ABSTRACT
Staphylococcus aureus, an invasive pathogen of humans and animals, requires a specialized ESS pathway to secrete proteins (EsxA, EsxB, EsxC, and EsxD) during infection. Expression of ess genes is required for S. aureus establishment of persistent abscess lesions following bloodstream infection; however, the mechanisms whereby effectors of the ESS pathway implement their virulence strategies were heretofore not known. Here, we show that EssE forms a complex with other members of the ESS secretion pathway and its substrates, promoting the secretion of EsxA, EsxB, EsxC, EsxD, and EssD. During bloodstream infection of mice, the S. aureus essE mutant displays defects in host cytokine responses, specifically in the production of interleukin-12 (IL-12) (p40/p70) and the suppression of RANTES (CCL5), activators of TH1 T cell responses and immune cell chemotaxis, respectively. Thus, essE-mediated secretion of protein effectors via the ESS pathway may enable S. aureus to manipulate host immune responses by modifying the production of cytokines.
IMPORTANCE Staphylococcus aureus and other firmicutes evolved a specialized ESS (EsxA/ESAT-6-like secretion system) pathway for the secretion of small subsets of proteins lacking canonical signal peptides. The molecular mechanisms for ESS-dependent secretion and their functional purpose are still unknown. We demonstrate here that S. aureus EssE functions as a membrane assembly platform for elements of the secretion machinery and their substrates. Furthermore, S. aureus EssE-mediated secretion contributes to the production or the suppression of specific cytokines during host infection, thereby modifying immune responses toward this pathogen.
KEYWORDS: ESS secretion, EssE, IL-12, MRSA, RANTES, effector
INTRODUCTION
Staphylococcus aureus, a commensal of humans and their domesticated animals (1, 2), is also an invasive pathogen that replicates via the formation of abscess lesions in tissues of infected hosts (3, 4). Abscess formation requires staphylococcal coagulases, secreted products associating with host prothrombin to generate a fibrin shield, thereby establishing a physical barrier between the pathogen and the host's immune defenses (5–7). S. aureus lesions attract large numbers of immune cells, predominantly neutrophils and lymphocytes, whose lysis and proliferation in the vicinity of staphylococcal abscess communities is associated with tissue destruction (8). Drainage of the ensuing purulent exudate ensures the spread of S. aureus in infected individuals or transmission to new hosts (9). Without surgical intervention or implementation of effective antibiotic therapy, infected hosts cannot clear S. aureus from deep-seated abscesses or from skeletal and internal organ lesions (10–12).
In addition to manipulating host hemostasis, S. aureus elaborates immune evasive strategies aimed at interfering with the chemotaxis of immune cells, the activation of complement, and opsonophagocytosis or the bactericidal activities of phagocytes (13). Earlier work identified the pathogen's ESS pathway (EsxA/ESAT-6-like secretion system), which is encoded by a cluster of contiguous genes on the staphylococcal chromosome (Fig. 1A) (14, 15). When induced for ess expression during growth in vertebrate blood or serum, S. aureus ESS secretes four small proteins, designated EsxA, EsxB, EsxC, and EsxD (14, 16, 17). Mutations that abrogate ess expression diminish the abundance of abscess lesions and their bacterial load compared to levels of lesions seeded by wild-type S. aureus (14, 18). Further, lesions derived from mutants with defects in ess expression are formed more rarely and cleared more frequently than abscesses populated with wild-type S. aureus (15).
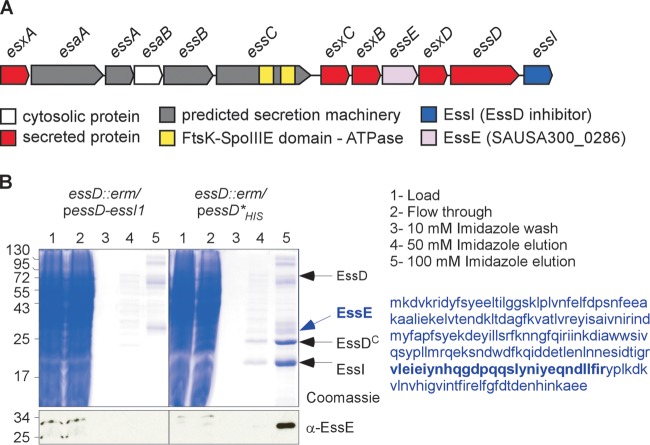
FIG 1.
EssE is a ligand of EssD. (A) Schematic representation of the ESS cluster in S. aureus. (B) Cultures of S. aureus strain USA300 essD::erm carrying plasmid pessD-essI1 or pessD*His to produce wild-type EssD or the nontoxic Leu546Pro variant with a C-terminal histidine tag were grown at 37°C and centrifuged, and sedimented bacteria were lysed to generate cleared lysates that were treated with DDM to solubilize membrane proteins for purification over Ni-NTA (lanes 1). The flowthrough containing unbound proteins (lanes 2), 10 mM imidazole wash (lanes 3), and the 50 and 100 mM imidazole elution fractions (lanes 4 and 5) were separated by SDS-PAGE and either stained with Coomassie blue or transferred to PVDF membrane for immunoblot analyses with the anti-EssE polyclonal serum. Numbers to the left indicate the mobility of molecular mass markers. Arrows point to bands corresponding to proteins identified by mass spectrometry. The sequence of EssE is shown in blue, and the region identified by mass spectrometry is in bold.
The mechanisms whereby the S. aureus ESS pathway implements its immune evasive strategies in the host were heretofore not known. Here, we show that essE, encoding a membrane-associated protein, is required for S. aureus secretion of EsxA, EsxB, EsxC, and EsxD. EssE forms a complex with EsxC and with other components of the ESS pathway, including EssC, EssD, and EssI. In the accompanying paper (19), we report that EssD is also secreted by the ESS pathway and that the protein bears a C-terminal nuclease domain (EssDC), whose activity is inhibited by EssI in the bacterial cytoplasm. Here, we report that interaction with EssE in the cytosol of S. aureus is important for EssD stability. Unlike wild-type S. aureus, essE mutants display defects in host cytokine responses, specifically the production of interleukin-12 (IL-12) (p40/p70) and the suppression of RANTES (CCL5), activators of TH1 T cell responses and T cell chemotaxis, respectively (11, 20). We propose that essE-mediated secretion of protein effectors via the ESS pathway may enable S. aureus to manipulate host immune responses by modifying the production of specific cytokines.
RESULTS
EssE copurifies with EssD.
S. aureus expression of essD is required for Esx protein secretion by the ESS pathway (18, 19). EssD is located in the bacterial membrane owing to the presence of a hydrophobic domain between residues 217 and 250 (18). Nevertheless, S. aureus also secretes EssD into the extracellular medium (19). We wondered whether retention of EssD at the plasma membrane could be caused by interaction with other protein ligands and used a biochemical approach to test this model. First, the essD open reading frame was extended at the 3′ end with 10 codons for C-terminal histidine residues (His10), enabling affinity chromatography purification on Ni-nitrilotriacetic acid (Ni-NTA)–Sepharose (19). Cloning of essD is associated with bacterial toxicity owing to the nuclease activity of EssD products (19). Fortuitously, we isolated essD*His, a mutant with a single codon substitution (Leu546Pro), abolishing nuclease activity of EssD*His, while restoring secretion of Esx proteins and of EssD*His (19).As a control for affinity chromatography, we expressed nontagged essD together with essI1, encoding a cytoplasmic polypeptide that binds to and inhibits the EssD nuclease (19). Ultracentrifuged lysates of S. aureus essD::erm carrying plasmids pessD-essI1 and pessD*His were subjected to solubilization with n-dodecyl-β-d-maltopyranoside (DDM) followed by Ni-NTA affinity chromatography to isolate products from plasmid-borne essD variants (Fig. 1B). Bound proteins were eluted with 10, 50, and 100 mM imidazole buffer, and aliquots of the eluate were analyzed by separation on SDS-PAGE gels and Coomassie blue staining (Fig. 1B). Compared to the EssD control, several proteins specifically eluted from Ni-NTA resin that had been charged with EssD*His lysate (Fig. 1B). Immunoblot analysis validated this interpretation, as anti-EssD identified EssD*His in the 100 mM imidazole eluate from Ni-NTA–Sepharose charged with EssD*His lysate but not in EssD-charged Ni-NTA–Sepharose eluate (data not shown). Gel slices with proteins marked by arrowheads in Fig. 1B were excised; proteins were cleaved with trypsin (C terminally of positively charged amino acids) and identified by mass spectrometry measurements of positively charged peptides, whose average masses were within the error rate for calculated masses assigned by in silico trypsin cleavage of translation products from the S. aureus genome (21). Mass spectrometry and bioinformatics analyses identified full-length EssD, its 24-kDa C-terminal nuclease domain (see below), EssE, and the EssD nuclease inhibitor EssI (20 kDa) (Fig. 1B). The EssD nuclease and EssI eluted in equimolar amounts, suggesting that the two proteins form an equimolar complex. The identity of EssD and EssI was also confirmed by Edman degradation (19). The 224-residue EssE is encoded by essE (SAUSA_0286), which is located between esxB and esxD within the ESS pathway gene cluster (Fig. 1A). Homology searches with EssE polypeptide sequence failed to identify domains with known or predicted biochemical functions. The essE open reading frame was cloned into pGEX-2TK to produce the translational hybrid glutathione S-transferase (GST)–EssE that was purified from cleared Escherichia coli lysates by affinity chromatography followed by thrombin treatment to remove GST. A rabbit was immunized with purified EssE to generate polyclonal antibodies specific for the recombinant polypeptide (anti-EssE serum). Anti-EssE was used to confirm the copurification of EssE from S. aureus extracts containing EssD*His but not from lysate with untagged EssD (Fig. 1B).
EssE is required for S. aureus Esx protein secretion.
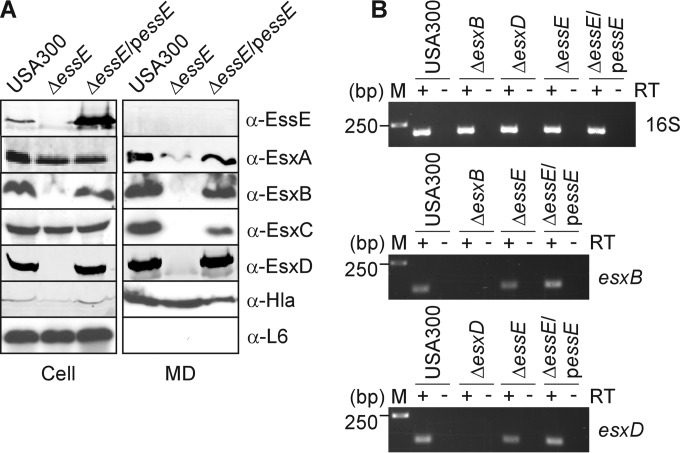
To avoid polar effects of essE mutations on the expression of flanking genes in the ESS cluster (Fig. 1), pKOR1 technology was used to generate an in-frame deletion of 194 codons in the essE gene of methicillin-resistant S. aureus (MRSA) isolate USA300 LAC (SAUSA300_0286) (22). For plasmid complementation studies, we constructed pessE for plasmid-borne, constitutive PhrpK-mediated expression of essE. Mid-log-phase cultures of the wild-type, ΔessE, and ΔessE/pessE variant S. aureus strains were fractionated to separate the extracellular medium and cell lysates (Fig. 2A, MD and cell, respectively). Proteins in each compartment were analyzed by SDS-PAGE and immunoblotting, which revealed that EssE was associated with S. aureus USA300 cells and was not secreted into the extracellular medium (Fig. 2A). The ΔessE mutant did not express the essE gene; however, EssE production was restored following transformation of ΔessE mutant cells with pessE (ΔessE/pessE strain) (Fig. 2A). As expected, S. aureus USA300 secreted EsxA, EsxB, EsxC, and EsxD into the extracellular medium (Fig. 2A). The ΔessE mutant did not secrete EsxB and EsxC and also displayed reduced secretion of EsxA (Fig. 2A). Additionally, the production of EsxB and EsxD was greatly diminished in ΔessE staphylococci (Fig. 2A). However, this was not due to loss of gene transcription, as shown by reverse transcription-PCR (RT-PCR) experiments; both esxB and esxD transcripts continued to be synthesized in the ΔessE mutant in a manner comparable to that of USA300 (Fig. 2B). All phenotypic defects of the ΔessE mutation on ESS secretion were ameliorated when mutant staphylococci were transformed with pessE (Fig. 2A and B). As controls, both wild-type and ΔessE mutant staphylococci secreted α-hemolysin (Hla) into the extracellular medium, whereas ribosomal protein L6 (L6) remained associated with the bacterial cells (Fig. 2A). Thus, the 194-codon in-frame deletion abolishes essE expression, causing specific defects in the secretion of Esx proteins by the S. aureus ESS pathway without impacting bacterial lysis or the secretion of Hla via the canonical Sec secretion pathway.
FIG 2.
EssE is required for the secretion of Esx proteins. (A) To assess production and secretion of proteins encoded by the ESS cluster, S. aureus cultures of strain USA300 or its isogenic ΔessE mutant with or without complementing plasmid (ΔessE/pessE strain) were grown to an A600 of 1.0 and centrifuged to separate the medium (MD) fraction from intact cells in the sediment. Cells were suspended in PBS buffer and lysed with lysostaphin to release all cellular content (Cell). Proteins in the medium and cell extracts were precipitated by the addition of TCA to a final concentration of 10% prior to separation by SDS-PAGE, followed by transfer for immunoblot analyses. Immunoblotting was performed using rabbit polyclonal antibodies specific for EssE, EsxA, EsxB, EsxC, EsxD, alpha-toxin (Hla), and ribosomal protein L6 (L6) proteins. Hla is a well-characterized protein secreted in an Ess-independent manner. L6 is a cytoplasmic protein. (B) To assess whether expression of esxB and esxD was affected following deletion of essE, S. aureus cultures were grown as described for panel A for extraction and purification of total mRNA. Reverse transcriptase (RT) was used to generate cDNA of esxB, esxD, and 16S transcripts. Expression of esxB and esxD in the USA300, ΔessE, and ΔessE/pessE strains was qualitatively assessed by running RT-PCR products on agarose gels. Total mRNA was also extracted from strains lacking esxB or esxD (ΔesxB or ΔesxD strain) to unambiguously identify the cognate transcripts. Amplification using 16S primers was performed as a calibration control. Numbers to the left of the gels indicate DNA markers and their corresponding lengths in base pairs. Positive and negative signs above gels indicate the inclusion or omission of RT during the experimental steps.
EssE impacts S. aureus processing of EssD.
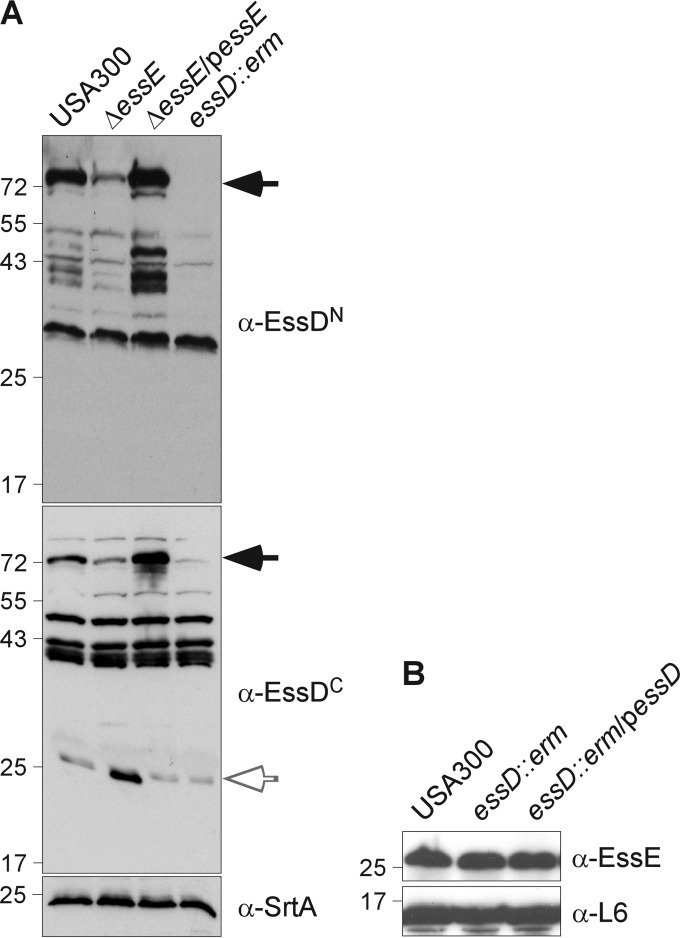
Earlier work reported that mutations in genes for substrates of ESS secretion (esxB, esxC, or esxD) resulted in defects of S. aureus essD expression (17, 18). The Esx secretion defect for mutants with defects in other ess genes had also been attributed to the loss of essD expression (17). To determine whether such a defect occurs in the essE mutant, we examined whole-cell lysates of wild-type S. aureus USA300 LAC as well as the ΔessE, ΔessE/pessE, and essD::erm mutants by immunoblotting with antibodies raised against amino acids 1 to 200 (anti-EssDN) or 432 to 617 (anti-EssDC) of the EssD polypeptide (617 residues). When samples were probed with anti-EssDN, a 72-kDa immunoreactive species was detected in cell lysates of the wild-type but not of the essD mutant staphylococci (Fig. 3A, black arrow). In addition to cross-reactive species that were also identified in essD mutant lysate, anti-EssDN detected immunoreactive species with ∼43-kDa mobility on SDS-PAGE gels (Fig. 3A). Immunoblotting with anti-EssDC identified a 72-kDa EssD (black arrow) as well as a smaller 24-kDa product (white arrow). These data suggest that the 72-kDa species represents full-length EssD, which is cleaved into the stable C-terminal fragment (24-kDa protein reactive with anti-EssDC). The abundance of full-length EssD (72 kDa) was diminished in the S. aureus ΔessE mutant while that of the C-terminal 24-kDa fragment was increased. Processing of the N-terminal EssD domain was not affected by the ΔessE mutation. Compared to wild-type staphylococci, the ΔessE/pessE variant produced similar amounts of full-length EssD and N- and C-terminal cleavage products, indicating that the observed defects on EssD processing are all attributable to essE expression.
FIG 3.
EssE is required to stabilize EssD. (A) S. aureus cultures of strain USA300 or its isogenic ΔessE, ΔessE/pessE, and essD::erm variants were grown to an A600 of 1.0, and lysostaphin was added to the whole culture. Proteins were TCA precipitated and separated by SDS-PAGE as described in the legend to Fig. 2A. Immunoblot analyses were performed using rabbit polyclonal antibodies specific for the N- and C-terminal domains of EssD (α-EssDN and α-EssDC) and membrane protein SrtA. Arrows point to various polypeptides that are specifically recognized by EssD antibodies. (B) Extracts of the USA300 strain or its isogenic essD::erm variant with or without complementing plasmid (pessD) were prepared as described for panel A and examined by immunoblotting for the presence of EssE and ribosomal protein L6.
To determine whether essD expression reciprocally affects the abundance of EssE, we analyzed S. aureus extracts by immunoblotting with antibodies specific for EssE (anti-EssE) (Fig. 3B). However, mutations in essD did not affect the abundance of EssE in staphylococcal cells. As a control, staphylococcal cell extracts were probed with antibodies specific for ribosomal protein L6 (anti-L6), validating that similar amounts of cell extracts had been analyzed in the immunoblot experiments.
Affinity chromatography of EssE.
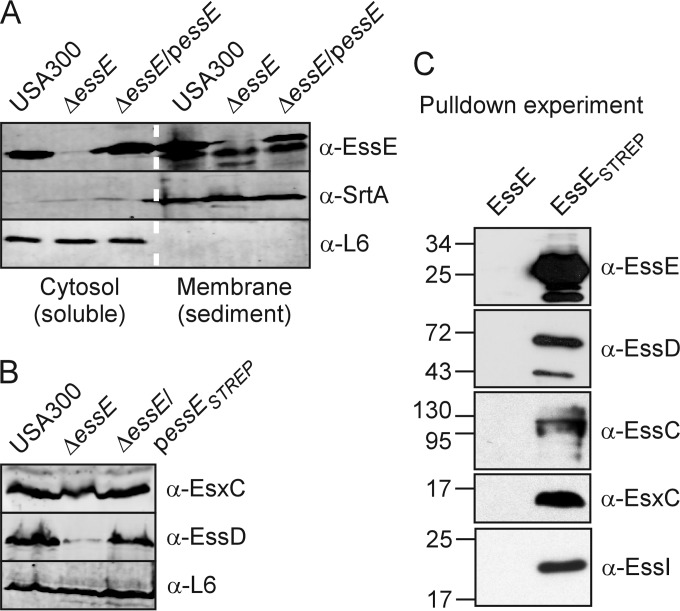
Several components of the ESS pathway encompass hydrophobic segments and are predicted to be localized in the bacterial membrane (14). When subjected to the PSORT (psort.hgc.jp) and TMHMM (cbs.dtu.dk/services/TMHMM) algorithms, EssE, however, was not identified as a membrane protein; it lacks segments with ≥10 consecutive hydrophobic amino acids (23). To determine the subcellular localization of EssE, the cell walls of staphylococci were removed with lysostaphin, bacterial cells were lysed in a French press, and lysates were ultracentrifuged (100,000 × g for 2 h). Soluble proteins were separated, and the supernatant (cytosol) from insoluble proteins sedimented with the membrane fraction (membrane) (Fig. 4A). Proteins in both fractions were precipitated with trichloroacetic acid (TCA), solubilized in sample buffer, and analyzed by immunoblotting. As a control, sortase A (SrtA), a membrane protein, was found predominantly in the membrane fraction, while L6 remained soluble in the cytosol (Fig. 4A). EssE was unambiguously identified by comparing cytosol and membrane fractions from the wild-type and ΔessE and ΔessE/pessE mutant strains (Fig. 4A). Further, EssE was observed with equal abundance in the cytosol and membrane fractions (Fig. 4A). Of note, the distribution of EssE between cytosol and membrane fractions was not affected by plasmid-borne expression of essE in ΔessE/pessE variant staphylococci (Fig. 4A).
FIG 4.
Identification of the EssE complex. (A) S. aureus cultures of strain USA300 or its isogenic ΔessE mutant with or without complementing plasmid (ΔessE/pessE strain) were grown to an A600 of 1.0. Cells were sedimented by centrifugation and lysed with lysostaphin, and lysates were subjected to ultracentrifugation (100,000 × g for 2 h). The supernatant and pellet fractions containing soluble proteins from the cytosol and membrane proteins from the sediment were separated, subjected to SDS-PAGE, and transferred to PVDF membranes for immunoblot analysis using antibodies against EssE as well as the membrane and cytosolic proteins SrtA and L6, respectively. (B) S. aureus cultures of strain USA300 or its isogenic ΔessE mutant with or without complementing plasmid producing EssE with a Strep-Tag (ΔessE/pessESTREP strain) were grown to an A600 of 1.0. Lysostaphin was added to the whole culture, and TCA was added to precipitate proteins and examine extracts as described in the legend to Fig. 2A. Immunoblot analyses were performed using anti-EssDN, anti-EsxC, and anti-L6. (C) S. aureus cells producing EssE without (EssE) or with the Strep-Tag (EssESTREP) were grown to an A600 of 2.0, sedimented, and lysed with lysostaphin. Cell lysates were suspended in DDM, and insoluble materials were removed by centrifugation at 100,000 × g. Detergent-soluble proteins were flowed by gravity over Strep-Tactin–Sepharose beads, washed extensively with PBS, and eluted with 2.5 mM desthiobiotin. Bound proteins in eluates were separated by SDS-PAGE and electrotransferred to PVDF membranes for identification by immunoblotting.
To enable biochemical analyses of EssE, we generated EssESTREP with an 8-amino-acid Strep-Tag extension at the C terminus of the polypeptide. Plasmid-borne expression of essESTREP by the ΔessE/pessESTREP variant restored the slightly reduced production of EsxC and the diminished abundance of EssD in the ΔessE mutant to a level observed in wild-type staphylococci (Fig. 4B). Further, the ΔessE/pessESTREP mutant secreted EsxA, EsxB, EsxC, and EsxD in a manner indistinguishable from that of wild-type and ΔessE/pessE variant staphylococci (data not shown). S. aureus ΔessE/pessE and ΔessE/pessESTREP strains were grown to an A600 of 2.0, and bacteria were sedimented by centrifugation. Following cell wall removal with lysostaphin, staphylococci were lysed using a French press, and membranes were sedimented by ultracentrifugation, incubated with 2% DDM on ice, and again subjected to ultracentrifugation. Proteins solubilized by 2% DDM were subjected to affinity chromatography on Strep-Tactin–Sepharose and eluted with desthiobiotin buffer. Immunoblot analysis of eluates revealed affinity chromatography purification of EssESTREP but not of EssE (Fig. 4C). Immunoblotting with a panel of rabbit antibodies raised against purified recombinant components of the S. aureus ESS pathway identified copurification of EsxC, EssC, EssD, and EssI during affinity chromatography of lysate from staphylococci with EssESTREP but not with EssE (Fig. 4C). We also examined chromatography eluates for EssESTREP copurification with EsxA, EsxB, and EsxD but failed to detect the substrates in the EssESTREP eluate (data not shown). Clearly, a protein complex can be isolated with tagged EssE. The secreted substrates EsxC and EssD are enriched in this complex, and at least one of four presumed secretion machinery components of the ESS pathway, the EssC ATPase (Fig. 1), is also recruited to this complex.
Mouse cytokine responses during bloodstream infection with wild-type and ΔessE mutant S. aureus.
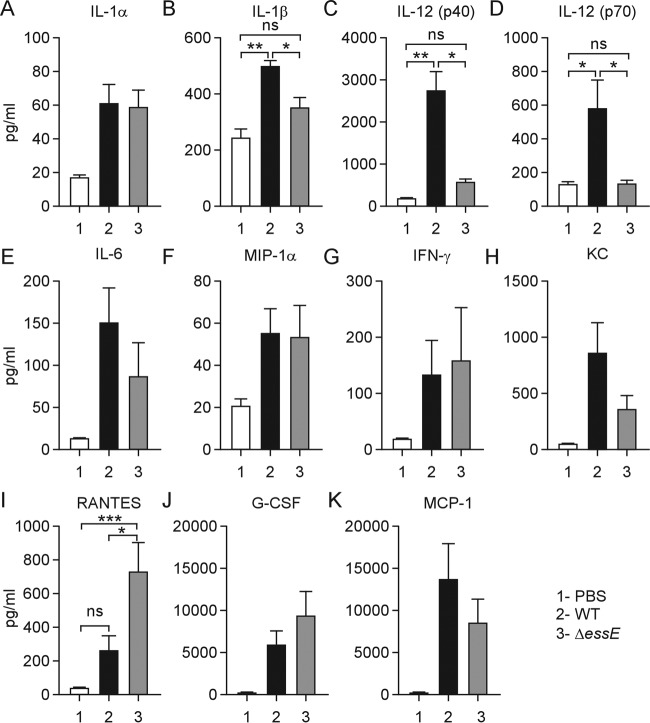
Earlier work reported that during mouse lung infection, S. aureus secretion of α-hemolysin, a pore-forming toxin, promotes inflammasome activation by providing access to pathogen-associated molecular patterns (PAMPs), thereby activating proinflammatory responses via the secretion of IL-1β (24, 25). We sought to study the impact of the ESS pathway on host cytokine responses in a mouse model of S. aureus bloodstream infection (10). Cohorts (n = 5) of C57BL/6 mice were infected by intravenous inoculation with 5 × 107 CFU of S. aureus USA300. Cytokine responses were analyzed in serum samples from cardiac blood drawn 12 h after infection. Compared to mock treatment in mice, S. aureus USA300 infection stimulated IL-1α, IL-1β, IL-6, IL-12 (p40/70), RANTES, keratinocyte-derived chemokine (KC), monocyte chemotactic protein 1 (MCP-1), macrophage inflammatory protein 1α (MIP-1α), granulocyte colony-stimulating factor (G-CSF), and gamma interferon (IFN-γ) signaling (Fig. 5). During infection with the ΔessE mutant, the serum concentrations of IL-1β and IL-12 (p40/70) were diminished, whereas bloodstream release of RANTES was increased (Fig. 5). These data suggest that the ESS pathway contributes to S. aureus activation of proinflammatory cytokine responses such as IL-1β secretion. As reported earlier, S. aureus bloodstream infection of mice promoted the secretion of IL-12 (p40/70) (26), a heterodimeric cytokine comprised of p40 and p35 (27, 28). IL-12 is produced in monocytes, macrophages, and dendritic cells to promote development and activation of cytotoxic T lymphocytes, natural killer cells, lymphokine-activated killer cells, and macrophages (26). Thus, the S. aureus ESS pathway appears to activate immune cells for the secretion of IL-12 (p40/70) as the increased secretion of this cytokine was abolished during mouse infection with the ΔessE mutant (Fig. 5). Of note, secretion of RANTES (CCL5), a chemoattractant for memory T lymphocytes and monocytes secreted by T cells, was enhanced during bloodstream infection with the ΔessE mutant (Fig. 5).
FIG 5.
Cytokine profiling of mice infected with S. aureus. C57BL/6 mice (n = 5) were infected by intravenous injection with 5 × 107 CFU of S. aureus USA300 or its ΔessE variant or were mock infected (PBS). Blood was collected at 12 h postinfection, and serum cytokine levels were determined by enzyme-linked immunosorbent assay. Data were averaged, and standard errors of the means were calculated and analyzed for significant differences with one-way ANOVA using Bonferroni's multiple-comparison test. The statistical analysis is shown only for the groups that exhibited a significant difference between the wild-type (WT) and ΔessE strain data sets (*, P < 0.05; **, P < 0.01; ***, P < 0.01).
Macrophage responses to infection with wild-type and ΔessE mutant S. aureus.
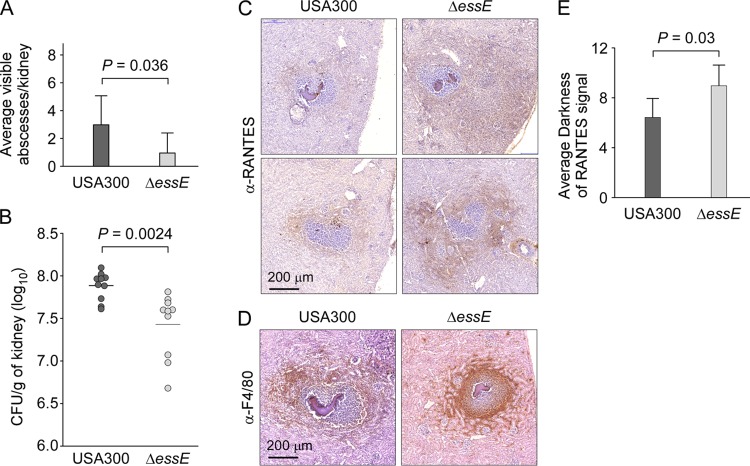
Our data suggest that during bloodstream infection, S. aureus may employ the ESS pathway to simultaneously promote the activation of proinflammatory cytokines while also restricting the trafficking of immune cells to sites of staphylococcal infection. To examine the impact of altered cytokine responses on staphylococcal disease, cohorts (n = 8 to 10) of C57BL/6 mice were infected by intravenous injection with 5 × 106 CFU of S. aureus USA300 or the ΔessE mutant. Five days after challenge, mice were euthanized and necropsied, and renal tissues were analyzed for the enumeration of abscess lesions in histopathology slides and quantification of bacterial load in tissue homogenates (Fig. 6A and B). USA300 infection caused an average of 3.0 surface abscesses per kidney while ΔessE mutant staphylococci produced an average of 0.95 surface abscesses (Fig. 6A) (P = 0.036). The average load of S. aureus USA300 per gram of kidney tissues was 7.9 log10 CFU, and it was 7.4 log10 CFU for the isogenic ΔessE mutant (Fig. 6B) (P = 0.0024), a reduction similar to that observed for other mutant alleles of the ESS cluster (15, 18). Immunohistochemical analysis of thin-sectioned tissue was used to assess the abundance of anti-F4/80-positive macrophages and of RANTES production in renal abscess lesions. Compared to lesions elicited by wild-type S. aureus, abscesses formed by the ΔessE mutant displayed increased staining with RANTES (Fig. 6C). Image analysis using Fiji software revealed that the average darkness staining for RANTES increased by 41% in the ΔessE mutant compared to that in USA300 (Fig. 6E) (P = 0.03). Immunohistochemistry using anti-F4/80 antibody revealed an infiltration of macrophages within abscess lesions formed by the ΔessE mutant (Fig. 6D). While macrophages were also recruited to lesions formed by USA300, the staining mostly surrounded abscesses (Fig. 6D). Thus, the ΔessE mutation not only affects Esx protein secretion and the production of cytokines during bloodstream infection but also impacts the ability of staphylococci to establish infectious lesions and to manipulate cellular immune responses of the host.
FIG 6.
essE mutants exhibit altered abscesses. Staphylococcal replication in kidneys was measured 5 days postinfection after injection of 5 × 106 CFU of USA300 or its isogenic essE mutant in mice (groups of 8 to 10). (A and B) Kidneys were removed during necropsy to enumerate surface abscesses and subsequently homogenized in 1% Triton X-100 for plating and enumeration of CFU. Statistical analyses were performed using an unpaired two-tailed Student's t test with Welch's correction (A) and a two-tailed Mann-Whitney test (B). (C to E) Kidney sections were taken at 12-μm intervals, and abscesses were first stained with H&E to identify abscesses with staphylococcal communities (dark purple and denser stain in the center of slides) surrounded by immune cells. Next, the sections were incubated with anti-RANTES antibody or F4/80 antibody, a macrophage-specific marker. Following incubation with a secondary HRP-conjugated antibody and development, brown staining reflected the presence of macrophages outside the fibrin cuffs and surrounding USA300 lesions while the staining penetrated ΔessE lesions. Representative images are shown. Anti–RANTES signals were quantified using Fiji software, and values are reported as average darkness. Statistical analysis was performed using a two-tailed Mann-Whitney test.
DISCUSSION
Type VII secretion systems (T7SS) were discovered in Mycobacterium tuberculosis to promote the secretion of T cell antigens (ESAT-6 and CFP-10) (29). Genes encoding ESAT-6/CFP-10 and their corresponding secretion machinery are located in a contiguous cluster now designated ESX-1 (30). The ESX-1 T7SS enables translocation of proteins from the bacterial cytoplasm across the phagolysosomal membrane into the cytosol of infected macrophages. Mycobacterial EccC, a transmembrane protein with three ATPases of the SpoIIIE-FtsK-like family, is thought to be responsible for recognizing WXG100 proteins as substrates for T7SS (31). Recent work proposed that binding of a discrete peptide signal in CFP10 (M. tuberculosis EsxB [Mt-EsxB]), a mycobacterial member of the WXG100 family, promotes EccC oligomerization and ATPase activity for transport of folded WXG100 dimers across the bacterial envelope (31).
Homologs of the mycobacterial T7SS that transport WXG100 proteins have also been described in firmicutes, including Bacillus anthracis (32), Bacillus subtilis (33), and Staphylococcus aureus (14). Firmicutes carry some (EsxA, EsxB, and EccC), but not all, of the mycobacterial ESX-1 components required for secretion of ESAT-6 and CFP-10 (34), and their pathways are designated type VIIb (29). In S. aureus, the ess (type VIIb) locus has been divided into four modules (1 to 4) (35). Modules 2 and 3 are variable among some of the different sequence type (ST) isolates of this pathogenic species (35). Module 2 encodes EccC; i.e., it harbors essC of S. aureus (14). Four essC alleles (essC1 to essC4) carry sequence variation in the terminal SpoIIIE-FtsK-like ATPase domain and are associated with specific groups of downstream genes, presumably encoding specific secretion substrates (35). The essC1 variant has the highest frequency (90/153 isolates) and was found in strains belonging to S. aureus clonal complexes (CCs) that are frequently associated with human invasive disease (CCs 1, 5, 7, 8, 9, 25, 51, and 88) (35). essC3 is found in CC30 and ST239 isolates, whereas essC4 and essC2 are restricted to CC22 and to CC15 and ST398 isolates, respectively (35). In the community acquired (CA)-MRSA isolate USA300 LAC (CC8), module 2 includes essC1 as well as the secretion substrate genes esxC, esxB, esxD, and essD in addition to essE (esaE), a gene of heretofore unknown function. These genes are absent from S. aureus isolates with secretion pathways comprised of essC2 to essC4 (35). Module 3 encompasses a complex arrangement of predicted genes, including members of the DUF600 family, which vary in number even for isolates from the same CC (35). Finally, module 4 includes two genes for hypothetical transmembrane proteins that are conserved among all S. aureus isolates. The requirements of the two module 4 genes for T7SS in S. aureus are, however, not known.
Here, we characterize EssE, which is located in the staphylococcal cytoplasm and forms a complex with EssD, EsxC, EssI (DUF600 protein), and EssC. EssE stabilizes EssD degradation and promotes the secretion of EsxA, EsxB, EsxC, EsxD, and EssD into the extracellular medium. Other components of the ESS secretion apparatus—EsaA, EssA, and EssB—were not observed in detergent-extracted EssE (15). Based on these observations, we propose a model whereby EssE may act as an assembly platform for the secretion of EsxC and EssD.
When analyzed in mouse models of bloodstream infection or pneumonia, mutations in ess genes of CC8 isolates S. aureus Newman, USA300 LAC, RN6390, and COL result in reduced virulence and diminished staphylococcal persistence (15, 18, 36). Similar phenotypes have been observed for essB variants of S. aureus ST398 isolates in mouse models for bloodstream infection and skin abscess formation (37). For ST398 isolates, expression of ess has been associated with neutrophil lysis and staphylococcal escape from phagocytic killing; however, it is not yet clear whether this phenotype is attributable to specific effectors secreted via the essC2 T7SS (37).
The molecular basis for S. aureus ess manipulation of host immune responses was heretofore not known. In M. tuberculosis, secretion of Mt-EsxA by the ESX-1 pathway into host cells activates cyclic GMP-AMP synthase (cGAS) and cyclic GMP-AMP (cGAMP) signaling via STING, the mammalian sensor for cytoplasmic DNA and bacterial cyclic dinucleotides (38), thereby stimulating type I interferon (IFN) responses (39–41). IFN activation is associated with disease progression in tuberculosis (TB) and other infectious diseases. ESX-1 activity also leads to inflammasome activation and secretion of the proinflammatory cytokine IL-1β (40). Finally, Mt-EsxA secretion has been shown to be required for formation of TB granulomas and the persistence of mycobacteria in these lesions (42). A recent model for cGAS and STING signaling postulates that Mt-EsxA may stimulate IFN signaling via pore formation and DNA transfer into the host cell cytosol (40, 41).
We show here that the S. aureus ESS pathway of the CC8 isolate USA300 LAC stimulates IL-12 (p40) and IL-12 (p35) production during host infection while limiting the secretion of RANTES (CCL5). IL-12 (p40) is shared between IL-12 (p70) and IL-23 and activates TH1 and TH17 responses, respectively, as well as gamma interferon signaling during bacterial infection, through the activation of pattern recognition receptors recognizing pathogen-associated molecular patterns (43, 44). Impaired TH1 and TH17 responses are associated with mucocutaneous S. aureus and Candida infections (45, 46). In mice and humans, the IL-12 heterodimer signals through the IL-12 receptor (IL-12R), which is comprised of IL-12Rβ1 and IL-12Rβ2 (47). IL-23, a complex formed from IL-23 (p19) and IL-12 (p40), signals via IL-23R and IL-12Rβ1 (48). IL-12 and IL-23 impact the development of TH1 cell and IL-17-producing T helper (TH17) cell responses, respectively (49). In a model of IL-12-mediated activation and recruitment of myeloid cells and other immune cells for staphylococcal abscess development, one would predict that mutational lesions abrogating IL-12 production or IL-12 receptor signaling may provide increased resistance toward S. aureus infection. The impact of IL-12/IL-23 signaling on the pathogenesis of S. aureus infection of mice has been studied in several challenge models even though the impact of IL-12 signaling on the outcome of staphylococcal diseases remained enigmatic. Following intravenous S. aureus challenge, IL-12p40−/− (IL-12/IL-23-deficient) mice displayed elevated mortality rates and increased staphylococcal loads in renal tissue 20 days after challenge (50). Intranasal challenge of S. aureus leads to lung infection; both the associated disease mortality and the bacterial burden are increased in IL-12p35−/− mice (51). Following intracerebral inoculation of staphylococci, the initial phases of brain abscess formation were not impacted by IL-12 (p35) deficiency; however, IL-12p35−/− mice displayed fewer clinical disease symptoms and healed more rapidly than wild-type mice (52). In humans, S. aureus infections result in a transient increase in antistaphylococcal antibody levels; but protective immunity is not observed, and recurrent infections occur frequently (53). S. aureus secretes enterotoxins and staphylococcal protein A (SpA) that function as T cell (54) and B cell superantigens (55), respectively. IL-12 secretion mediated by the ESS pathway may further compromise the induction of a humoral immune response because IL-12 skewing of gamma interferon-producing TH1 cells impedes TH2 polarization and thereby the host's ability to produce antibodies (56).
RANTES (CCL5) is expressed by many cells, including T lymphocytes, macrophages, and platelets, and may be induced by NF-κB activation following stimulation by CD40L or IL-15 (57). Abscess lesions of essE strain-infected mice stained heavily for RANTES in a manner that correlated with an increased recruitment of macrophages to these lesions. Since RANTES is upregulated when animals are infected with the essE variant, we postulate that during infection with wild-type S. aureus, the ESS pathway acts to limit macrophage trafficking to abscess lesions. In this regard, it seems noteworthy that S. aureus also secretes the LukED pore-forming toxins that specifically target myeloid cells expressing CCR5 (58). CCL5 is a ligand of CCR5, a seven-transmembrane G-protein-coupled receptor that modulates diverse signaling cascades upon ligand interaction. Thus, S. aureus may disable CCL5-CCR5 activity by both blocking RANTES production (ESS-dependent) and killing CCL5 target cells. Future work will need to address the contribution of ESS-mediated IL-12 production in inflammatory myeloid cells and inhibition of CCL5 production toward the establishment of persistent infection in mice.
MATERIALS AND METHODS
Bacterial cultures.
S. aureus cultures were grown in tryptic soy broth (TSB) or agar (TSA). Chloramphenicol was added to a final concentration of 20 μg/ml for plasmid selection. Anhydrotetracycline was used at 50 ng/ml for pKOR1-mediated allelic replacements of target genes (22). To assay for protein production and secretion, staphylococcal cultures were grown with vigorous shaking at 37°C for 2.5 h in TSB supplemented with 0.2% horse serum. Escherichia coli cultures were grown in Luria-Bertani medium at 37°C (17). Ampicillin was used at 100 μg/ml for plasmid selection in E. coli.
Bacterial strains and plasmids.
USA300 LAC, a clone of the American community-acquired methicillin-resistant S. aureus (CA-MRSA) epidemic strain (59), was used as the wild-type S. aureus strain. E. coli DH5α was used for cloning experiments. USA300 variant essD::erm, essD::erm/pessD-essI1, and essD::erm/pessD*His strains are described elsewhere (19). The USA300 variant ΔessE strain was generated by allelic replacement with plasmid pKOR1 to fuse the first 15 and the last 15 codons of essE, resulting in a deletion of 194 codons. Briefly, two 1-kbp DNA sequences flanking essE upstream and downstream of the deletion were amplified from USA300 template DNA with primers EssE1 (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTGAAGATGATGTTAAAAAGCTTATTAC-3′) and EssE2 (5′-AAAGATCTTAATTCTTCGTAAGAAAAATAATC-3′) as well as EssE3 (5′-AAAGATCTGGATTCGATACTGATGAAAATC-3′) and EssE4 (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTGGATTAATACAAGATAAAACTACC-3′). PCR products were cut with BglII and ligated at the restriction site. The ligation product was cloned into pKOR1 using the Gateway BP Clonase II enzyme from Invitrogen (catalog number 11789-020). Procedures for cloning and allelic replacement were performed as previously described (17). The complementing plasmid pessE carries the coding sequence of essE under transcriptional control of the constitutive hprK promoter in pWWW412 (60). pessE was assembled by PCR amplification of the essE coding sequence using USA300 template and primers EssE5 (5′-AACATATGAAAGATGTTAAGCGAATAG-3′) and EssE6 (5′-AAGGATCCTTACTCCTCTGCTTTATTAATATG-3′). The PCR product was cut with NdeI and BamH1 and cloned into the corresponding sites of pWWW412. pessESTREP expresses a variant of essE from the hprK promoter of pWWW412. To assemble pessESTREP, the essE gene was amplified with EssE5 and EssE7 (5′-AAGGATCCTTACTTCTCGAACTGTGGGTGGCTCCACTCCTCTGCTTTATTAATATG-3′), thereby extending the open reading frame by eight codons to yield EssESTREP with eight additional amino acids at the C terminus (Trp-Ser-His-Pro-Gln-Phe-Glu-Lys). For protein production and purification from lysates of E. coli, the essE coding sequence was PCR amplified with the primers EssE8 (AACATATGAAAGATGTTAAGCGAATAG) and EssE9 (AAGGATCCTTACTCCTCTGCTTTATTAATATG), cut with BamHI and EcoRI, and ligated to pGEX-2TK (GE Healthcare) restricted with the same enzymes. Isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible expression from the Ptac promoter of pGEX-2TK-EssE in E. coli generates a hybrid protein comprised of glutathione S-transferase (GST) linked via a thrombin cleavage site to EssE.
RNA extraction and qualitative cDNA PCR analysis.
RNA was extracted from 10-ml cultures grown to an absorbance of 1.0 at 600 nm (A600). Cultures were centrifuged (9,000 × g for 10 min), and the bacterial sediment was suspended in 1 ml of TRIzol containing 0.5 ml of glass beads (0.1-mm diameter). Samples were beat four times in a FastPrep-24 (MP Biomedicals) homogenizer, 200 μl of chloroform was added, and tubes were shaken for 30 s, incubated at 25°C for 3 min, and again centrifuged (12,000 × g for 15 min). Isopropanol (500 μl) was added to 500 μl of the carefully recovered upper phase and incubated at 25°C for 10 min, and samples were again centrifuged (12,000 × g for 10 min). Sediments were washed with 75% ethanol, dried, and rehydrated with 50 μl of diethylpyrocarbonate-treated water, and contaminating DNA was removed using a Turbo DNA-free kit (Ambion) to obtain 100 ng/μl RNA per sample. An iScript cDNA synthesis kit (Bio-Rad) was used to generate cDNA. Reverse transcriptase (RT) was omitted in control experiments. cDNA and control samples were used for qualitative PCR analysis using GoTaq Flexi DNA polymerase (Promega). Primer pairs for amplification of esxB-, esxD-, and 16S-specific transcripts were CGCTGAGTATATCGAAGGTAGTG and CCATCGGTTGTACTAATTCTTCTTG (esxB), CATTAGTGGTCTCAAAGGTCCA and GTAAAGCTTGGCAAATTCCGT (esxD), and GAAAGCCACGGCTAACTACG and CATTTCACCGCTACACATGG (16S), respectively.
Fractionation of bacterial cultures and immunoblot analysis.
To assess protein secretion, S. aureus cultures were grown to an A600 of 1.0, and 50-ml aliquots were centrifuged (9,000 × g for 10 min). The extracellular medium was removed with the supernatant and separated from the bacterial sediment. Staphylococci were washed and suspended in 50 ml of 50 mM Tris-HCl (pH 7.0) and 10 mM MgCl2 (TM medium), incubated with 20 μg/ml lysostaphin at 37°C for 60 min, and centrifuged (100,000 × g for 40 min at 4°C) to sediment membranes (17). Soluble proteins from the cytoplasm were collected with the supernatant and separated from insoluble membrane proteins. Membrane sediment was suspended in 50 ml of TM medium. Proteins in each fraction (extracellular medium, cytoplasm, or membrane) were precipitated with 10% trichloroacetic acid (TCA), incubated for 30 min on ice, and centrifuged (13,000 × g for 10 min). The supernatants were discarded, and protein precipitates were washed with 1 ml of ice-cold acetone and again sedimented by centrifugation (13,000 × g for 10 min). Acetone supernatants were discarded, and protein sediments were dried prior to solubilization in sample buffer (0.5 M Tris-HCl [pH 8.0], 4% SDS, 10% glycerol, 5% 2-mercaptoethanol, 0.01% bromophenol blue) and heated at 90°C for 10 min.
In an alternate protocol, proteins from lysostaphin-digested staphylococci were subjected directly to TCA precipitation (without fractionation of cytoplasm and membrane), washed in acetone, and solubilized in sample buffer. Protein samples were separated on 15% SDS-PAGE gels, electrotransferred to polyvinylidene difluoride (PVDF) membrane, and analyzed by immunoblotting with rabbit polyclonal antibodies raised against purified proteins. Immunoreactive products were revealed by chemiluminescent detection after incubation with an anti-rabbit horseradish peroxidase (HRP)-conjugated secondary antibody (Cell Signaling Technology).
Protein purification from S. aureus.
Cells from 8 liters of S. aureus culture that had grown to an A600 of 2.0 were sedimented by centrifugation (9,000 × g for 10 min). Cells were suspended in buffer A (50 mM Tris-HCl [pH 7.5], 150 mM NaCl), 20 μg/ml lysostaphin was added, and mixed samples were incubated for 1 h at 37°C. The resulting protoplasts were lysed in a French press at 14,000 lb/in2 (ThermoSpectronic, Rochester, NY). Unbroken cells were removed by centrifugation (5,000 × g for 15 min). Crude lysates were separated with the supernatant from sedimented protoplasts and peptidoglycan and subjected to ultracentrifugation (100,000 × g for 2 h at 4°C). Soluble proteins in the staphylococcal cytoplasm were separated from the membrane sediment and subjected via gravity flow to chromatography on Strep-Tactin–Sepharose (IBA) or Ni-nitrilotriacetic acid (Ni-NTA)–Sepharose with a packed volume of 2 ml preequilibrated with buffer A. Columns were washed with 20× bed volumes of buffer A and eluted with either 4 ml of 2.5 mM desthiobiotin in buffer A (Strep-Tactin–Sepharose) or stepwise gradients of 50 to 500 mM imidazole in buffer A (Ni-NTA–Sepharose). For affinity chromatography of membrane-associated proteins, the sediment of staphylococcal membranes (100,000 × g for 2 h) was extracted with 40 ml of 2% n-dodecyl-β-d-maltoside (DDM) in buffer A for 2 h at 4°C. Extracts were again subjected to ultracentrifugation (100,000 × g for 2 h at 4°C), and DDM-solubilized proteins were subjected via gravity flow to chromatography on Strep-Tactin–Sepharose (IBA) or Ni-nitrilotriacetic acid (Ni-NTA)–Sepharose with a packed volume of 2 ml preequilibrated with buffer A. Columns were washed with 20× bed volumes of buffer A and eluted with either 4 ml of 2.5 mM desthiobiotin in buffer A (Strep-Tactin–Sepharose) or stepwise gradients of 50 to 500 mM imidazole in buffer A (Ni-NTA–Sepharose). Aliquots of eluted fractions were mixed with an equal volume of sample buffer, separated on 15% SDS-PAGE gels, and analyzed by Coomassie blue staining or immunoblotting. The identification of proteins was performed by mass spectrometry of tryptic digests at the Taplin Mass Spectrometry Facility at Harvard Medical School.
EssE antiserum.
A New Zealand White rabbit was purchased from Harlan Sprague Dawley and used to generate the polyclonal EssE antiserum. EssE antigen was purified via GST-EssE affinity chromatography and cleaved with thrombin, and EssE was isolated as described previously (18). Purified EssE (100 μg) was emulsified with complete Freund's adjuvant (Difco), and the emulsion was injected subcutaneously into the rabbit. At 21-day intervals, two booster immunizations with 100 μg of EssE emulsified with incomplete Freund's adjuvant were performed. Serum was obtained from blood samples, mixed with 0.02% sodium azide, aliquoted, and frozen for long-term storage at −80°C or stored at 4°C for reiterative use.
Animal challenge.
Cohorts of female, 6-week-old C57BL/6 mice (Jackson Laboratory) were anesthetized by intraperitoneal injection with an aqueous solution containing 65 mg/ml ketamine and 6 mg/ml xylazine per kg of body weight. Anesthetized mice were infected by intravenous injection with 0.5 × 107 to 5 × 107 CFU of staphylococci into the retro-orbital venous plexus (10). For inoculation, S. aureus cultures were grown in TSB with rotation at 37°C to an A600 of 0.4. Bacteria were sedimented by centrifugation (5,000 × g for 10 min), washed, and suspended in phosphate-buffered saline (PBS) in a volume of 100 μl. Control animals were mock infected by injection with 100 μl of PBS without bacteria. To analyze cytokines in blood at 12 h postinfection, cohorts of mice (n = 5) were euthanized by CO2 inhalation, blood was retrieved by cardiac puncture, and serum samples were prepared. A Bio-Plex Pro Mouse Cytokine 23-Plex assay kit (M60009RDPD; Bio-Rad) was used for the analysis of serum cytokines. The multiplex data were analyzed with one-way analysis of variance (ANOVA) using Bonferroni's multiple-comparison test. Five days after infection, cohorts of mice (n = 10) were euthanized and necropsied, and both kidneys were removed. For each animal, tissues from the right kidney were homogenized, and serial dilutions of homogenates were spread on agar plates for enumeration of the staphylococcal load. Bacterial loads were analyzed with a two-tailed Mann-Whitney test to measure statistical significance. The left kidneys were flash frozen in 22-oxacalcitriol (OCT) medium for cryo-sectioning in 12-μm intervals using a Thermo Scientific Microm HM550 microtome. Sections were stained with hematoxylin and eosin (H&E) using a Rapid Chrome frozen section staining kit from Thermo Scientific, and samples were visualized on a dissecting microscope. An unpaired two-tailed Student t test with Welch's correction was used to evaluate differences in abscess enumeration. Sections directly above or below those used for abscess enumeration were subjected to immunohistochemistry (IHC) staining. Slides were fixed with acetone for 30 min at −20°C and blocked for 1 h with a solution containing 1.5% bovine serum albumin (BSA) and human IgG. Slides were washed in PBS and incubated for 1 h with anti-F4/80 antibody (anti-F4/80), a marker specific for macrophages, or anti-RANTES antibody (anti-RANTES). Slides were washed in PBS and incubated for 1 h with an HRP-conjugated secondary antibody, washed again in PBS, and incubated for 1 min with diaminobenzidine-hydrogen peroxide solution to reveal areas of antibody binding. The reaction was quenched by plunging slides into PBS solutions. Slides were counterstained with hematoxylin and mounted, and images were analyzed using a Panoramic Viewer from 3D Histech, Ltd. Fiji image software was used to quantify staining of IHC images. Specifically, average darkness was determined by taking the entire image and separating the diaminobenzidine staining from the hematoxylin counterstain via color deconvolution. Statistical analysis was performed using a two-tailed Mann-Whitney test.
Ethics statement.
The preparation of rabbit antibodies and mouse challenge experiments were performed according to protocols that were reviewed, approved, and performed under the regulatory supervision of The University of Chicago's Institutional Animal Care and Use Committee (IACUC). Animal experiments were conducted in accordance with recommendations detailed in the Guide for the Care and Use of Laboratory Animals (61). Animal care was managed by The University of Chicago Animal Resource Center, which is accredited by the American Association for Accreditation of Laboratory Animal Care and acts in compliance with NIH guidelines of laboratory animal care and use.
ACKNOWLEDGMENTS
We thank members of our laboratory for suggestions, discussion, and comments on the manuscript.
M.A. was supported by a National Institute of Allergy and Infectious Diseases (NIAID) Biodefense Training Grant in Host-Pathogen Interactions at the University of Chicago (T32 AI065382) and was a recipient of an American Heart Association Award (11PRE7600117). R.J.O. was supported by a Molecular Cell Biology Training Grant at the University of Chicago (T32 GM007183). This work was supported by grants AI075258 and AI110937 from NIAID to D.M. Work in the laboratory of O.S. is supported by NIAID grants AI038897 and AI052474.
Footnotes
For a companion article on this topic, see https://doi.org/10.1128/JB.00528-16.
REFERENCES
- 1.Wertheim HF, Vos MC, Ott A, van Belkum A, Voss A, Kluytmans JA, van Keulen PH, Vandenbroucke-Grauls CM, Meester MH, Verbrugh HA. 2004. Risk and outcome of nosocomial Staphylococcus aureus bacteraemia in nasal carriers versus non-carriers. Lancet 364:703–705. doi: 10.1016/S0140-6736(04)16897-9. [DOI] [PubMed] [Google Scholar]
- 2.Fitzgerald JR. 2012. Livestock-associated Staphylococcus aureus: origin, evolution and public health threat. Trends Microbiol 20:192–198. doi: 10.1016/j.tim.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 3.Lowy FD. 1998. Staphylococcus aureus infections. N Engl J Med 339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 4.David MZ, Daum RS. 2010. Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin Microbiol Rev 23:616–687. doi: 10.1128/CMR.00081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Friedrich R, Panizzi P, Fuentes-Prior P, Richter K, Verhamme I, Anderson PJ, Kawabata S, Huber R, Bode W, Bock PE. 2003. Staphylocoagulase is a prototype for the mechanism of cofactor-induced zymogen activation. Nature 425:535–539. doi: 10.1038/nature01962. [DOI] [PubMed] [Google Scholar]
- 6.Cheng AG, McAdow M, Kim HK, Bae T, Missiakas DM, Schneewind O. 2010. Contribution of coagulases towards Staphylococcus aureus disease and protective immunity. PLoS Pathog 6:e1001036. doi: 10.1371/journal.ppat.1001036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomer L, Emolo C, Thammavongsa V, Kim HK, McAdow ME, Yu W, Kieffer M, Schneewind O, Missiakas D. 2016. Antibodies against a secreted product of I trigger phagocytic killing. J Exp Med 213:293–301. doi: 10.1084/jem.20150074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thammavongsa V, Missiakas DM, Schneewind O. 2013. Staphylococcus aureus conversion of neutrophil extracellular traps into deoxyadenosine promotes immune cell death. Science 342:863–866. doi: 10.1126/science.1242255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng AG, DeDent AC, Schneewind O, Missiakas DM. 2011. A play in four acts: Staphylococcus aureus abscess formation. Trends Microbiol 19:225–232. doi: 10.1016/j.tim.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng AG, Kim HK, Burts ML, Krausz T, Schneewind O, Missiakas DM. 2009. Genetic requirements for Staphylococcus aureus abscess formation and persistence in host tissues. FASEB J 23:3393–3404. doi: 10.1096/fj.09-135467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ziegler C, Goldmann O, Hobeika E, Geffers R, Peters G, Medina E. 2011. The dynamics of T cells during persistent Staphylococcus aureus infection: from antigen-reactivity to in vivo anergy. EMBO Mol Med 3:652–666. doi: 10.1002/emmm.201100173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu C, Bayer AS, Cosgrove SE, Daum RS, Fridkin SK, Gorwitz RJ, Kaplan SL, Karchmer AW, Levine DP, Murray BE, Rybak MJ, Talan DA, Chambers HF. 2011. Clinical practice guidelines by the Infectious Diseases Society of America for the treatment of methicillin-resistant Staphylococcus aureus infections in adults and children: executive summary. Clin Infect Dis 52:285–292. doi: 10.1093/cid/cir034. [DOI] [PubMed] [Google Scholar]
- 13.Spaan AN, Surewaard BGJ, Nijland R, van Strijp JAG. 2013. Neutrophils versus Staphylococcus aureus: a biological tug of war. Annu Rev Microbiol 67:629–650. doi: 10.1146/annurev-micro-092412-155746. [DOI] [PubMed] [Google Scholar]
- 14.Burts ML, Williams WA, DeBord K, Missiakas DM. 2005. EsxA and EsxB are secreted by an ESAT-6-like system that is required for the pathogenesis of Staphylococcus aureus infections. Proc Natl Acad Sci U S A 102:1169–1174. doi: 10.1073/pnas.0405620102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burts ML, DeDent AC, Missiakas DM. 2008. EsaC substrate for the ESAT-6 secretion pathway and its role in persistent infections of Staphylococcus aureus. Mol Microbiol 69:736–746. doi: 10.1111/j.1365-2958.2008.06324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen YH, Anderson M, Hendrickx AP, Missiakas D. 2012. Characterization of EssB, a protein required for secretion of ESAT-6 like proteins in Staphylococcus aureus. BMC Microbiol 12:219. doi: 10.1186/1471-2180-12-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anderson M, Aly KA, Chen YH, Missiakas D. 2013. Secretion of atypical protein substrates by the ESAT-6 secretion system of Staphylococcus aureus. Mol Microbiol 90:734–743. doi: 10.1111/mmi.12395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anderson M, Chen YH, Butler EK, Missiakas DM. 2011. EsaD, a secretion factor for the Ess pathway in Staphylococcus aureus. J Bacteriol 193:1583–1589. doi: 10.1128/JB.01096-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ohr RJ, Anderson M, Schneewind O, Missiaks DM. 2017. EssD, a nuclease effector of the Staphylococcus aureus ESS pathway. J Bacteriol 199:e00528-16. doi: 10.1128/JB.00528-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McNicholas S, Talento AF, O'Gorman J, Hannan MM, Lynch M, Greene CM, Humphreys H, Fitzgerald-Hughes D. 2014. Cytokine responses to Staphylococcus aureus bloodstream infection differ between patient cohorts that have different clinical courses of infection. BMC Infect Dis 14:580. doi: 10.1186/s12879-014-0580-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. 2006. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 1:2856–2860. [DOI] [PubMed] [Google Scholar]
- 22.Bae T, Schneewind O. 2006. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 55:58–63. doi: 10.1016/j.plasmid.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 23.Eisenberg D, Schwarz E, Komaromy M, Wall R. 1984. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J Mol Biol 179:125–142. doi: 10.1016/0022-2836(84)90309-7. [DOI] [PubMed] [Google Scholar]
- 24.Bubeck Wardenburg J, Schneewind O. 2008. Vaccine protection against Staphylococcus aureus pneumonia. J Exp Med 205:287–294. doi: 10.1084/jem.20072208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kebaier C, Chamberland RR, Allen IC, Gao X, Broglie PM, Hall JD, Jania C, Doerschuk CM, Tilley SL, Duncan JA. 2012. Staphylococcus aureus α-hemolysin mediates virulence in a murine model of severe pneumonia through activation of the NLRP3 inflammasome. J Infect Dis 205:807–817. doi: 10.1093/infdis/jir846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schindler D, Gutierrez MG, Beineke A, Rauter Y, Rohde M, Foster S, Goldmann O, Medina E. 2012. Dendritic cells are central coordinators of the host immune response to Staphylococcus aureus bloodstream infection. Am J Pathol 181:1327–1337. doi: 10.1016/j.ajpath.2012.06.039. [DOI] [PubMed] [Google Scholar]
- 27.Gately MK, Renzetti LM, Magram J, Stern AS, Adorini L, Gubler U, Presky DH. 1998. The interleukin-12/interleukin-12-receptor system: role in normal and pathologic immune responses. Annu Rev Immunol 16:495–521. doi: 10.1146/annurev.immunol.16.1.495. [DOI] [PubMed] [Google Scholar]
- 28.Kobayashi M, Fitz L, Ryan M, Hewick RM, Clark SC, Chan S, Loudon R, Sherman F, Perussia B, Trinchieri G. 1989. Identification and purification of natural killer cell stimulatory factor (NKSF), a cytokine with multiple biologic effects on human lymphocytes. J Exp Med 170:827–845. doi: 10.1084/jem.170.3.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abdallah AM, Gey van Pittius NC, Champion PA, Cox J, Luirink J, Vandenbroucke-Grauls CM, Appelmelk BJ, Bitter W. 2007. Type VII secretion–mycobacteria show the way. Nat Rev Microbiol 5:883–891. doi: 10.1038/nrmicro1773. [DOI] [PubMed] [Google Scholar]
- 30.Houben D, Demangel C, van Ingen J, Perez J, Baldeón L, Abdallah AM, Caleechurn L, Bottai D, van Zon M, de Punder K, van der Laan T, Kant A, Bossers-de Vries R, Willemsen P, Bitter W, van Soolingen D, Brosch R, van der Wel N, Peters PJ. 2012. ESX-1-mediated translocation to the cytosol controls virulence of mycobacteria. Cell Microbiol 14:1287–1298. doi: 10.1111/j.1462-5822.2012.01799.x. [DOI] [PubMed] [Google Scholar]
- 31.Rosenberg OS, Dovala D, Li X, Connolly L, Bendebury A, Finer-Moore J, Holton J, Cheng Y, Stroud RM, Cox JS. 2015. Substrates control multimerization and activation of the multi-domain ATPase motor of type VII secretion. Cell 161:501–512. doi: 10.1016/j.cell.2015.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garufi G, Butler E, Missiakas D. 2008. ESAT-6-like protein secretion in Bacillus anthracis. J Bacteriol 190:7004–7011. doi: 10.1128/JB.00458-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sysoeva TA, Zepeda-Rivera M, Huppert LA, Burton BM. 2014. Dimer recognition and secretion by the ESX secretion system in Bacillus subtilis. Proc Natl Acad Sci U S A 111:7653–7658. doi: 10.1073/pnas.1322200111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pallen MJ. 2002. The ESAT-6/WXG100 superfamily—and a new Gram-positive secretion system? Trends Microbiol 10:209–212. doi: 10.1016/S0966-842X(02)02345-4. [DOI] [PubMed] [Google Scholar]
- 35.Warne B, Harkins CP, Harris SR, Vatsiou A, Stanley-Wall N, Parkhill J, Peacock SJ, Palmer T, Holden MT. 2016. The Ess/type VII secretion system of Staphylococcus aureus shows unexpected genetic diversity. BMC Genomics 17:222. doi: 10.1186/s12864-016-2426-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kneuper H, Cao ZP, Twomey KB, Zoltner M, Jäger F, Cargill JS, Chalmers J, van der Kooi-Pol MM, van Dijl JM, Ryan RP, Hunter WN, Palmer T. 2014. Heterogeneity in ess transcriptional organization and variable contribution of the Ess/type VII protein secretion system to virulence across closely related Staphylococcus aureus strains. Mol Microbiol 93:928–943. doi: 10.1111/mmi.12707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y, Hu M, Liu Q, Qin J, Dai Y, He L, Li T, Zheng B, Zhou F, Yu K, Fang J, Liu X, Otto M, Li M. 2016. Role of the ESAT-6 secretion system in virulence of the emerging community-associated Staphylococcus aureus lineage ST398. Sci Rep 6:25163. doi: 10.1038/srep25163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao D, Wu J, Wu YT, Du F, Aroh C, Yan N, Sun L, Chen ZJ. 2013. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science 341:903–906. doi: 10.1126/science.1240933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dey B, Dey RJ, Cheung LS, Pokkali S, Guo H, Lee JH, Bishai WR. 2015. A bacterial cyclic dinucleotide activates the cytosolic surveillance pathway and mediates innate resistance to tuberculosis. Nat Med 21:401–406. doi: 10.1038/nm.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wassermann R, Gulen MF, Sala C, Perin SG, Lou Y, Rybniker J, Schmid-Burgk JL, Schmidt T, Hornung V, Cole ST, Ablasser A. 2015. Mycobacterium tuberculosis differentially activates cGAS- and inflammasome-dependent intracellular immune responses through ESX-1. Cell Host Microbe 17:799–810. doi: 10.1016/j.chom.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 41.Watson RO, Bell SL, MacDuff DA, Kimmey JM, Diner EJ, Olivas J, Vance RE, Stallings CL, Virgin HW, Cox JS. 2015. The cytosolic sensor cGAS detects Mycobacterium tuberculosis DNA to induce type I interferons and activate autophagy. Cell Host Microbe 17:811–819. doi: 10.1016/j.chom.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cambier CJ, Falkow S, Ramakrishnan L. 2014. Host evasion and exploitation schemes of Mycobacterium tuberculosis. Cell 159:1497–1509. doi: 10.1016/j.cell.2014.11.024. [DOI] [PubMed] [Google Scholar]
- 43.O'Garra A, Arai N. 2000. The molecular basis of T helper 1 and T helper 2 cell differentiation. Trends Cell Biol 10:542–550. doi: 10.1016/S0962-8924(00)01856-0. [DOI] [PubMed] [Google Scholar]
- 44.van de Vosse E, van Dissel JT, Ottenhoff TH. 2009. Genetic deficiencies of innate immune signalling in human infectious disease. Lancet Infect Dis 9:688–698. doi: 10.1016/S1473-3099(09)70255-5. [DOI] [PubMed] [Google Scholar]
- 45.Maródi L, Cypowyj S, Tóth B, Chernyshova L, Puel A, Casanova JL. 2012. Molecular mechanisms of mucocutaneous immunity against Candida and Staphylococcus species. J Allergy Clin Immunol 130:1019–1027. doi: 10.1016/j.jaci.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O'Shea JJ, Plenge R. 2012. JAK and STAT signaling molecules in immunoregulation and immune-mediated disease. Immunity 36:542–550. doi: 10.1016/j.immuni.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trinchieri G. 2003. Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat Rev Immunol 3:133–146. doi: 10.1038/nri1001. [DOI] [PubMed] [Google Scholar]
- 48.Oppmann B, Lesley R, Blom B, Timans JC, Xu Y, Hunte B, Vega F, Yu N, Wang J, Singh K, Zonin F, Vaisberg E, Churakova T, Liu M, Gorman D, Wagner J, Zurawski S, Liu Y, Abrams JS, Moore KW, Rennick D, de Waal-Malefyt R, Hannum C, Bazan JF, Kastelein RA. 2000. Novel p19 protein engages IL-12p40 to form a cytokine, IL-23, with biological activities similar as well as distinct from IL-12. Immunity 13:715–725. doi: 10.1016/S1074-7613(00)00070-4. [DOI] [PubMed] [Google Scholar]
- 49.Teng MW, Bowman EP, McElwee JJ, Smyth MJ, Casanova JL, Cooper AM, Cua DJ. 2015. IL-12 and IL-23 cytokines: from discovery to targeted therapies for immune-mediated inflammatory diseases. Nat Med 21:719–729. doi: 10.1038/nm.3895. [DOI] [PubMed] [Google Scholar]
- 50.Dodson KW, Pinkner JS, Rose T, Magnusson G, Hultgren SJ, Waksman G. 2001. Structural basis of the interaction of the pyelonephritic E. coli adhesin to its human kidney receptor. Cell 105:733–743. doi: 10.1016/S0092-8674(01)00388-9. [DOI] [PubMed] [Google Scholar]
- 51.Hilliard JJ, Datta V, Tkaczyk C, Hamilton M, Sadowska A, Jones-Nelson O, O'Day T, Weiss WJ, Szarka S, Nguyen V, Prokai L, Suzich J, Stover CK, Sellman BR. 2015. Anti-alpha-toxin monoclonal antibody and antibiotic combination therapy improves disease outcome and accelerates healing in a Staphylococcus aureus dermonecrosis model. Antimicrob Agents Chemother 59:299–309. doi: 10.1128/AAC.03918-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Held J, Preuße C, Döser A, Richter L, Heppner FL, Stenzel W. 2013. Enhanced acute immune response in IL-12p35−/− mice is followed by accelerated distinct repair mechanisms in Staphylococcus aureus-induced murine brain abscess. J Infect Dis 208:749–760. doi: 10.1093/infdis/jit126. [DOI] [PubMed] [Google Scholar]
- 53.Kim HK, Thammavongsa V, Schneewind O, Missiakas D. 2012. Recurrent infections and immune evasion strategies of Staphylococcus aureus. Curr Opin Microbiol 15:92–99. doi: 10.1016/j.mib.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McCormick JK, Yarwood JM, Schlievert PM. 2001. Toxic shock syndrome and bacterial superantigens: an update. Annu Rev Microbiol 55:77–104. doi: 10.1146/annurev.micro.55.1.77. [DOI] [PubMed] [Google Scholar]
- 55.Silverman GJ, Goodyear CS. 2006. Confounding B-cell defences: lessons from a staphylococcal superantigen. Nat Rev Immunol 6:465–475. doi: 10.1038/nri1853. [DOI] [PubMed] [Google Scholar]
- 56.Zundler S, Neurath MF. 2015. Interleukin-12: Functional activities and implications for disease. Cytokine Growth Factor Rev 26:559–568. doi: 10.1016/j.cytogfr.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 57.Chenoweth MJ, Mian MF, Barra NG, Alain T, Sonenberg N, Bramson J, Lichty BD, Richards CD, Ma A, Ashkar AA. 2012. IL-15 can signal via IL-15Rα, JNK, and NF-κB to drive RANTES production by myeloid cells. J Immunol 188:4149–4157. doi: 10.4049/jimmunol.1101883. [DOI] [PubMed] [Google Scholar]
- 58.Alonzo F III, Kozhaya L, Rawlings SA, Reyes-Robles T, DuMont AL, Myszka DG, Landau NR, Unutmaz D, Torres VJ. 2013. CCR5 is a receptor for Staphylococcus aureus leukotoxin ED. Nature 493:51–55. doi: 10.1038/nature11724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Diep BA, Gill SR, Chang RF, Phan TH, Chen JH, Davidson MG, Lin F, Lin J, Carleton HA, Mongodin EF, Sensabaugh GF, Perdreau-Remington F. 2006. Complete genome sequence of USA300, an epidemic clone of community-acquired methicillin-resistant Staphylococcus aureus. Lancet 367:731–739. doi: 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
- 60.Bubeck Wardenburg J, Williams WA, Missiakas D. 2006. Host defenses against Staphylococcus aureus infection require recognition of bacterial lipoproteins. Proc Natl Acad Sci U S A 103:13831–13836. doi: 10.1073/pnas.0603072103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.National Research Council Committee for the Update of the Guide for the Care and Use of Laboratory Animals. 2011. Guide for the care and use of laboratory animals, 8th ed National Academies Press, Washington, DC. [Google Scholar]