supercoiled plasmids in the same experiments were mutated to Ampr as before, suggesting that either the relaxed plasmid was no longer an AID target or the hypermutable region was shifted to a new region. To test the latter possibility, we sequenced the entire 3.5-kb plasmid DNA of 40 Kanr clones (each from two separate experiments) that were derived from transformation with relaxed AID-treated plasmid, but no mutations were found in the total 144-kb DNA that was sequenced (Table 2). Thus, our data collectively indicate that AID is able to act on supercoiled, but not relaxed, double-stranded DNA.
Fig. 3.
Treatment of pKM2 with Topo I. (A) M, marker. Lane 1, plasmid treated with Topo I (before purification of DNA); lane 2, the same treated plasmid (after purification of DNA); and lane 3, supercoiled plasmid (not treated with Topo I). (B) SnaB1. M, marker. Lane 1, supercoiled plasmid, 100%; lane 2, plasmid treated with SnaB1, 100%; and lanes 3–7, supercoiled plasmid, 20%, 10%, 3%, 1%, and 0.5% of amount shown in lanes 1 and 2.
Discussion
These findings clearly show that AID can target double-stranded DNA when it is supercoiled. The pattern of C-deaminations is very similar to the observed pattern of SHM in vivo. Of the 129 mutations in region 1, 64 are in WRC/GYW motifs, which are known hotspots of SHM. This 50% proportion is significantly higher than the mutation frequency in the hotspots that would be expected if AID targeting were random (22%). Also, in hypermutable region 2, about one-half of the mutations are located in WRC/GYW motifs (Fig. 2B). If the mutations were random, the mutation frequency in the motifs in region 2 would be 14%. Thus, as has been found for SHM in vivo (20) and single-stranded DNA in vitro (9), AID prefers the SHM hotspots in supercoiled DNA in vitro as well. The proportion of targeted hotspots is similar in the supercoiled plasmid and in single-stranded DNA as analyzed in ref. 9 in which the proportion of mutations in hotspots was 57% and the frequency of C hotspots in the sequence was 25% (9). The pattern of mutations in the supercoiled and single-stranded DNAs in vitro mimics endogenous Ig gene SHM in vivo (in which about one-half of the C or G mutations are in hotspots; ref. 20) and in an Ig transgene (whose sequence had 25% of Cs and Gs in hotspots and in which 63% of the C or G mutations were in hotspots; ref. 18).
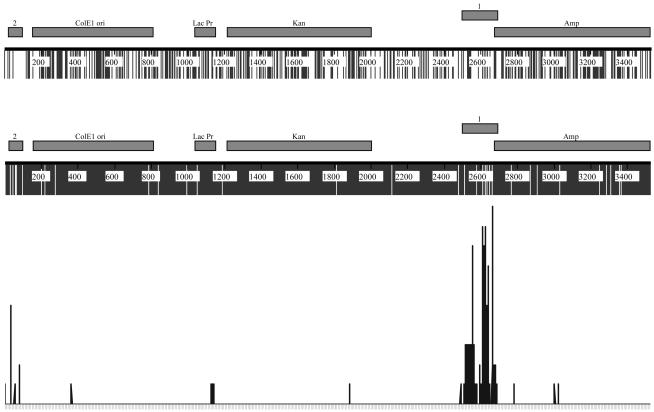
The distribution of mutations in the pKM2 plasmid is nonrandom, which raises the question of why regions 1 and 2 are hypermutable. To determine whether the frequency of WRC/GYW motifs or the base composition played a major role, we compared these parameters across the plasmid sequence (Fig. 4). The sequence of the plasmid shows that the frequency of the WRC/GYW motif in the hypermutable regions 1 and 2 is actually lower than in most of the rest of the plasmid (Fig. 4 Top). Thus, the targeting of the hypermutable regions is not due to an excess of known mutation hotspots. Concerning the base composition, the hypermutable regions have the lowest G+C content in the plasmid (Fig. 4 Middle). Hypermutable region 1 has 36% G+C and hypermutable region 2 has 25% G+C, suggesting that a low G+C vs. A+T content may play a role in the AID-deamination activity. However, the A+T-rich region is not sufficient to attract AID because in the relaxed plasmid the A+T-rich sequence is not a target for AID. However, the higher A+T content will make the hypermutable regions more deformable in the supercoiled state.
Fig. 4.
Alignment of SHM hotspots (Top), cytosines (Middle), and mutations (C→ T and G→ A) (Bottom) in the pKM2 plasmid. The horizontal boxes in Top and Middle are plasmid regions, as shown in Fig. 1 A. The bottom shows mutations recorded in Figs. 1 A and 2 of the entire plasmid sequence of supercoiled pKM2 treated with AID. The lowest mark represents one point mutation, and the highest mark represents 10 mutations at a given position.
The two A+T-rich hypermutable regions in the supercoiled plasmid have been shown to be negatively supercoiled (21). In vivo, negative supercoiling is created during transcription elongation upstream of the RNA polymerase (22, 23). We postulate that this DNA topology may provide access for AID to both DNA strands. By analogy, negatively supercoiled DNA shows increased accessibility to DNA methyltransferase (24). The DNA topology in the transcribed Ig genes may resemble the topology of the hypermutable regions in the plasmid pKM2. The hypermutable regions are unlikely to be single-stranded because they can be digested to completion by restriction enzymes (Fig. 3B) (e.g., the start codon of the Ampr gene is completely cut by SnaBI). Because 20% of all supercoiled plasmids have mutations (Table 2) and 14% of plasmids that were not selected for Amp-resistance have the mutated SnaB1 site (Fig. 2 A), ≈3% of the SnaB1 sites must be double-stranded. This level of DNA not cut by SnaB1 would have been detected (Fig. 3b).
It is possible that short nonpalindromic sequences between short-inverted repeats are stably or occasionally extruded as single-stranded DNA in the hypermutable region (25). If such structures existed and the nonpalindromic sequences between the palindromes contained Cs, these Cs could be targets for AID. We have analyzed the hypermutable region for such structures. There are only two sequences in the hypermutable region 1 that would fit these criteria: one is an 8-bp inverted repeat that is separated by 11 nonrepetitious base pairs, and the other is an 8-bp inverted repeat that is separated by three nonrepetitious base pairs. The sequences are as follows: GAGTCCACGTTC*TTTAATAGTGGACTC and (directly 3′ to this) TTGTTCCAAAC*TGGAACAA. Both sequences contain a C in the nonrepetitious region (indicated by asterisk) that is mutated in one and three (hotspot) cases, respectively. (The sequence is shown on the top line on the left of Fig. 2 A). There is also one palindrome in hypervariable region 2, which is a 10-bp inverted repeat that is separated by the following seven nonrepetitious base pairs: AAAAGGATCTAGGTGAAGATCCTTTT (Fig. 2B top line). No mutation has been found in any of the Gs in the nonrepetitious sequence. However, there are four Cs in the inverted repeats in region 1 and one C in region 2 that are mutated (Fig. 2), which should be protected from AID if the repeats were annealed and only single-stranded DNA were an AID target. Thus, because regions in potential stems are mutated and no additional similar short-inverted repeats are found in the hypermutable regions, it appears to be unlikely that periodically opened single-stranded DNA is the only target for AID. However, possible extruded single-strand loops cannot be ruled out as targets for AID, in addition to double-stranded DNA that has been twisted and may expose the amino-group of cytosine for AID deamination. The clusters of consecutive mutations are shorter in vivo than in vitro (18). It is likely that in vivo supercoiled regions near the RNA polymerase are kept short by the counteracting effects of nucleosomes and topoisomerases and/or that the mutable region is restricted by DNA binding proteins which partially block AID function. When supercoils are introduced by transcription, their structure is transient and dynamic. Targeting by AID may be aided by its association with the RNA polymerase, as postulated in our model of SHM (26) and recently reported to exist in mutating B cells by Shimizu and coworkers (27).
Given the findings with the pKM2 plasmid, it is perhaps unexpected that in vivo AID targets in E. coli that express AID are mutated preferably in the nontranscribed strand (10, 11). However, in these experiments, the transcribed strand was also found to be several-fold more highly mutated with than without AID. It is possible that single-stranded DNA, as it exists in the nontranscribed strand, is more accessible to AID than supercoiled DNA. If this situation were the case in vertebrate cells undergoing SHM, one would expect an imbalance of top/bottom strand mutations. Although, on average, mutations from C and G occur with equal frequencies, the ratios of transitions from C (top strand) and G (bottom strand) vary considerably in different mutated Ig genes (28). It is likely that the DNA sequence would influence the balance between mutations in the nontranscribed strand versus supercoiled regions. It is also possible that C→U deaminations in the nontranscribed strand are more readily corrected by base excision repair to the original C.
Acknowledgments
We thank M. Goodman, R. Bransteitter, and P. Pham (University of Southern California, Los Angeles) for the baculovirus AID expression plasmid; C.-A. Reynaud and J.-C. Weill (Institut National de la Santé et de la Recherche Médicale, Paris) for anti-AID antibodies; and A. Bhagwat for the uracil–DNA glycosylase-deficient E. coli. We also thank A. Bransteitter for advice on the production of the AID protein; D. Mydlowski and B. Eisfelder (University of Chicago Immunology Applications Facility) for the production and amplification of the AID-producing insect cells; W. Buikema (University of Chicago DNA Sequencing Facility) for advice; V. Volgina (University of Chicago) for advice on protein and antibody purification; S. Pylawka (University of Chicago) for AID assays of purified protein; and S. Longerich, N. Michael, P. Engler, T. E. Martin (University of Chicago), and J. Widom (Northwestern University, Evanston, IL) for critical reading and creative discussions of the manuscript. This work was supported by National Institutes of Health Grant AI47380. H.M.S. is a Young Investigator of the Cancer Research Foundation.
Abbreviations: AID, activation-induced cytidine deaminase; SHM, somatic hypermutation; CSR, class-switch recombination; Ampr, ampicillin resistance; Topo I, topoisomerase I; Kanr, kanamycin resistance.
References
- 1.Muramatsu, M., Sankaranand, V., Anant, S., Sugai, M., Kinoshita, K., Davidson, N. & Honjo, T. (1999) J. Biol. Chem. 274, 18470–18476. [DOI] [PubMed] [Google Scholar]
- 2.Muramatsu, M., Kinoshita, K., Fagarasan, S., Yamada, S., Shinkai, Y. & Honjo, T. (2000) Cell 102, 553–563. [DOI] [PubMed] [Google Scholar]
- 3.Revy, P., Muto, T., Levy, Y., Geissman, F., Plebani, A., Sanal, O., Catalan, N., Forveille, M., Dufourcq-Lagelouse, R., Gennery, A., et al. (2000) Cell 102, 565–575. [DOI] [PubMed] [Google Scholar]
- 4.Diaz, M. & Storb, U. (2003) DNA Repair 2, 623–627. [DOI] [PubMed] [Google Scholar]
- 5.Bransteitter, R., Pham, P., Scharff, M. & Goodman, M. (2003) Proc. Natl. Acad. Sci. USA 100, 4102–4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dickerson, S., Market, E., Besmer, E. & Papavasiliou, F. N. (2003) J. Exp. Med. 197, 1291–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu, K., Huang, F.-T. & Lieber, M. (2004) J. Biol. Chem. 279, 6496–6500. [DOI] [PubMed] [Google Scholar]
- 8.Chaudhury, J., Tian, M., Khuong, C., Chua, K., Pinaud, E. & Alt, F. (2003) Nature 422, 726–730. [DOI] [PubMed] [Google Scholar]
- 9.Pham, P., Bransteitter, R., Petruska, J. & Goodman, M. (2003) Nature 424, 103–107. [DOI] [PubMed] [Google Scholar]
- 10.Ramiro, A., Stavropoulos, P., Jankovic, M. & Nussenzweig, M. (2003) Nat. Immunol. 4, 452–456. [DOI] [PubMed] [Google Scholar]
- 11.Sohail, A., Klapacz, J., Samaranayake, M., Ullah, A. & Bhagwat, A. (2003) Nucleic Acids Res. 31, 2990–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Betz, A., Milstein, C., Gonzalez-Fernandes, R., Pannell, R., Larson, T. & Neuberger, M. (1994) Cell 77, 239–248. [DOI] [PubMed] [Google Scholar]
- 13.Peters, A. & Storb, U. (1996) Immunity 4, 57–65. [DOI] [PubMed] [Google Scholar]
- 14.Storb, U., Peters, A., Kim, N., Shen, H. M., Bozek, G., Michael, N., Hackett, J., Klotz, E., Loeb, L. & Martin, T. (1999) Cold Spring Harbor Symp. Quant. Biol. 64, 227–234. [DOI] [PubMed] [Google Scholar]
- 15.Rada, C., Williams, G., Nilsen, H., Barnes, D., Lindahl, T. & Neuberger, M. (2002) Curr. Biol. 12, 1748–1755. [DOI] [PubMed] [Google Scholar]
- 16.Faili, A., Aoufouchi, S., Gueranger, Q., Zober, C., Leon, A., Bertocci, B., Weill, J.-C. & Reynaud, C.-A. (2002) Nat. Immunol. 3, 815–820. [DOI] [PubMed] [Google Scholar]
- 17.Klapacz, J. & Bhagwat, A. (2002) J. Bacteriol. 184, 6866–6872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michael, N., Martin, T. E., Nicolae, D., Kim, N., Padjen, K., Zhan, P., Nguyen, H., Pinkert, C. & Storb, U. (2002) Immunity 16, 123–134. [DOI] [PubMed] [Google Scholar]
- 19.Ta, V., Nagaoka, H., Catalan, N., Durandy, A., Fischer, A., Imai, K., Nonoyama, S., Tashiro, J., Ikegawa, M., Ito, S., Kinoshita, K., Muramatsu, M. & Honjo, T. (2003) Nature Immunol. 4, 843–848. [DOI] [PubMed] [Google Scholar]
- 20.Shapiro, G., Aviszus, K., Ikle, D. & Wysocki, L. (1999) J. Immunol. 163, 259–268. [PubMed] [Google Scholar]
- 21.Kowalski, D., Natale, D. & Eddy, M. (1988) Proc. Natl. Acad. Sci. USA 85, 9464–9468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu, L. F. & Wang, J. (1987) Proc. Natl. Acad. Sci. USA 84, 7024–7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Collins, I., Weber, A. & Levens, D. (2001) Mol. Cell. Biol. 21, 8437–8451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bestor, T. (1987) Nucleic Acids Res. 15, 3835–3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lilley, D. (1980) Proc. Natl. Acad. Sci. USA 77, 6468–6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Storb, U. & Stavnezer, J. (2002) Curr. Biol. 12, R725–R727. [DOI] [PubMed] [Google Scholar]
- 27.Nambu, Y., Sugai, M., Gonda, H., Lee, C., Katakai, T., Agata, Y., Yokota, Y. & Shimizu, A. (2003) Science 302, 2137–2140. [DOI] [PubMed] [Google Scholar]
- 28.Milstein, C., Neuberger, M. & Staden, R. (1998) Proc. Natl. Acad. Sci. USA 95, 8791–8794. [DOI] [PMC free article] [PubMed] [Google Scholar]