Abstract
Activation-induced cytidine deaminase is required for the DNA cleavage step of Ig class switch recombination (CSR). However, its molecular mechanism is controversial. RNA-editing hypothesis postulates that activation-induced cytidine deaminase deaminates cytosine in an unknown mRNA to generate a new mRNA encoding an endonuclease for CSR and thus predicts that DNA cleavage depends on de novo protein synthesis. On the other hand, DNA deamination hypothesis proposes that DNA cleavage is initiated by cytosine deamination in DNA, followed by uracil removal by uracil DNA glycosylase. By using the chromatin immunoprecipitation assay to detect γ-H2AX focus formation as a marker for DNA cleavage, we found that cycloheximide inhibited DNA cleavage in the Ig heavy-chain locus during CSR. Requirement of protein synthesis in the DNA cleavage step of CSR strengthens the RNA-editing hypothesis.
Activation-induced cytidine deaminase (AID) is essential for all three types of DNA alteration, namely somatic hypermutation, gene conversion, and class switch recombination (CSR) in B cells activated by antigen stimulation (1-5). AID is expressed in activated B cells, especially in germinal centers of lymphoid follicles (6). Although AID is involved in the DNA cleavage step in CSR (7, 8), its molecular mechanism is still controversial. The RNA-editing hypothesis postulates that AID converts unknown mRNA precursors to novel mRNAs encoding putative endonucleases to cleave the target DNA (6). This hypothesis was originally proposed based on evolutionary conservation of AID with apoB mRNA-editing catalytic polypeptide 1 (APOBEC-1). This enzyme deaminates cytosine (C) at nucleotide 6666 of apoB 100 mRNA encoding a cholesterol carrier protein, converting it to apoB 48 mRNA encoding a triglyceride carrier (9). The AID and APOBEC-1 loci are tightly linked on mouse and human chromosomes (2). In addition to these evolutionary conservations, AID has functional similarities with APOBEC-1, including dimer formation, requirement of cofactors for their function, and shuttling between the cytoplasm and nucleus with the N-terminal nuclear localization and C-terminal nuclear export signal (10-12). Furthermore, AID-induced CSR depends on de novo protein synthesis in agreement with the requirement of the RNA-editing hypothesis (13).
On the other hand, the DNA deamination hypothesis predicts that AID directly deaminates C to uracil (U) in the target DNA, followed by strand breakage by using the base excision repair pathway, including uracil DNA glycosylase or uracil N-glycosylase (UNG), and apurinic/apyrimidinic-endonuclease (14). Overexpression of AID enhanced mutagenesis probably by inducing cytosine deamination in a number of genes in Escherichia coli (15). This hypothesis is further supported by marked reduction in the class switching efficiency in UNG-/- B cells (16). Furthermore, in vitro DNA deamination activity of AID is consistent with this hypothesis (17-20).
Double-strand breakage (DSB) in the Sμ region and another S region is essential to initiate CSR. DSB in interphase DNA is under surveillance by a set of proteins including ataxia-telangiectasia mutated (ATM)/ATM- and Rad3-related (ATR) and DNA-dependent protein kinase catalytic subunit (DNA-PKcs) that carry protein kinase activity (21). Such a protein complex phosphorylates histone H2AX in the DNA domain next to DSB soon after the DNA lesion (γ-H2AX focus formation) (22, 23). Phosphorylated H2AX (γ-H2AX) recruits many proteins involved in DNA repair. DSBs in CSR are finally joined by the nonhomologous end-joining (NHEJ) mechanism (24-26). Because γ-H2AX covers hundreds of kilobases of DNA adjacent to DSB, this accumulation serves as a good marker for DNA cleavage sites (7, 22, 23).
Because the RNA-editing hypothesis assumes involvement of de novo protein synthesis in AID-dependent DNA cleavage, it is critical to test whether the S region cleavage during CSR depends on protein synthesis. Here, we report evidence that shows requirement of protein synthesis for the S region cleavage in CSR, using the chromatin immunoprecipitation (ChIP) assay with anti-γ-H2AX antibodies. These results support the RNA-editing hypothesis.
Materials and Methods
Spleen Cell and Retroviral Infection. Splenocytes prepared from AID-/- mice were activated for 2 days in the presence of lipopolysaccharide (LPS) (50 μg/ml) and IL-4 (15 ng/ml) before retroviral infection (3). Plat-E (27) cells were transfected with either pFB-IRES-GFP (empty vector) or pFB-mAID-IRES-GFP [mouse AID (mAID)] retroviral expression constructs to obtain the recombinant viruses for infection of splenocytes (28). After 1.5 days of infection, spleen cells were harvested and stained with biotinylated anti-IgG1, followed by incubation with allphycocyanin-labeled streptavidin for flow cytometric analysis and subsequent cell sorting. For ChIP analysis, GFP+ live cells were sorted from vector and mAID virus-infected splenocytes. In the case of mAID virus-infected cells, GFP+IgG1- and GFP+IgG1+ were separately collected and analyzed by γ-H2AX ChIP.
CH12 Cells and Derived Cell Line. The AER cell line is a derivative of CH12F3-2, which contains the transgene AIDER-IRES-GFP under the control of the tet-off promoter (13). AIDER [AID fused with the hormone-binding domain of the estrogen receptor (ER)] was activated by the addition of estrogen analogue 4-hydroxytamoxifen (OHT) in the culture at a concentration of 1 μM (13). Expression of AIDER was monitored by GFP expression in AER cells. The CH12F3-2 line and its derivatives were induced to switch to IgA with soluble CD40L (CD8α-CD40L fusion) containing culture supernatant (50% final), 5 ng/ml recombinant mouse IL-4 (eBioscience, San Diego; or PeproTech, Rocky Hill, NJ) and 1.0 ng/ml recombinant human transforming growth factor (TGF-β1) (R & D Systems) as described (29). Switching efficiency was assayed by fluorescence-activated cell sorter (FACS, Becton Dickinson) for surface IgA expression. The P13 mutant of AID, which does not have the ability to induce class switching, has been described (10). A fusion construct of P13 with estrogen receptor (P13ER) was also prepared, and a derivative of the CH12F3-2 line that expresses P13ER was produced. No IgA-positive cells were detected upon OHT induction in the P13ER-expressing CH12 line.
ChIP Assay. ChIP assay Kit (Upstate Biotechnology, Lake Placid, NY) was used for the entire study according to the method recommended by the manufacturer, with minor modifications. In brief, 1-5 × 106 cells were fixed in the presence of 1% formaldehyde for 10 min at room temperature. The reaction was stopped by the addition of glycine at a final concentration 0.125 M. A soluble chromatin fraction containing fragmented DNA of 500-2,000 bp was obtained after cell lysis and sonication. The fraction was diluted ten times and precleared by using Protein A agarose slurry. Precleared lysates were aliquotted, and chromatin immunoprecipitation (30) was done by incubating with 4 μg of anti-γ-H2AX polyclonal antibodies (Upstate Biotechnology) or rabbit-IgG as a control. Ten percent of precleared lysate was saved for each sample to determine the input chromatin amount. Immunoprecipitated DNA was used for the template of PCR with or without serial dilution.
The primers that we used were as follows: Sμ region, Sμ-5′F (5′-GCT TCT AAA ATG CGC TAA ACT GAG GTG ATT-3′) and Sμ-5′R2 (5′-GTT TAG CTC TAT TCA ACC TAG-3′); Cμ region, Cμ-5′ (5′-CTG TCG CAG AGA TGA ACC CCA-3′) and Cμ-3′ (5′-ATC CTT TGT TCT CGA TGG TCA CCG G-3′); Sγ1 region, Sγ1F2 (5′-CTA GGA GTG TAG GGG ACC AAG CTG AGC A-3′) and Sγ1R2 (5′-AGC TCA TCC CCT ACA CCC TAA CCT G-3′). Primer sequences for other loci are available on request. Each set of the ChIP experiment presented showed similar results in more than three independent experiments.
γ-H2AX Immunostaining. CH12F3-2 cells were layered on a slide glass by cytospin and fixed for 5 min with 1% paraformaldehyde in PBS at room temperature, followed by another 5-min incubation in methanol at -20°C. Blocking was done with 8% BSA/PBS for 20 min and staining was done with an anti-γ-H2AX monoclonal antibody (Upstate Biotechnology) and a FITC-labeled F(ab′)2 fragment of goat anti-mouse IgG1 (Southern Biotechnology Associates). Nuclei were stained by 4′,6-diamidino-2-phenylindole (DAPI, Molecular Probes).
Results and Discussion
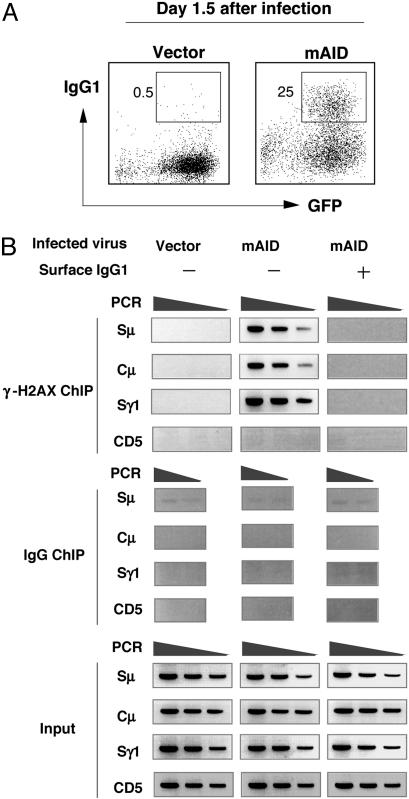
AID-Dependent γ-H2AX Focus Formation at the IgH Locus. We first tried to establish the ChIP assay for CSR-associated γ-H2AX focus formation in the IgH locus. To examine whether the ChIP assay for the γ-H2AX association with the IgH locus is AID-dependent, AID-/- spleen B cells retrovirally infected with or without AID cDNA insert were analyzed for surface IgG1 expression by FACS and for γ-H2AX focus formation by the ChIP assay. AID-/- spleen B cells infected by vector alone did neither switch isotype nor form γ-H2AX foci in the IgH locus (Sμ, Cμ, and Sγ1) (Fig. 1A and B). When AID was expressed in AID-/- spleen cells, the γ-H2AX ChIP signal appeared in the IgH locus of nonswitched (IgG1-) cells that should include B cells in the process of CSR. However, IgG1+ cells that had completed CSR did not show the γ-H2AX focus formation in the IgH locus. A non-Ig locus, such as CD5, did not show accumulation of γ-H2AX by the ChIP assay. The results support the idea that γ-H2AX focus formation in the IgH locus is an AID-dependent process and a transient intermediate step of CSR.
Fig. 1.
AID-dependent γ-H2AX ChIP signal was predominant on preswitching cells. (A) Spleen cells from AID-/- mice were retrovirally infected with either mAID-IRES-GFP construct or vector only. Percentages of IgG1+ cells 1.5 days postinfection were indicated in the FACS profile. (B) GFP+ cells were sorted to obtain IgG1- and IgG1+ populations for γ-H2AX ChIP. PCR of Sμ,Cμ, and Sγ1 regions was carried out with 2-fold serial dilutions of ChIP DNA and 5% of input DNAs. PCR of the 3′-flanking region of Sγ core repeat is shown.
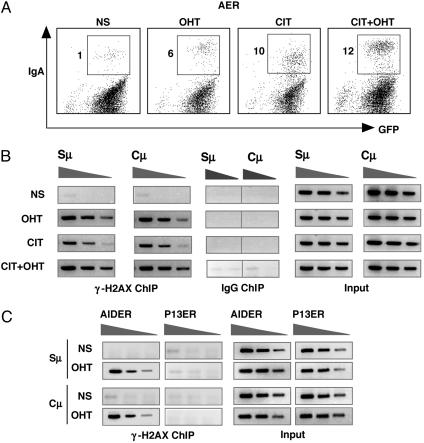
Specificity of γ-H2AX ChIP for IgH Locus. To carry out protein synthesis inhibition experiments, we used a derivative of the CH12F3-2 cell line (AER) that expressed AIDER, AID fused with the hormone-binding domain of the estrogen receptor (ER) (13). Although the AIDER-IRES-GFP transgene was actively expressed by a tetracycline (Tet)-repressible promoter, the AIDER protein produced remained inactive before addition of tamoxifen (OHT) and stable for a short period of cycloheximide treatment. As shown before (13), the addition of OHT alone induced CSR in AER cells although the efficiency was slightly less than by stimulation with CD40L, IL-4, and TGF-β1 (CIT) (Fig. 2A). Stimulation with both OHT and CIT further enhanced CSR in AER cells by induction of endogenous AID expression and probably more efficient transcription of the target. We then carried out ChIP experiments under these activation conditions. We observed that the OHT or CIT activation induced more or less similar levels of γ-H2AX accumulation at the Sμ region (Fig. 2B), indicating that activation of AIDER by the addition of OHT alone could induce DNA breakage. Accumulation of γ-H2AX at the Sμ region was increased by further addition of CIT stimulation in parallel with enhanced CSR although the ChIP assay efficiency is not strictly quantitative. Another negative control is the absence of the γ-H2AX focus formation in the IgH locus in a similar cell line expressing a loss-of-function mutant (P13) of AIDER (10) (Fig. 2C). Protein expression levels of AIDER and P13ER were comparable by Western blotting analysis (data not shown).
Fig. 2.
Association of γ-H2AX at the IgH locus by AIDER activation. (A) AER cells expressing AIDER were stimulated as indicated, and surface IgA expression was examined at day 1 by FACS. NS, not stimulated. (B) γ-H2AX ChIP analysis was done by using similarly stimulated cells for a period of 6 h. PCR amplification of Sμ and Cμ was shown by using 2-fold diluted ChIP samples and 5% of input DNA. (C) Two CH12F3-2-derived lines containing AIDER and its nonfunctional mutant (P13ER) were treated by OHT, and γ-H2AX ChIP was carried out after 6 h of stimulation as above.
These results confirmed that the γ-H2AX ChIP assay is a faithful recapitulation of the γ-H2AX focus formation at the IgH locus, which was previously detected by immunostaining coupled with fluorescence in situ hybridization (FISH) (7), and provides a rapid analysis of AID-induced DNA breakage events associated with CSR.
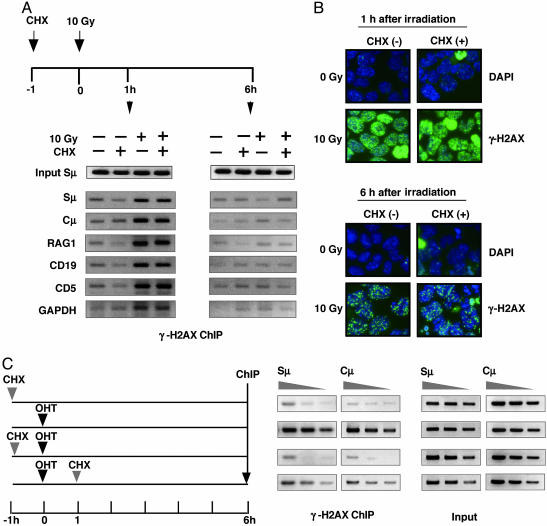
Requirement of de Novo Protein Synthesis for γ-H2AX Focus Formation at the IgH Locus. Using this assay, we examined whether the γ-H2AX focus formation at the IgH locus is sensitive to a protein synthesis inhibitor, cycloheximide. First, to exclude the possibility that cycloheximide acts as a general inhibitor of γ-H2AX focus formation, we tested the effect of cycloheximide on the γ-H2AX focus formation induced by γ-ray irradiation. Cycloheximide did not affect the level of γ-H2AX focus formation at the IgH locus as well as non-Ig genes by ChIP assay at 1 and 6 h after irradiation (Fig. 3A). The majority of DSBs induced by γ-ray irradiation was repaired by 6 h. Similar results were obtained by the immunostaining assay (Fig. 3B). These results are consistent with a previous report (22).
Fig. 3.
γ-H2AX ChIP of IgH locus is inhibited by cycloheximide. (A) Radiation-induced γ-H2AX focus formation is insensitive to cycloheximide treatment. CH12F3-2 cells were first cultured in the presence and absence of cycloheximide (CHX) (0.2 μg/ml) for 1 h and then exposed to either 0 Gy or 10 Gy γ-radiation. Cells were collected for γ-H2AX ChIP analysis 1 h and 6 h after irradiation, as depicted in the scheme. ChIP PCR was performed for several non-Ig loci indicated together with Sμ and Cμ. (B) Anti-γ-H2AX staining pattern of the irradiated cells used for ChIP analysis. Substantial loss of ChIP signal at 6 h corresponded to residual focus intensity as repair of radiation-induced breaks was equally operative in the presence and absence of CHX. The apparent difference in signals of A and B at 6 h after irradiation is due to the fact that anti-γ-H2AX immunostaining detects foci at any loci, whereas the γ-H2AX ChIP assay measures foci at specific genes. (C) AER cells were stimulated by OHT, and CHX (0.2 μg/ml) was added to the culture either 1 h before or after the OHT addition as illustrated in the diagram. γ-H2AX ChIP analysis was done after 6 h of OHT treatment. IgH locus (Sμ and Cμ)-specific PCR was performed by using 2-fold serially diluted ChIP samples and 5% of input DNA.
Then, to test whether AID-induced DSB in the IgH locus depends on de novo protein synthesis, AIDER expressed in AER cells was activated by OHT in the absence or presence of cycloheximide, followed by γ-H2AX ChIP analysis at indicated time points (Fig. 3C). Pretreatment with cycloheximide abolished the γ-H2AX association with the Sμ and Cμ region DNA, which was induced by activated AIDER. Addition of cycloheximide at the same time with OHT (data not shown) or 1 h after OHT showed reduced inhibitory effects. We confirmed that germ-line transcripts of Sμ and Sα regions were not significantly inhibited by the cycloheximide treatment (ref. 13 and data not shown). These observations not only confirmed the previous report that de novo protein synthesis is required for CSR (13) but also demonstrated that new protein synthesis is required for generation of AID-dependent DNA breakage.
Is DNA Deamination by AID Physiological? AID has been shown to deaminate C in DNA in vitro as well as in E. coli (14, 15, 17-20). Interestingly, bona fide RNA-editing enzyme APOBEC-1 can also deaminate C in E. coli DNA (31) although APOBEC-1 cannot induce somatic hypermutation or CSR in B cells (32, 33). Furthermore, the amount of AID protein used in in vitro reactions to detect DNA deamination is far in excess of that of in vivo reaction. It is also important to note that chromatin DNA may be a much less efficient substrate for deamination compared with naked DNA, as shown in uracil DNA glycosylase activity of UNG (34). In addition, U removal activity of UNG is shown to be dispensable for CSR (35). It remains to be shown whether DNA deamination activity of AID is required for CSR.
Supporting Evidence for the RNA-Editing Hypothesis. The requirement of de novo protein synthesis for AID-dependent DNA cleavage fulfills one of the predictions of the RNA-editing hypothesis. γ-H2AX ChIP analysis showed that DSB is introduced within a few hours after AID activation (35). The results indicate that protein synthesis required for DSB must take place very quickly. This observation is also suggestive of RNA editing because modification of preexisting mRNA can produce a novel protein much faster than transcriptional induction of new mRNA.
AID was recently shown to be a nucleo-cytoplasmic shuttle protein (12) like APOBEC-1, which recognizes apoB 100 mRNA through the APOBEC-1 complementation factor in the nucleus, edits this mRNA, and carries the edited mRNA (apoB 48) to the cytoplasm for translation (36). AID is suggested to form a complex with at least two proteins each specific for CSR and somatic hypermutation, although the molecular structure of AID cofactors is not known (10, 37). In addition to the previous evolutionary conservation between AID and APOBEC-1, recent cell biological and biochemical similarities strengthened the RNA-editing hypothesis. Furthermore, dispensability of U removal activity of UNG in CSR (35) forces us to reconsider the current model of DNA deamination in class switching.
Acknowledgments
We thank Drs. S. Fagarasan and T. Doi for critical comments and for reading the manuscript and Ms. A. Takano for technical support. This investigation was supported by Center of Excellence Grant 12CE2006 from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Abbreviations: AID, activation-induced cytidine deaminase; CSR, class switch recombination; APOBEC-1, apoB mRNA-editing catalytic polypeptide 1; UNG, uracil N-glycosylase; DSB, double-strand breakage; ChIP, chromatin immunoprecipitation; AIDER, AID fused with the hormone-binding domain of the estrogen receptor; OHT, 4-hydroxytamoxifen; mAID, mouse AID.
References
- 1.Honjo, T., Kinoshita, K. & Muramatsu, M. (2002) Annu. Rev. Immunol. 20, 165-196. [DOI] [PubMed] [Google Scholar]
- 2.Stavnezer, J., Kinoshita, K., Muramatsu, M. & Honjo, T. (2004) in Molecular Biology of B Cells, eds. Honjo, T., Alt, F. W. & Neuberger, M. (Elsevier Academic, London), pp. 307-319.
- 3.Muramatsu, M., Kinoshita, K., Fagarasan, S., Yamada, S., Shinkai, Y. & Honjo, T. (2000) Cell 102, 553-563. [DOI] [PubMed] [Google Scholar]
- 4.Revy, P., Muto, T., Levy, Y., Geissmann, F., Plebani, A., Sanal, O., Catalan, N., Forveille, M., Dufourcq-Labelouse, R., Gennery, A., et al. (2000) Cell 102, 565-575. [DOI] [PubMed] [Google Scholar]
- 5.Arakawa, H., Hauschild, J. & Buerstedde, J. M. (2002) Science 295, 1301-1306. [DOI] [PubMed] [Google Scholar]
- 6.Muramatsu, M., Sankaranand, V. S., Anant, S., Sugai, M., Kinoshita, K., Davidson, N. O. & Honjo, T. (1999) J. Biol. Chem. 274, 18470-18476. [DOI] [PubMed] [Google Scholar]
- 7.Petersen, S., Casellas, R., Reina-San-Martin, B., Chen, H. T., Difilippantonio, M. J., Wilson, P. C., Hanitsch, L., Celeste, A., Muramatsu, M., Pilch, D. R., et al. (2001) Nature 414, 660-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Catalan, N., Selz, F., Imai, K., Revy, P., Fischer, A. & Durandy, A. (2003) J. Immunol. 171, 2504-2509. [DOI] [PubMed] [Google Scholar]
- 9.Anant, S. & Davidson, N. O. (2001) Curr. Opin. Lipidol. 12, 159-165. [DOI] [PubMed] [Google Scholar]
- 10.Ta, V. T., Nagaoka, H., Catalan, N., Durandy, A., Fischer, A., Imai, K., Nonoyama, S., Tashiro, J., Ikegawa, M., Ito, S., et al. (2003) Nat. Immunol. 4, 843-848. [DOI] [PubMed] [Google Scholar]
- 11.Barreto, V., Reina-San-Martin, B., Ramiro, A. R., McBride, K. M. & Nussenzweig, M. C. (2003) Mol. Cell 12, 501-508. [DOI] [PubMed] [Google Scholar]
- 12.Ito, S., Nagaoka, H., Shinkura, R., Begum, N., Muramatsu, M., Nakata, M. & Honjo, T. (2004) Proc. Natl. Acad. Sci. USA 101, 1975-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doi, T., Kinoshita, K., Ikegawa, M., Muramatsu, M. & Honjo, T. (2003) Proc. Natl. Acad. Sci. USA 100, 2634-2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petersen-Mahrt, S. K., Harris, R. S. & Neuberger, M. S. (2002) Nature 418, 99-103. [DOI] [PubMed] [Google Scholar]
- 15.Ramiro, A. R., Stavropoulos, P., Jankovic, M. & Nussenzweig, M. C. (2003) Nat. Immunol. 4, 452-456. [DOI] [PubMed] [Google Scholar]
- 16.Rada, C., Williams, G. T., Nilsen, H., Barnes, D. E., Lindahl, T. & Neuberger, M. S. (2002) Curr. Biol. 12, 1748-1755. [DOI] [PubMed] [Google Scholar]
- 17.Dickerson, S. K., Market, E., Besmer, E. & Papavasiliou, F. N. (2003) J. Exp. Med. 197, 1291-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pham, P., Bransteitter, R., Petruska, J. & Goodman, M. F. (2003) Nature 424, 103-107. [DOI] [PubMed] [Google Scholar]
- 19.Chaudhuri, J., Tian, M., Khuong, C., Chua, K., Pinaud, E. & Alt, F. W. (2003) Nature 422, 726-730. [DOI] [PubMed] [Google Scholar]
- 20.Sohail, A., Klapacz, J., Samaranayake, M., Ullah, A. & Bhagwat, A. S. (2003) Nucleic Acids Res. 31, 2990-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuhne, C., Tjornhammar, M. L., Pongor, S., Banks, L. & Simoncsits, A. (2003) Nucleic Acids Res. 31, 7227-7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rogakou, E. P., Pilch, D. R., Orr, A. H., Ivanova, V. S. & Bonner, W. M. (1998) J. Biol. Chem. 273, 5858-5868. [DOI] [PubMed] [Google Scholar]
- 23.Downs, J. A., Lowndes, N. F. & Jackson, S. P. (2000) Nature 408, 1001-1004. [DOI] [PubMed] [Google Scholar]
- 24.Casellas, R., Nussenzweig, A., Wuerffel, R., Pelanda, R., Reichlin, A., Suh, H., Qin, X. F., Besmer, E., Kenter, A., Rajewsky, K. & Nussenzweig, M. C. (1998) EMBO J. 17, 2404-2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manis, J. P., Dudley, D., Kaylor, L. & Alt, F. W. (2002) Immunity 16, 607-617. [DOI] [PubMed] [Google Scholar]
- 26.Manis, J. P., Gu, Y., Lansford, R., Sonoda, E., Ferrini, R., Davidson, L., Rajewsky, K. & Alt, F. W. (1998) J. Exp. Med. 187, 2081-2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morita, S., Kojima, T. & Kitamura, T. (2000) Gene Ther. 7, 1063-1066. [DOI] [PubMed] [Google Scholar]
- 28.Nagaoka, H., Muramatsu, M., Yamamura, N., Kinoshita, K. & Honjo, T. (2002) J. Exp. Med. 195, 529-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakamura, M., Kondo, S., Sugai, M., Nazarea, M., Imamura, S. & Honjo, T. (1996) Int. Immunol. 8, 193-201. [DOI] [PubMed] [Google Scholar]
- 30.Weinmann, A. S. & Farnham, P. J. (2002) Methods 26, 37-47. [DOI] [PubMed] [Google Scholar]
- 31.Harris, R. S., Petersen-Mahrt, S. K. & Neuberger, M. S. (2002) Mol. Cell 10, 1247-1253. [DOI] [PubMed] [Google Scholar]
- 32.Eto, T., Kinoshita, K., Yoshikawa, K., Muramatsu, M. & Honjo, T. (2003) Proc. Natl. Acad. Sci. USA 100, 12895-12898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fugmann, S. D., Rush, J. S. & Schatz, D. G. (2004) Eur. J. Immunol. 34, 844-849. [DOI] [PubMed] [Google Scholar]
- 34.Beard, B. C., Wilson, S. H. & Smerdon, M. J. (2003) Proc. Natl. Acad. Sci. USA 100, 7465-7470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Begum, N., Kinoshita, K., Kakazu, N., Muramatsu, M., Nagaoka, H., Shinkura, R., Biniszkiewicz, D., Boyer, L. A., Jaenisch, R. & Honjo, T. Science, in press. [DOI] [PubMed]
- 36.Chester, A., Somasekaram, A., Tzimina, M., Jarmuz, A., Gisbourne, J., O'Keefe, R., Scott, J. & Navaratnam, N. (2003) EMBO J. 22, 3971-3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shinkura, R., Ito, S., Begum, N. A., Nagaoka, H., Muramtsu, M., Kinoshita, K., Sakakibara, Y., Hijikata, H. & Honjo, T. (2004) Nat. Immunol. 5, 707-712. [DOI] [PubMed] [Google Scholar]