Abstract
Astrocytoma is the most common malignant brain tumor in humans. Loss of the p53 signaling pathway and up-regulation of the ras signaling pathway are common during tumor progression. We have shown previously that mice mutant for Trp53 and Nf1 develop astrocytoma, progressing to glioblastoma, on a C57BL/6J strain background. In contrast, here we present data that mice mutant for Trp53 and Nf1 on a 129S4/SvJae background are highly resistant to developing astrocytoma. Through analysis of F1 progeny, we demonstrate that susceptibility to astrocytoma is linked to chromosome 11, and that the modifier gene(s) responsible for differences in susceptibility is closely linked to Nf1 and Trp53. Furthermore, this modifier of astrocytoma susceptibility is itself epigenetically modified. These data demonstrate that epigenetic effects can have a strong effect on whether cancer develops in the context of mutant ras signaling and mutant p53, and that this mouse model of astrocytoma can be used to identify modifier phenotypes with complex inheritance patterns that would be unidentifiable in humans. Because analysis of gene function in the mouse is often performed on a mixed C57BL/6,129 strain background, these data also provide a powerful example of the potential of these strains to mask interesting gene functions.
Astrocytoma is a characteristically diffuse tumor of the central nervous system (CNS). Because of its diffuse infiltration, it often cannot be completely resected, leading to a very poor prognosis for patients. Astrocytoma, together with glioblastoma (the highest grade of astrocytoma), accounts for more than three-quarters of all gliomas, making it the most common malignant brain tumor (1). The 5-year survival rate for glioblastoma is <3%. A better understanding of the genetic risk factors associated with astrocytoma will give insight into the mechanism of astrocytoma initiation and progression and will lead to better screening methods and new targets for therapy.
Data from human populations pointing to genetic risk factors for astrocytoma are sparse. Malmer et al. (2) have examined the increased family risk of developing low- vs. high-grade glioma and favor the view that autosomal recessive genes affect astrocytoma risk, although the role of a common environment in familial risk cannot be excluded. Several familial cancer syndromes show an increased risk for astrocytoma, including neurofibromatosis type 1 (NF1) (3, 4) and Li-Fraumeni syndrome (LFS) (5). NF1 patients have a mutation in the NF1 gene (6) (Nf1 in the mouse) and are predisposed to neurofibromas and optic gliomas, with an increased risk for malignant peripheral nerve sheath tumors and diffuse astrocytoma/glioblastoma (3, 4). Studies of NF1 families have demonstrated a role for modifier genes unlinked to NF1 in the severity of the disease with respect to the numbers of neurofibromas and the presence or absence of optic gliomas (7). Studies of NF1 patients have also shown that patients with optic glioma are more likely to develop CNS tumors such as astrocytoma (8, 9), although this observation is as likely due to the particular mutant allele of NF1 (10) as to genetic or environmental risk factors. Astrocytomas are also frequently associated with mutations in Tp53 in sporadic cases (11, 12) or in LFS patients carrying germline mutations in the Tp53 gene (5) (Trp53 in the mouse). Although there are no clear data in patients that genetic background affects the risk of particular tumors in LFS patients, data from the mouse show that the strain background can affect the incidence of teratomas and mammary tumors in Trp53 mutant mice (13, 14). The relatively low penetrance of astrocytoma in the population makes association studies to look at genetic risk factors difficult. The identification of these risk factors is best accomplished in inbred animal models and the results then tested in human populations.
Because of the difficulty of identifying genetic risk factors in humans with astrocytoma, we have used a mouse model of the disease to examine the genetic basis of astrocytoma susceptibility. Although several mouse models now exist that recapitulate the pathology of astrocytoma (15), we have chosen the Nf1;Trp53cis (NPcis) mutant mouse model, because it can be propagated by a simple breeding strategy and is maintained on an inbred strain background to facilitate the dissection of modifier genetics. In this model, Nf1 and Trp53 are mutated on the same chromosome (chr) 11 of the mouse and are tightly linked so that they are inherited as a single mutation in genetic crosses. The mice on a C57BL/6J (B6) inbred strain background develop astrocytomas spontaneously with an average latency of 6 months and show loss of the WT copies of Nf1 and Trp53 (16). Because astrocytomas were not observed in the original characterization of NPcis mice on a C57BL/6J,129S4/SvJae mixed background (17, 18), we hypothesized that modifier genes affect susceptibility to astrocytoma between the B6 and 129S4/SvJae (129) strains. To address this possibility directly, we have generated this model on an inbred 129 background and examined the genetics of susceptibility to astrocytoma.
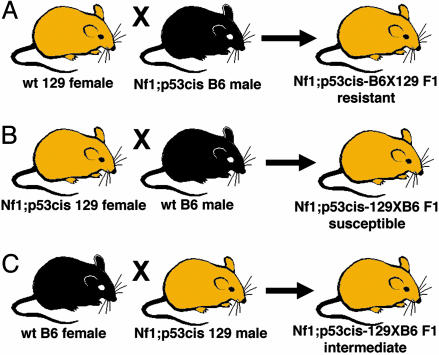
Materials and Methods
Generation of NPcis Mice on Different Genetic Backgrounds. NPcis mice on a B6 background were generated as described (16) and maintained by crossing to WT B6 purchased from The Jackson Laboratory. The NPcis-B6 mice described here are backcross generation 8-11 onto B6. The NPcis-129 mice were generated by crossing Nf1+/- 129 mice (19) to Trp53+/- 129 mice (20) to generate Nf1;Trp53trans 129 mice that were then crossed to WT 129 mice, maintained as a colony at Massachusetts Institute of Technology (MIT) to generate the NPcis mice. F1 progeny between NPcis-B6 and A/J (A), DBA/2J (DB), and CBA/J (CB) were generated by crossing NPcis-B6 mice to inbred strains purchased from The Jackson Laboratory. NPcis-B6 × 129 F1 progeny were generated from an NPcis-B6 male inbred 16 generations onto B6 crossed to WT 129 females (Fig. 3, cross A). NPcis-129XB6 F1 progeny were generated by crossing either male or female NPcis-129 mice to WT B6 (Fig. 3, cross B and C). All mice were maintained on a 9% fat diet. Twelve of the mice in this study (all F1s between B6 and 129) were moved from MIT to National Cancer Institute-Frederick (NCI-Frederick) at ≈6 months of age to complete the aging study. We have not observed any change in phenotype of these strains at NCI-Frederick, so these mice were pooled with the mice aged entirely at MIT. All mice in this study were cared for according to the policies of the Animal Care and Use Committees of MIT and NCI-Frederick.
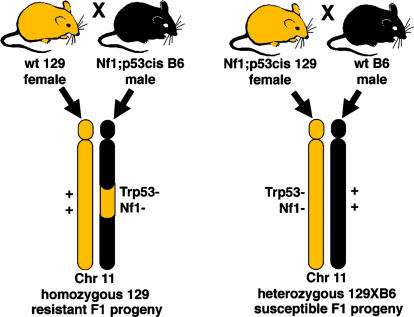
Fig. 3.
F1 crosses between B6 and 129. F1 progeny were generated in three different crosses. (A) An NPcis-B6 male was bred to a WT 129 female to give NPcis-B6X129 F1 progeny that were resistant to astrocytoma. (B) NPcis-129 females were bred to WT B6 males to give NPcis-129XB6 F1 progeny that were susceptible to astrocytoma. (C)An NPcis-129 male was bred to a WT B6 female to give NPcis-129XB6 F1 progeny that developed an intermediate astrocytoma phenotype.
Genotyping of NPcis Mice. Mice were genotyped for Trp53 mutations by PCR on tail-clip DNA as described (20). Mutations in Nf1 were assayed by PCR as described (19) or by using the primers Nf1 WT 5′-TTCTGGCCTTATTGGACACC-3′, Nf1 common 5′-GCACAAAAGAGGCACTGGAT-3′, Nf1 mutant 5′-GGAGAGGCTTTTTGCTTCCT-3′, with an annealing temperature of 60°C. We added 0.1% BSA and 1% polyvinylpyrrolidone (catalogue no. PVP-40, Sigma-Aldrich) to improve specificity of the PCR reaction (21). For analysis of simple sequence length polymorphisms (SSLPs), tail DNA was amplified with ResGen MapPairs primer sets (Invitrogen) for 35 cycles and run on a 3% Metaphor agarose (Cambrex, East Rutherford, NJ), 1× Tris-borate-EDTA-buffered gel. The markers used were D11Mit20, D11Mit271, D11Mit140, D11Mit349, D11Mit5, D11Mit341, D11Mit285, and D11Mit258. Chromosomal locations of SSLP markers, Trp53, and Nf1 were taken from National Center for Biotechnology Information (NCBI) Mouse Build 32, version 1 (www.ncbi.nlm.nih.gov/genome/guide/mouse).
Phenotyping of NPcis Mice. Mice were aged until tumors developed as described (16) and processed for hematoxylin and eosin-stained sections. The midline sagittal section and a parasagittal section through the eye were examined independently by K.M.R. and by two veterinary neuropathologists, R.T.B. and C.D.S. A consensus diagnosis was determined by K.M.R. based on the World Health Organization (WHO) grading criteria for diffuse astrocytoma (22) and given the prefix GEM to indicate that the tumors arise in genetically engineered mouse models.
Statistical Analysis of Tumor Spectrum. Statistical analysis was performed as described (16). Differences in astrocytoma incidence were compared pairwise between strains by χ2 test in Microsoft excel X.
Single Nucleotide Polymorphism (SNP) Analysis. Chr 11 SNPs with a B6 genotype were downloaded from the NCBI Single Nucleotide Polymorphism Database (dbSNP) (Build 120) (www.ncbi.nlm.nih.gov/SNP). We selected 597 SNPs located between D11Mit271 (45.4 Mb, NCBI Build 32) and D11Mit285 (82.6 Mb, NCBI Build 32) polymorphic for B6 and 129. PCR primers were designed for 161 DNA fragments containing the 597 SNPs. reveal analysis (Spectrumedix, State College, PA) was used to identify PCR fragments that were polymorphic between B6 and 129. Of the 89 DNA fragments found to be polymorphic or uninterpretable by reveal analysis, 48 fragments were chosen for sequencing. SNPs were confirmed by sequencing in both directions along the DNA strand and genotyped on the B6, 129, A, CB, and DB strains. Sequencing primers used are reported in Table 4. The Celera SNP database (www.celeradiscoverysystems.com) was searched for SNPs between D11Mit271 (44.2 Mb, release R3.6) and D11Mit285 (96.5 Mb, release R3.6). Two search criteria were used: (i) 129S1/SvImJ (129S1) ≠ B6 and 129S1 ≠ A and 129S1 ≠ DB and (ii) 129X1/SvJ (129X1) ≠ B6 and 129X1 ≠ A and 129X1 ≠ DB. Results were manually checked for genotyping ambiguities. The genotype with the higher count number was taken as the more accurate genotype. Results of the search are reported in Table 5, which is published as supporting information on the PNAS web site.
Results
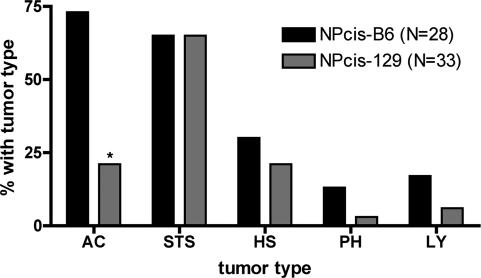
To test the difference in susceptibility to astrocytoma between the B6 and 129 strains, we have regenerated the NPcis mice on an inbred 129 background by crossing Nf1+/- 129 and Trp53+/- 129 mice to regenerate the cis mutant chr (16-18). NPcis mice on a 129 background develop significantly fewer astrocytomas compared with NPcis-B6 mice (Fig. 1). Soft-tissue sarcomas were seen frequently on both the B6 and 129 backgrounds, as observed on the B6,129 mixed background (17, 18). We did not observe significant differences in the incidence of other common tumor types, lymphoma, histiocytic sarcoma, or pheochromocytoma. NPcis-129 mice developed fewer astrocytomas in the population than NPcis-B6 mice, and the astrocytomas were lower grade in NPcis-129 mice. On the B6 background, 89% of astrocytomas were classified as GEM WHO III, whereas 29% of astrocytomas on the 129S4/SvJae background were classified as GEM WHO III, with the remainder classified as GEM WHO II (Fig. 2). We have generated cell lines from NPcis-129 astrocytomas that show loss of the WT copies of Trp53 and Nf1 (data not shown).
Fig. 1.
Tumor spectrum of NPcis-B6 and NPcis-129 mice. The graph shows the major tumor types observed. The difference in astrocytoma incidence in the two strains is statistically significant (P = 0.001, indicated by asterisk). Tumor types are astrocytoma (AC), soft-tissue sarcoma (STS), histiocytic sarcoma (HS), pheochromocytoma (PH), and lymphoma (LY).
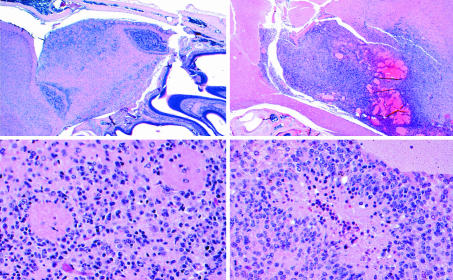
Fig. 2.
Histology of astrocytoma in the resistant NPcis-129 strain compared with the susceptible F1 progeny of NPcis-129 crossed to WT B6. (Left) One of the two GEM WHO III tumors found in NPcis-129 mice (n = 33). The tumor is confined to a focal area of the ventral olfactory bulb (Upper Left). Atypical nuclei diffusely infiltrate the olfactory bulb (Lower Left) and mitotic figures are rare (not shown). (Right) One of three GEM WHO IV tumors found in F1 NPcis-129XB6 progeny (n = 22). This tumor is characterized by infiltrative boundaries (Upper Right), a high mitotic index, and pseudopallisading tumor cells around areas of necrosis (Lower Right).
To determine whether the genes modifying astrocytoma formation in NPcis mice act dominantly in the B6 or 129 strain, we generated F1 progeny between the B6 and 129 strains carrying the NPcis mutation. We found upon analyzing these F1 progeny that susceptibility to astrocytoma does not follow simple Mendelian inheritance, in that some F1 progeny were resistant to astrocytoma and others were susceptible (Fig. 3, Table 1). We have examined the data to look for epigenetic effects with respect to inheritance of the strain background from the mother or father, inheritance of the NPcis mutant chr from the B6 or 129 strain, and inheritance of the NPcis mutant chr from the mother or father.
Table 1. Inheritance of mutant chr 11 affects astrocytoma susceptibility.
| Cross (Fig. 3) | Cross | n | With astrocytoma | Without astrocytoma | P, χ2 |
|---|---|---|---|---|---|
| A | WT 129 female × NPcis B6 male | 19 | 6 (32%) | 13 (68%) | |
| B | NPcis 129 female × WT B6 male | 22 | 16 (73%) | 6 (27%) | |
| C | WT B6 female × NPcis 129 male | 23 | 12 (52%) | 11 (48%) | |
| A+B | 129 female × B6 male | 41 | 22 (54%) | 19 (46%) | |
| C | B6 female × 129 male | 23 | 12 (52%) | 11 (48%) | 0.9 |
| B+C | NPcis 129 × WT B6 | 45 | 28 (62%) | 17 (38%) | |
| A | NPcis B6 × WT 129 | 19 | 6 (32%) | 13 (68%) | 0.03 |
| A+C | WT female × NPcis male | 42 | 18 (43%) | 24 (57%) | |
| B | NPcis female × WT male | 22 | 16 (73%) | 6 (27%) | 0.02 |
Maternal or paternal imprinting of a modifier from B6 or 129 could alter the expression pattern of the modifier gene(s) and affect its ability to act on astrocytoma susceptibility. Similarly, strain differences inherited specifically from the mother, such as mitochondria or other cytoplasmic maternal factors, could potentially affect developmental pathways leading to indirect effects on astrocytoma susceptibility. We examined whether astrocytoma susceptibility changed when B6 alleles were inherited from the mother or father. Table 1 shows that astrocytoma susceptibility is not affected by imprinting of strain-specific genes or by strain-specific maternal factors. Mice that inherit B6 from either the maternal (Fig. 3, cross C) or the paternal side (Fig. 3, crosses A and B) show a similar incidence of astrocytoma. In both cases, F1 progeny were significantly more susceptible than the NPcis 129 parental strain (P = 0.03 for B6 coming from the mother, and P = 0.01 for B6 coming from the father).
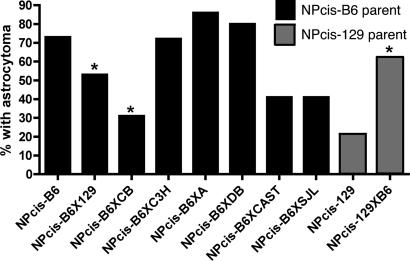
The Nf1 and Trp53 mutant chr, or conversely WT chr 11, could affect tumorigenesis differently, depending on whether it is inherited from B6 or 129. For example, particular alleles of Nf1 or Trp53 could be less effective at blocking tumorigenesis in the heterozygous state, or the WT chr from one strain might be lost more easily than the other strain to initiate tumorigenesis, as has been shown for strain effects on APCMin/+ mice (23). Additionally, genes tightly linked to Nf1 and Trp53 on chr 11 might affect tumorigenesis differently when inherited from B6 or 129. We examined the effect of inheriting the mutant chr from either 129 or B6. Table 1 shows that F1 progeny inheriting the NPcis mutant chr from 129 (Fig. 3, crosses B and C) are significantly more susceptible to developing astrocytoma than F1 progeny inheriting the mutant chr from the B6 strain (Fig. 3, cross A) (P = 0.03). F1 progeny inheriting the mutant chr from 129 and the WT chr 11 from B6 are significantly more susceptible to astrocytoma than the NPcis-129 parental strain (Fig. 4) (P = 0.0009), demonstrating a dominant effect of the WT B6 chr 11 on astrocytoma susceptibility. The F1 progeny inheriting the mutant chr from B6 and the WT chr 11 from 129 are significantly more resistant to astrocytoma than the NPcis-B6 parent (Fig. 4) (P = 0.02), demonstrating a dominant effect of the WT 129 chr 11 on astrocytoma resistance. These data suggest that the WT copy of chr 11 acts dominantly to modify susceptibility to astrocytoma.
Fig. 4.
Astrocytoma incidence in F1 progeny of different inbred strains. NPcis-B6X129 F1 progeny (cross A, Fig. 3) show reduction in astrocytoma incidence compared to the NPcis-B6 parental strain (P = 0.02). NPcis-129XB6 F1 progeny (cross B and C, Fig. 3), show increase in astrocytoma incidence compared to the NPcis-129 parental strain (P = 0.0009). NPcis-B6XCB F1 progeny show reduction in astrocytoma incidence compared to the NPcis-B6 parental strain (P = 0.04). Previously published data (16) from B6XC3H/HeJ, B6XCAST/EiJ, and B6XSJL/J are shown for comparison. Of the seven strains tested, only 129 and CB show significant resistance to astrocytoma compared to B6. Statistically significant changes relative to the parental strain are indicated by asterisks. NPcis-B6 n = 28, NPcis-B6X129 n = 19, NPcis-B6XCB n = 26, NPcis-B6XC3H n = 25, NPcis-B6XA n = 20, NPcis-B6XDB n = 20, NPcis-B6XCAST n = 22, NPcis-B6XSJL n = 34, NPcis-129 n = 33, and NPcis-129XB6 n = 45. (Adapted from ref. 16.)
Because the data in Table 1 point to a role of chr 11 in modifying astrocytoma susceptibility, we determined whether imprinting on this chr affects astrocytoma formation, given that one copy of chr 11 is lost during tumor initiation, leaving only the maternal or paternal copy. This is distinct from the analysis described above, in that it does not require the modifier to be polymorphic between B6 and 129, but only that it be linked to Nf1 and Trp53. Table 1 shows that F1 progeny inheriting the NPcis chr from the mother (Fig. 3, cross B) are more susceptible to astrocytoma than F1 progeny inheriting the mutant chr from the father (Fig. 3, cross A and C) (P = 0.02). We found no significant differences in the incidence of astrocytoma between male and female F1 progeny. It is not clear from this analysis whether the imprinting effect is at the same gene locus as the B6 or 129 modifying locus, but both loci are linked to chr 11.
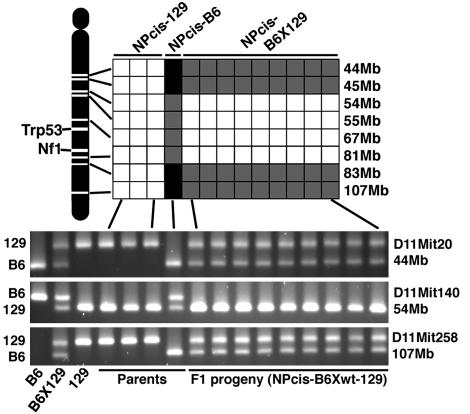
To further analyze the differences on chr 11 between the different F1 progeny, we genotyped SSLP markers along the length of chr 11. The original Nf1 and Trp53 mutations used to generate the NPcis-B6 animals were made on a 129 strain background (19, 20). During the inbreeding of mutations onto B6, the region around Nf1 and Trp53 is continually selected and retains 129 sequence. The NPcis-B6 male founder used to generate the F1 progeny in cross A of Fig. 3 retains 129 sequence from 54 through 81 Mb on chr 11 (Fig. 5). The NPcis F1 progeny in cross A inherit this congenic region and are homozygous for 129 sequence around Trp53 and Nf1.
Fig. 5.
SSLP characterization of chr 11 in NPcis parents and F1 progeny. SSLP markers spanning the region around Trp53 and Nf1 were genotyped for differences between B6 and 129. (Upper) A summary of the results indicating homozygous 129 (white), homozygous B6 (black), and heterozygous B6X129 (gray). (Lower) Three SSLP examples. Trp53 is located at 69 Mb and Nf1 is located at 73 Mb. In the congenic region the NPcis-B6 parent is heterozygous for 129 and B6 sequences, and the F1 progeny are homozygous for 129. Outside the congenic region the NPcis-B6 parent is homozygous for B6 sequences, and the F1 progeny are heterozygous for 129 and B6 sequences. The first three lanes of the gel (Lower) are inbred and F1 control DNA samples to show the size of the SSLP fragments for each strain.
We performed a survey of six additional inbred strains to look at the effects on astrocytoma susceptibility in F1 progeny. Three of these F1 strain combinations (B6XC3H/HeJ, B6XCAST/EiJ, and B6XSJL/J) have been published (16) and are included in Fig. 4 for comparison. As we described previously, NPcis-B6XCAST/EiJ and NPcis-B6XSJL/J F1 progeny develop fewer astrocytomas overall but develop astrocytomas at the same age as NPcis-B6 mice and show accelerated tumor latency, developing other tumors before they live long enough to develop astrocytoma. Therefore, the reduction of astrocytoma in these two F1 groups is not necessarily due to resistance to astrocytoma. We found that the CB strain had a similar effect to the 129 strain when crossed to NPcis-B6 mice. In both cases, the F1 progeny are significantly more resistant to astrocytoma than the NPcis parental strain. The NPcis-B6XCB F1 progeny showed no significant differences in any other tumor type, developing soft-tissue sarcomas, pheochromocytomas, histiocytic sarcomas, and lymphomas similarly to the NPcis-B6 parental line, and there was no significant change in the survival curves between the F1 and the B6 parental strain. Furthermore, the astrocytomas that developed in NPcis-B6XCB mice were delayed relative to NPcis-B6 mice. Thus, similar to the modifying effect observed in the 129 strain, the CB strain modifier effect is limited to increasing resistance to astrocytoma and is not seen in five other strains examined.
Because both 129 and CB can dominantly repress astrocytomas in NPcis-B6 mice, whereas A and DB cannot, the polymorphisms responsible for this effect are expected to be polymorphic between 129 and B6, 129 and A, and 129 and DB. If the dominant effects of 129 and CB are due to the same locus, the polymorphisms may also have a common haplotype in 129 and CB. We analyzed SNPs from both the NCBI dbSNP database and the Celera SNP database to determine whether there are SNPs within the region shown in Fig. 5 that fit these criteria. We selected 597 SNPs from the congenic region between D11Mit271 and D11Mit285 in NCBI dbSNP, contained on 161 PCR fragments. Of these 161 PCR fragments, 72 were found to not contain polymorphisms between B6 and 129 by reveal analysis of hybrid stability. Of the remaining 89, 48 were sequenced, and of 159 SNPs represented, 13 were found to be polymorphic for B6 and 129. Of the 13 SNPs, 8 were polymorphic for 129 and A, and 6 were polymorphic for 129 and DB. Two SNPs were found that met all criteria, in that they were polymorphic for 129 and B6, 129 and A, and 129 and DB, and were not polymorphic for 129 and CB (Table 2)
Table 2. NCBI, sequenced SNPs polymorphic for B6 and 129.
| ss# | Gene | N Mb* | C Mb* | B6 | A | DB | 129 | CB |
|---|---|---|---|---|---|---|---|---|
| 12733678 | Vamp2 | 68.7 | 73.0 | G | G | G | A | G |
| 12733665 | Vamp2 | 68.7 | 73.0 | A | A | A | G | A |
| 12733667 | Vamp2 | 68.7 | 73.0 | G | G | G | A | G |
| 12732038 | Chrnb1 | 69.4 | C | C | T | T | ||
| 5069192 | Trpv1† | 72.8 | 77.8 | C | C | C | G | G |
ss#, NCBI dbSNP submitted sequence identification no.
N Mb, physical location in NCBI; C Mb, location in Celera.
The Trpv1 SNP and one of the SNPs in Trpv3 in Table 3 are the same SNP.
The Celera database was searched directly for SNPs that are polymorphic for 129 and B6, 129 and A, and 129 and DB. There are two 129 substrains represented in the Celera database, 129X1 and 129S1. Of the 46,736 SNPs within the congenic region between D11Mit271 and D11Mit285, 16,220 SNPs were polymorphic between either B6 and 129X1 or B6 and 129S1. Of the 16,220 SNPs, 854 were polymorphic for A and DB. The SNPs covered intergenic regions and 117 gene products. Identified genes varied between those with a single candidate SNP and those with as many as 41 candidate SNPs. The genes with many SNPs fitting the candidate criteria likely represent regions of common haplotypes between B6, A, and DB, polymorphic with 129. Because of potential differences between our 129 strain and 129X1 and 129S1, these 854 candidate SNPs will need to be confirmed by direct sequencing of 129 and CB. The candidate genes with >10 candidate SNPs are listed in Table 3. The complete summary of the candidate SNPs identified from NCBI dbSNP and Celera are included as Tables 4 and 5.
Table 3. Celera, genes with >10 candidate SNPs.
| Gene ID | Gene | N Mb* | C Mb* | No. SNPs |
|---|---|---|---|---|
| mCG140133 | Trim11 | 58.6 | 60.7 | 11 |
| mCG23374 | NcoR1 | 61.9 | 66.3 | 41 |
| mCG14821 | 1700019l23Rik | 62.5 | 66.9 | 15 |
| mCG11237 | Vamp2 | 68.7 | 73.0 | 11 |
| mCG21169 | Rabep1 | 70.4 | 75.4 | 31 |
| mCG6663 | Ankfy1 | 72.3 | 77.3 | 12 |
| mCG140764 | Trpv3† | 72.8 | 77.8 | 11 |
| mCG48633 | Msi2h | 87.9 | 95.0 | 28 |
N Mb, physical location in NCBI; C Mb, location in Celera.
The Trpv1 SNP in Table 2 and one of the SNPs in Trpv3 are the same SNP.
Discussion
The identification of modifier genes or susceptibility and resistance genes in human cancer is difficult, particularly in the case of less common cancers such as astrocytoma, where familial clustering is more difficult to find. We demonstrate here that modifier genes on mouse chr 11 affect susceptibility to astrocytoma in the presence of mutations in the ras and p53 signaling pathways, two of the major pathways mutated in human astrocytoma. Furthermore, these modifier genes show complex inheritance patterns in mice, suggesting that their identification in humans would be especially difficult. We use phenotype data from several inbred strains to identify potential candidate genes on chr 11. By identifying the genes responsible for these effects and understanding their mechanism, it may be possible to implicate them in human astrocytoma.
The data presented here show that susceptibility to astrocytoma depends on the mode of inheritance of the NPcis mutant chr. Because NPcis-B6 mice were inbred from a B6,129 background and the original mutations were engineered on a 129 background (19, 20), the NPcis mice inbred onto B6 carry regions of 129 sequence surrounding the two mutations on chr 11. Our data suggest that a major modifier affecting resistance to astrocytoma lies within these 129-retained regions in the NPcis B6 inbred mice. We have mapped the region of 129 sequence in the NPcis-B6 parent of our F1 crosses and found that the region is >27 and <38 Mb in length surrounding Nf1 and Trp53 (Fig. 5). According to our model (Fig. 6), NPcis-129 mice are homozygous for 129 alleles at all loci, whereas NPcis-B6 mice are heterozygous for B6 and 129 at a small number of loci, including the region around Nf1 and Trp53 on chr 11. The B6 modifier allele(s) acts dominantly in this model to increase susceptibility to astrocytoma. In the case of the F1 progeny inheriting the NPcis chr from the 129 line, the progeny inherit the modifier allele from the WT B6 parent and are susceptible (Table 1). In the case of the F1 progeny inheriting the NPcis chr from the B6 line, the progeny do not inherit the dominant modifier allele and are resistant. The model predicts that B6, A, and DB all carry similar dominant susceptibility alleles at the modifier locus, whereas 129 and CB carry recessive resistance alleles. The dominant effect seen in NPcis B6XCB mice is due to loss of the B6 susceptibility allele and maintenance of two recessive resistance alleles at the modifier locus. Alternatively, the modifier in the CB background may be unlinked to the modifier on chr 11 but acts dominantly to suppress astrocytoma in the F1 progeny.
Fig. 6.
Model for inheritance of resistance to astrocytoma. The F1 progeny showing different susceptibility to astrocytoma differ in the strain background around the Trp53 and Nf1 loci. NPcis-B6 mice are inbred from 129 and carry a 129 congenic region on chr 11 (Left), whereas NPcis-129 mice are inbred 129 along the entire length of chr 11 (Right). F1 progeny differ in whether they are homozygous for 129 around Nf1 and Trp53 or heterozygous for B6 and 129. According to this model, the B6 alleles on chr 11 confer susceptibility to astrocytoma in a dominant manner.
We have tested whether candidate SNPs exist within the congenic region that are consistent with our phenotype data, using in silico haplotype mapping (24, 25) and sequencing. Importantly, no candidate SNPs were identified in Trp53 or Nf1. Many of the candidates identified are expressed in the brain, and several are implicated in CNS stem cell biology or astrocytoma. NcoR1 has been shown to inhibit the differentiation of neural stem cells into astrocytes (26). The Msi2h gene product has been implicated in the proliferation and maintenance of neural stem cells (27). Changes in neural stem cells could favor the development of astrocytoma by increasing the number of cells available to become tumorigenic. Alternatively, genes affecting neural stem cell differentiation could affect how well astrocytes maintain their differentiated state and resist transformation. Sparc is up-regulated in diffuse astrocytomas (28); however, because the 129, A, and DB share a common genotype, Sparc is not the best candidate for the astrocytoma modifier.
In addition to the observed strain-origin effect in F1s, we have also observed a parent-origin effect of NPcis inheritance in F1s (Table 1). Because the parent-origin effect is specific to chr 11 and not to the inheritance of B6 or 129, we argue that strain-specific differences in mitochondria from the mother or other cytoplasmic maternal factors are not responsible for this effect. This suggests that imprinting of genes on chr 11 modulates the susceptibility to astrocytoma. Imprinted genes have been identified on chr 11 in the mouse, supporting this possibility (29, 30). The F1 data suggest that inheritance of the WT chr 11 from the father increases susceptibility, whereas inheritance of the WT chr 11 from the mother decreases susceptibility. It remains to be seen whether this effect is due to silencing of a maternal susceptibility allele or to silencing of a paternal resistance allele, and whether this effect acts on the same locus as the strain-origin effect. Whereas the strain-origin effect points to a locus linked to Nf1 and Trp53, due to 129 sequences found in NPcis-B6 mice, the imprinted modifier could be anywhere on chr 11, affecting tumorigenesis through loss or reduplication of monoallelic expression. Interestingly, Grb10 is a tyrosine kinase receptor adaptor protein imprinted on chr 11 and paternally expressed in the mouse brain (31, 32). It is likely that many imprinted genes exist along chr 11, and our experiments do not localize the imprinted modifier because large regions of WT chr 11 are likely lost during tumor initiation. It remains to be seen whether Grb10 or another monoallelically expressed gene on chr 11 can specifically alter tumor progression.
The modifier genes acting on astrocytoma susceptibility in this mouse model do not affect the development of other tumor types or tumor latency. This suggests that the modifiers act not on the overall tumor suppressor function of Nf1 and/or Trp53 but rather on the effect of one or both of these genes on the CNS. The modifier genes could be acting within the precursor cell to the astrocytoma, the tumor cell as it progresses, or in the surrounding normal tissue to support tumor growth and survival. Because the WT copies of Nf1 and Trp53 are lost during the initiation of tumorigenesis in NPcis mice (16, 17), modifier alleles linked to these genes on the WT copy of chr 11 may be affecting tumorigenesis during initiation steps or by acting in surrounding normal tissue to alter tumor progression. We observe that the tumors in NPcis 129 mice are lower grade in addition to being fewer in number; therefore, we favor an effect of the modifier on tumor progression as well as tumor initiation.
Much of the characterization of mutant phenotypes in the mouse occurs on 129 substrains or B6,129 mixed strain backgrounds. This is due to the predominant use of 129 embryonic stem cells for gene targeting. These data demonstrate the importance of examining phenotypes on multiple strain backgrounds. The role of mutations in the ras and p53 pathways in astrocytoma is well established; however, mouse models generated by mutation in these pathways fail to develop astrocytoma on the 129 inbred and B6,129 mixed backgrounds. It is only when the mutations are moved to B6 that the importance of these pathways in murine astrocytoma is appreciated.
Supplementary Material
Acknowledgments
We thank M. Perella, D. Crowley, A. Caron, and K. Mercer for technical assistance; E. Frazier for assistance with figures; and M. McLaughlin, N. Copeland, and K. Hunter for helpful discussions. This work was supported in part by grants from the Leukemia and Lymphoma Society, the American Association for Cancer Research, the American Cancer Society, the Department of the Army, and the Ludwig Foundation, and by federal funds from the National Cancer Institute under contract NO1-CO-12400 to Science Applications International Corporation-Frederick. T.J. is an Investigator of the Howard Hughes Medical Institute.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: NF1, neurofibromatosis type 1; SSLP, simple sequence length polymorphism; NPcis, Nf1;Trp53cis; B6, C57BL/6J; 129, 129S4/SvJae; A, A/J; DB, DBA/2J; CB, CBA/J; 129S1, 129S1/SvImJ; 129X1, 129X1/SvJ; SNP, single nucleotide polymorphism; chr, chromosome; WHO, World Health Organization; NCBI, National Center for Biotechnology Information; dbSNP, Single Nucleotide Polymorphism Database.
Data deposition: The SNP data reported in this paper have been deposited in the NCBI Single Nucleotide Polymorphism Database (dbSNP) (dbSNP ID nos. 28476647-28476655; see also Table 4, which is published as supporting information on the PNAS web site).
References
- 1.Central Brain Tumor Registry of the United States (2002) CBTRUS: 2002-2003 Primary Brain Tumors in the United States-Statistical Report (Central Brain Tumor Registry of the United States, Chicago).
- 2.Malmer, B., Henriksson, R. & Gronberg, H. (2002) Neuroepidemiology 21, 279-286. [DOI] [PubMed] [Google Scholar]
- 3.Blatt, J., Jaffe, R., Deutsch, M. & Adkins, J. C. (1986) Cancer 57, 1225-1229. [DOI] [PubMed] [Google Scholar]
- 4.Sorensen, S. A., Mulvihill, J. J. & Nielsen, A. (1986) N. Engl. J. Med. 314, 1010-1015. [DOI] [PubMed] [Google Scholar]
- 5.Malkin, D. (1994) Biochem. Biophys. Acta 1198, 197-213. [DOI] [PubMed] [Google Scholar]
- 6.Gutmann, D., Wood, D. & Collins, F. (1991) Proc. Natl. Acad. Sci. USA 88, 9658-9662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Easton, D., Ponder, M., Huson, S. & Ponder, B. (1993) Am. J. Hum. Genet. 53, 305-313. [PMC free article] [PubMed] [Google Scholar]
- 8.Friedman, J. M. & Birch, P. (1997) Neuropediatrics 28, 131-132. [DOI] [PubMed] [Google Scholar]
- 9.Szudek, J., Evans, D. G. & Friedman, J. M. (2003) Hum. Genet. 112, 289-297. [DOI] [PubMed] [Google Scholar]
- 10.Ars, E., Kruyer, H., Morell, M., Pros, E., Serra, E., Ravella, A., Estivill, X. & Lazaro, C. (2003) J. Med. Genet. 40, e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.von Deimling, A., Eibl, R. H., Ohgaki, H., Louis, D. N., von Ammon, K., Petersen, I., Kleihues, P., Chung, R. Y., Wiestler, O. D. & Seizinger, B. R. (1992) Cancer Res. 52, 2987-2990. [PubMed] [Google Scholar]
- 12.Fults, D., Brockmeyer, D., Tullous, M. W., Pedone, C. A. & Cawthon, R. M. (1992) Cancer Res. 52, 674-679. [PubMed] [Google Scholar]
- 13.Harvey, M., McArthur, M., Montgomery Jr., C., Bradley, A. & Donehower, L. (1993) FASEB J. 7, 938-942. [DOI] [PubMed] [Google Scholar]
- 14.Kuperwasser, C., Hurlbut, G. D., Kittrell, F. S., Dickinson, E. S., Laucirica, R., Medina, D., Naber, S. P. & Jerry, D. J. (2000) Am. J. Pathol. 157, 2151-2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weiss, W. A., Israel, M., Cobbs, C., Holland, E., James, C. D., Louis, D. N., Marks, C., McClatchey, A. I., Roberts, T., Van Dyke, T., et al. (2002) Oncogene 21, 7453-7463. [DOI] [PubMed] [Google Scholar]
- 16.Reilly, K. M., Loisel, D. A., Bronson, R. T., McLaughlin, M. E. & Jacks, T. (2000) Nat. Genet. 26, 109-113. [DOI] [PubMed] [Google Scholar]
- 17.Cichowski, K., Shih, T., Schmitt, E., Santiago, S., Reilly, K., McLaughlin, M., Bronson, R. & Jacks, T. (1999) Science 286, 2172-2176. [DOI] [PubMed] [Google Scholar]
- 18.Vogel, K., Klesse, L., Velasco-Miguel, S., Meyers, K., Rushing, E. & Parada, L. (1999) Science 286, 2176-2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacks, T., Shih, T. S., Schmitt, E. M., Bronson, R. T., Bernards, A. & Weinberg, R. A. (1994) Nat. Genet. 7, 353-361. [DOI] [PubMed] [Google Scholar]
- 20.Jacks, T., Remington, L., Williams, B. O., Schmitt, E. M., Halachmi, S., Bronson, R. T. & Weinberg, R. A. (1994) Curr. Biol. 4, 1-7. [DOI] [PubMed] [Google Scholar]
- 21.Xin, Z., Velten, J. P., Oliver, M. J. & Burke, J. J. (2003) BioTechniques 34, 820-824, 826. [DOI] [PubMed] [Google Scholar]
- 22.Kleihues, P. & Cavenee, W. (2000) Pathology and Genetics of Tumours of the Nervous System (International Agency for Research on Cancer, Lyon, France).
- 23.Shoemaker, A. R., Moser, A. R., Midgley, C. A., Clipson, L., Newton, M. A. & Dove, W. F. (1998) Proc. Natl. Acad. Sci. USA 95, 10826-10831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grupe, A., Germer, S., Usuka, J., Aud, D., Belknap, J. K., Klein, R. F., Ahluwalia, M. K., Higuchi, R. & Peltz, G. (2001) Science 292, 1915-1918. [DOI] [PubMed] [Google Scholar]
- 25.Park, Y. G., Clifford, R., Buetow, K. H. & Hunter, K. W. (2003) Genome Res. 13, 118-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hermanson, O., Jepsen, K. & Rosenfeld, M. G. (2002) Nature 419, 934-939. [DOI] [PubMed] [Google Scholar]
- 27.Sakakibara, S., Nakamura, Y., Yoshida, T., Shibata, S., Koike, M., Takano, H., Ueda, S., Uchiyama, Y., Noda, T. & Okano, H. (2002) Proc. Natl. Acad. Sci. USA 99, 15194-15199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rempel, S. A., Golembieski, W. A., Ge, S., Lemke, N., Elisevich, K., Mikkelsen, T. & Gutierrez, J. A. (1998) J. Neuropathol. Exp. Neurol. 57, 1112-1121. [DOI] [PubMed] [Google Scholar]
- 29.Hatada, I., Sugama, T. & Mukai, T. (1993) Nucleic Acids Res. 21, 5577-5582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miyoshi, N., Kuroiwa, Y., Kohda, T., Shitara, H., Yonekawa, H., Kawabe, T., Hasegawa, H., Barton, S. C., Surani, M. A., Kaneko-Ishino, T. & Ishino, F. (1998) Proc. Natl. Acad. Sci. USA 95, 1102-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blagitko, N., Mergenthaler, S., Schulz, U., Wollmann, H. A., Craigen, W., Eggermann, T., Ropers, H. H. & Kalscheuer, V. M. (2000) Hum. Mol. Genet. 9, 1587-1595. [DOI] [PubMed] [Google Scholar]
- 32.Hikichi, T., Kohda, T., Kaneko-Ishino, T. & Ishino, F. (2003) Nucleic Acids Res. 31, 1398-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.