ABSTRACT
FtsZ, a bacterial tubulin homologue, is a cytoskeletal protein that assembles into protofilaments that are one subunit thick. These protofilaments assemble further to form a “Z ring” at the center of prokaryotic cells. The Z ring generates a constriction force on the inner membrane and also serves as a scaffold to recruit cell wall remodeling proteins for complete cell division in vivo. One model of the Z ring proposes that protofilaments associate via lateral bonds to form ribbons; however, lateral bonds are still only hypothetical. To explore potential lateral bonding sites, we probed the surface of Escherichia coli FtsZ by inserting either small peptides or whole fluorescent proteins (FPs). Among the four lateral surfaces on FtsZ protofilaments, we obtained inserts on the front and back surfaces that were functional for cell division. We concluded that these faces are not sites of essential interactions. Inserts at two sites, G124 and R174, located on the left and right surfaces, completely blocked function, and these sites were identified as possible sites for essential lateral interactions. However, the insert at R174 did not interfere with association of protofilaments into sheets and bundles in vitro. Another goal was to find a location within FtsZ that supported insertion of FP reporter proteins while allowing the FtsZ-FPs to function as the sole source of FtsZ. We discovered one internal site, G55-Q56, where several different FPs could be inserted without impairing function. These FtsZ-FPs may provide advances for imaging Z-ring structure by superresolution techniques.
IMPORTANCE One model for the Z-ring structure proposes that protofilaments are assembled into ribbons by lateral bonds between FtsZ subunits. Our study excluded the involvement of the front and back faces of the protofilament in essential interactions in vivo but pointed to two potential lateral bond sites, on the right and left sides. We also identified an FtsZ loop where various fluorescent proteins could be inserted without blocking function; these FtsZ-FPs functioned as the sole source of FtsZ. This advance provides improved tools for all fluorescence imaging of the Z ring and may be especially important for superresolution imaging.
KEYWORDS: green fluorescent protein, mEos, mMaple, superresolution, tubulin, Venus
INTRODUCTION
FtsZ (filamentous temperature sensitive Z), a bacterial homologue of tubulin, assembles the cytoskeletal framework of the Z ring, which constricts to divide the cell. FtsZ forms protofilaments (pfs), which are considered the building blocks of the Z ring. The Z ring serves as a scaffold for recruitment of downstream cell division proteins, which are mostly involved in remodeling the peptidoglycan wall, generating the septum and eventually the new poles. FtsZ by itself can generate a constriction force when reconstituted in liposomes and is thought to provide a primary force to constrict the septum (1, 2). However, recent work suggested that peptidoglycan remodeling limits the rate of constriction and may contribute to the force (3; also see related discussions at the F1000Prime website [4, 5] and in reference 6).
The FtsZ protein comprises a globular domain that is structurally homologous to tubulin. This globular domain can be divided further into two subdomains, which fold independently (7, 8). In Escherichia coli FtsZ, the N-terminal subdomain includes amino acids (aa) 12 to 195, and the C-terminal subdomain includes aa 196 to 316 (8). Following the tubulin-like globular domain, FtsZ has a C-terminal (Ct) tail that tethers it to the membrane. As previously defined (9), the Ct tail consists of a Ct linker, a 50-aa peptide (aa 317 to 366) that is intrinsically disordered and serves as an entropic spring (9), followed by a 17-aa peptide (aa 367 to 383) that binds the membrane proteins FtsA and ZipA, thus tethering FtsZ to the membrane. The final 4 aa do not participate in this binding but may affect protofilament bundling (10).
A major advance in imaging the Z ring was the use of green fluorescent protein (GFP) for fusion to E. coli FtsZ (11). This opened the door for observing FtsZ protein localization and dynamics in living cells. FtsZ-GFP and GFP-FtsZ (with GFP fused at the carboxyl [C] and amino [N] termini, respectively) could localize to the FtsZ ring when expressed at levels lower than that of wild-type FtsZ (wtFtsZ). However, neither fusion could function as the sole source of FtsZ. This suggested that these FtsZ fusions could copolymerize and therefore localize midcell with wtFtsZ, but the fusions apparently interfered with some functions when the proteins were expressed at high levels. Thus, to avoid abnormalities, most groups used FtsZ-GFP as a dilute label. In a later study, Osawa and Erickson found that FtsZ with a C-terminal yellow fluorescent protein (YFP) could function as the sole source of FtsZ after it generated a suppressor mutation somewhere in the E. coli genome (12).
Several superresolution techniques have been applied to bacteria to study the FtsZ ring. Structured illumination microscopy (SIM) and stimulated emission depletion (STED) microscopy give resolutions of ∼100 nm or better, versus the 250-nm resolution of conventional light microscopy. Strauss et al. (13) observed by SIM that the Z rings from both Bacillus subtilis and Staphylococcus aureus do not have a continuous uniform density but are patchy structures with bright segments alternating with gaps. Rowlett and Margolin confirmed the patchy structure of the E. coli Z ring by three-dimensional (3-D) SIM (14). STED microscopy of B. subtilis was able to resolve that the Z ring occasionally separated into a helix with a small pitch that was usually not resolved by conventional fluorescence microscopy (15). This study also imaged irregular and discontinuous helices away from the Z ring.
Another superresolution technique, photoactivated localization microscopy (PALM), can provide an even higher resolution. Several studies have already applied PALM to study the Z rings of the Gram-negative bacteria E. coli and Caulobacter crescentus (16–19). However, a limitation of these studies was that the photoactivatable fluorescent protein (PAFP) was fused to the C terminus of FtsZ, where it could be used only as a dilute label.
In contrast to E. coli, in which FtsZ-GFP could be used only as a dilute label, for B. subtilis Levin et al. (20) produced a strain in which ftsZ-gfp was incorporated into the genome, replacing the ftsZ gene. Strauss et al. (13) created a similar strain and used it for SIM imaging at 30°C. In Streptococcus pneumoniae, Fleurie et al. incorporated ftsZ-gfp into the genome, where it functioned as the sole source of FtsZ (21). Jacq et al. extended this discovery to develop the fully functional, genomically expressed FtsZ-spDendra2 protein, which they used for PALM analysis of S. pneumoniae (19).
The problems of classical N- and C-terminal fusions for supporting cell division suggested a search for a better site. An alternative approach is to insert the fluorescent protein (FP) in frame in a loop of a parent domain. An early report of this insertional “sandwich” fusion was by Ehrmann et al. (22), whose fusion allowed for the topology of the membrane protein MalF (parent domain) to be probed precisely by insertion of alkaline phosphatase (this insertion domain was not an FP). Later studies inserted FPs internally to achieve better function than that with fusion to the N or C terminus (23–25). More recently, Bendezú et al. tested surface-exposed loops of the bacterial actin homolog MreB for insertion of red fluorescent protein (RFP) (26). They found one site that allowed MreB-RFP to replace genomically expressed MreB, and the protein was judged to be fully functional (26).
Our goal was to probe loops on the surfaces of E. coli FtsZ pfs to identify sites where an inserted FP would give a fully functional FtsZ-FP. This would provide an improved tool for imaging the Z ring, especially if we could identify a PAFP that could be used for PALM. A second goal was to identify sites where the inserted FP or a smaller peptide could block a region of FtsZ's lateral surface but allow FtsZ to remain functional. Because these inserts would sterically block protein interactions at the site, this would explore the surface of the pf for potential sites for lateral bonds between FtsZ pfs.
RESULTS
FtsZ with mVenus inserted in the loop at G55-Q56 is fully functional.
To identify sites on FtsZ where an FP might be inserted to give a fully functional construct, we first identified sites on a multiple-sequence alignment where FtsZ proteins of some species showed insertion or deletion of 1 or more aa. The precise locations of the inserts were refined to aa that were exposed within a loop on the surface in the crystal structure of Pseudomonas FtsZ (Protein Data Bank [PDB] entry 2VAW) (27). The sites tested were in loops on the front, back, and lateral surfaces. (The front is the surface equivalent to the outside of a microtubule, the top is the site of GDP, and left and right correspond to the lateral interfaces in a microtubule. See reference 2 for a pf model defining these faces.) Subsequently, we tested additional loop sites in the globular domain and sites within the C-terminal linker, and we made one insert each on the top and bottom that would block assembly of the pf.
We cloned each FtsZ-FP fusion into the pJSB2 plasmid, and we used the E. coli JKD7-1/pKD3 system, originally developed by Dai and Lutkenhaus (28), to test for complementation. JKD7-1 cells, which are genomic ftsZ null cells, are supported by the expression of wtFtsZ from a rescue plasmid, pKD3. pKD3 is temperature sensitive for replication, and after several generations at 42°C, wtFtsZ is depleted. The mutant FtsZ protein to be tested is produced from the arabinose-inducible plasmid pJSB2 (29, 30). In the presence of arabinose (in induction medium) and at the restrictive temperature of 42°C, the pJSB2 plasmid becomes the only source of FtsZ available for cell division. In the present study, we judged a variant form of FtsZ to be capable of complementation when the number of CFU on the induction plate at the restrictive temperature (cells expressing FtsZ solely from pJSB) was at least 80% of that on the repression plate at the permissive temperature (cells expressing wtFtsZ solely from pKD3, with expression from pJSB2 inhibited by the absence of arabinose). This indicated that the FtsZ protein was fully functional for cell division. Note, however, that some of these constructs gave aberrant Z rings and elongated cells when examined in detail, especially during rapid growth at 37°C. For the purposes of this paper, “fully complemented” means that the construct gave full colony formation in the complementation assay and did not need a suppressor mutation.
In a previous study, Osawa and Erickson (12) further investigated FtsZ mutants that were noncomplementing in the simple assay, i.e., those that produced no colonies when 1,000 cells were plated. When 1,000,000 cells were plated, some FtsZ mutants generated 100 to 500 colonies. These were attributed to suppressor mutations that arose in the E. coli genome. (To verify and identify the genomic mutations responsible for suppressor strains, we sequenced the genomes of 13 suppressor strains. Eleven showed a unique single-nucleotide polymorphism that we interpreted to be responsible for the suppressor phenotype. The genes were in various metabolic pathways, mostly with no obvious connection to cell division. We concluded that a wide variety of metabolic pathways can feed back to and facilitate cell division by a slightly compromised FtsZ protein. Details of this study are in the thesis of Kiani Gardner, which is available at http://dukespace.lib.duke.edu/dspace/handle/10161/7189.) These particular FtsZ mutants were therefore functional for cell division in the suppressor strain. In the present study, we first tested all FtsZ linker substitutions for simple complementation. Those that did not provide complementation were tested for the generation of suppressor strains. We considered FtsZ mutants nonfunctional if they did not provide complementation and did not generate suppressor strains and partially functional if they could function in a suppressor strain.
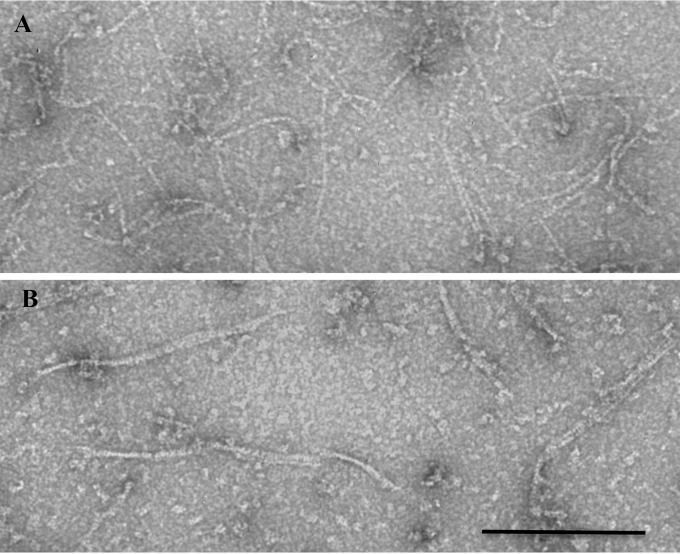
One of the first insert sites that we tested was between G55 and Q56. We initially tested unmodified Venus, a YFP, and this was completely nonfunctional; it neither provided complementation nor formed suppressor strains. We then expressed the protein and tested its assembly in vitro. Negative-stain electron microscopy (EM) showed that FtsZ-Venus formed pf pairs or ribbons, unlike the single pfs of unmodified FtsZ (Fig. 1). Reasoning that the lateral association might be caused by the known dimerizing tendency of GFP, we made a new construct containing the monomerizing mutation A206K (31, 32). The new construct, mVenus (we now refer to the original, dimerizing Venus as dVenus), assembled normal-looking, one-stranded protofilaments in vitro (Fig. 1) (though there were fewer than those of the wt—we do not understand the basis for this discrepancy), and it provided complementation in vivo. This construct therefore functions fully for cell division as the sole source of FtsZ.
FIG 1.
Negative-stain EM images of FtsZ-FP insertions. (A) FtsZ-55-mVenus-56. (B) FtsZ-55-dVenus-56. FtsZ was present at 1 mg/ml in HMK100 buffer. Bar, 200 nm.
Various GFP constructs fused to the C terminus of FtsZ have long been known to function as dilute labels in the presence of 70% or more wtFtsZ. Our lab previously found that dVenus fused at the C terminus could generate suppressor strains in which it functioned as the sole source of FtsZ. In light of the success of mVenus inserted at G55-Q56, we tested mVenus fused at the C terminus. This construct still did not provide complementation; however, it formed suppressor strains at a higher frequency than that with C-terminal dVenus.
Crystal structures of GFP variants show that the first 3 to 5 and the last 11 aa are disordered. Shimozono et al. showed that truncation of the first 5 and last 11 aa of a cyan fluorescent protein (CFP)-YFP pair gave a functional fluorescence resonance energy transfer (FRET) pair, demonstrating that these extensions are not needed for fluorescence (33, 34). These extensions are probably flexible and provide linkers that might allow the FP insert to minimally disrupt the folding of FtsZ. However, to be conservative, we added additional linkers of 5 aa at the start of mVenus (GSTLE) as well as at its end (LEGST). Figure 2 shows a model of the FtsZ-mVenus construct based on PDB files.
FIG 2.
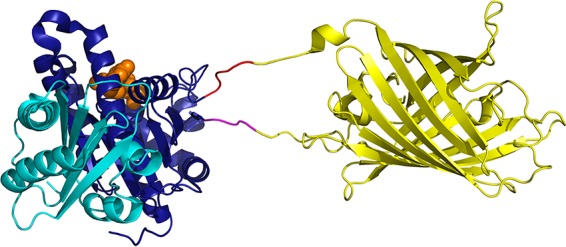
Structural model of FtsZ with YFP inserted between G55 and Q56. The FtsZ structure is based on PDB file 2VAW (Pseudomonas aeruginosa FtsZ [27]). The N-terminal subdomain is shown in dark blue, the C-terminal subdomain is shown in cyan, and GDP is shown by orange spheres. YFP (PDB entry 1YFP) is shown in yellow. The 3- and 11-aa tails of YFP were added as extended peptides, and the 5-aa linkers are shown in magenta and red. The figure was prepared by the PyMOL Molecular Graphics System, version 1.8 (Schrödinger, LLC).
To explore the need for the linkers, we truncated 2 and 10 aa from the N and C termini, respectively, of mVenus, leaving the 5-aa linkers in place. This 3mVenus229 construct also provided complementation. When this construct was further modified by reducing each linker from 5 aa to 2 aa (LE-3mVenus229-LE), it no longer functioned.
We tested 17 other sites for insertion of mVenus into loops on the lateral surfaces of FtsZ. None of these insertions provided complementation or generated suppressor strains (Table 1).
TABLE 1.
Sites tested for complementation with insertion of mVenus
| Insertion site construct | Location of inserta | Function as sole FtsZ sourceb | Dominant negative effectc |
|
|---|---|---|---|---|
| Small-colony formation | No colony formation | |||
| mVenus-M1-FtsZ | Amino terminus | S | ||
| E35-mVenus-G36 | Back | N | ||
| G55-mVenus-Q56 | Back | C, G | No DN effect | — |
| G63-mVenus-I64 | Left | N | No DN effect | — |
| G71-mVenus-A72 | Top | N | No DN effect | — |
| L123-mVenus-G124 | Left bottom | N | ||
| G124-mVenus-I125 | Left bottom | N | ||
| M206-mVenus-N207 | Bottom (T7 loop) | N | 1× | 4× |
| S218-mVenus-E219 | Front | N | ||
| G221-mVenus-Y222 | Front | N | ||
| E233-mVenus-D234 | Back right | N | ||
| E250-mVenus-D251 | Front | N | ||
| D251-mVenus-I252 | Front | N | ||
| S255-mVenus-G256 | Front | N | ||
| F268-mVenus-D269 | Back | N | ||
| D288-mVenus-N289 | Front | N | 3× | — |
| D299-mVenus-P300 | Back | N | 3× | — |
| N303-mVenus-D304 | Back right | N | ||
| D304-mVenus-E305 | Back right | N | 3× | — |
| G316-mVenus-M317 | C-terminal linker | S | 5× | — |
| Q333-mVenus-P334 | C-terminal linker | S | No DN effect | — |
| A366-mVenus-K367 | C-terminal linker | S | 3× | — |
| FtsZ-D383-mVenus | Carboxyl terminus | S | ||
“Front,” “back,” “right,” and “top” refer to the subunit oriented as in a microtubule, where “front” is the outside surface and “top” is the location of GDP; see Fig. 6 for the front view.
C, full complementation (note that this means that the FtsZ construct was fully functional for colony formation; some constructs produced aberrant Z rings and longer cells, indicating defects in the Z ring that were not lethal); S, function in suppressor strains; N, did not provide complementation or generate suppressor strains; G, was successfully incorporated into the genome, replacing the genomically expressed FtsZ.
Dominant negative (DN) effects were tested for some inserts. The number indicates the expression level of mutant FtsZ compared to that of wtFtsZ within BW27783 that caused small or no colonies, as determined in a previous study (29). 3×, expression with 0.0005% arabinose; 4×, expression with 0.0015% arabinose; 5×, expression with 0.005% arabinose; no DN effect, colonies were not reduced in size at 0.005% arabinose, in which the mutant FtsZ protein was present at ∼5× the level for wtFtsZ; —, small colonies were still present at the 5× level.
Screening for other functional fluorophores.
A major interest for fluorescence microscopy is the use of PAFPs for superresolution light microscopy. Because G55-Q56 was an insertion site that gave a fully functional protein with the mVenus insert, we tested other fluorophores at this site (Table 2). Dreiklang, which is closely related to YFP and Venus, did not provide complementation but did generate suppressor strains. Brakemann et al. described Dreiklang as largely monomeric, but it did not have the A206K monomerizing mutation (35). We made this mutation (producing mDreiklang) and found that mDreiklang provided complementation. We therefore designated the original construct dDreiklang. We believe that its dimerization is less than that of dVenus, because dDreiklang could generate suppressor strains while dVenus could not. In addition, mCerulean (36), another Aequorea victoria-based FP, nearly provided complementation; the complementation plate had a few colonies, but they made up less than 80% of the total CFU plated.
TABLE 2.
Testing of constructs with various fluorescent proteins (FPs) at G55-Q56 for function as the sole FtsZ sourcea
| FP inserted at G55-Q56 | Complementation |
|---|---|
| G55-dVenus-Q56 | N |
| GSTLE-mVenus-LEGST | C, G |
| GSTLE-3mVenus229-LEGST (1) | C |
| LE-3mVenus229-LE (2) | N |
| mCerulean | S, G |
| mCherry | S |
| mNeonGreen | S, G |
| dDreiklang | S |
| mDreiklang | C |
| mEos2 | S |
| mEos3.1 | S |
| mEos3.2 | S |
| mEos4b | S |
| mMaple | S |
| mMaple2 | S |
| mMaple3 | S |
| mKikGR | N |
C, full complementation; S, function in suppressor strains; N, neither C nor S; G, successfully incorporated into the genome, replacing the genomically expressed FtsZ. Unless indicated otherwise, FP constructs contained the full protein sequence and additional 5-aa linkers at the N (GSTLE) and C (LEGST) termini. This is specifically indicated for the mVenus constructs, as we also tested variations in the linkers, as follows: (1), 3mVenus229 was missing the first 2 and last 10 aa of the mVenus sequence but retained the 5-aa linkers, and (2), LE-3mVenus229-LE was truncated by an additional 3 aa in each of the linkers.
We tested four variants of mEos (mEos2, -3.1, -3.2, and -4b), a promising PAFP from coral (37–39). Although mEos2 was described as weakly dimerizing (39), it was able to generate suppressor strains in which it was functional. None of the mEos constructs provided complementation, but all generated suppressor strains in which they could function as the sole source of FtsZ. Among the mEos constructs, mEos3.1 was most promising because it gave the best-looking Z rings.
mMaple is a recently derived PAFP with optimal photophysical characteristics (40). It is, however, weakly dimerizing, so Wang et al. engineered mMaple2 and -3, which reduced dimerization while retaining most of the desirable photophysical characteristics (41). The mMaple and mMaple2 and -3 constructs all generated suppressor strains, but when cells were grown in LB at 37°C, the cells were filamentous, the cell growth was very slow, and the Z rings were aberrant.
We used gene replacement to attempt to incorporate several of the FP constructs into the genome, replacing genomic ftsZ and putting the FtsZ-FP-encoding gene under its normal genomic control (42). This was successful for mVenus, mCerulean, and mNeonGreen. mNeonGreen is especially promising because of its brightness, photostability, and extremely short maturation time (less than 10 min versus 30 to 120 min for other FPs) (43). Thus, a large fraction of mNeonGreen should be fluorescent even under rapid growth conditions. Based on our complementation results, mVenus and mCerulean were most likely incorporated into genomic ftsZ without generating a second-site mutation. Because mNeonGreen needed to generate a suppressor strain when it was expressed from the pJSB2 plasmid, the genomic incorporation may have been accompanied by a suppressor mutation. However, this strain gave cells of normal size, shape, and Z-ring appearance and should be an ideal tool for future studies using fluorescence microscopy. We sequenced the genomic region of the three FtsZ-FP-encoding genomic inserts and confirmed that the ftsZ gene had no mutation.
We found that mCherry (44) generated suppressor strains, while mKikGR (45) was completely nonfunctional (it neither provided complementation nor generated suppressor strains).
Better phenotypes from genomic expression.
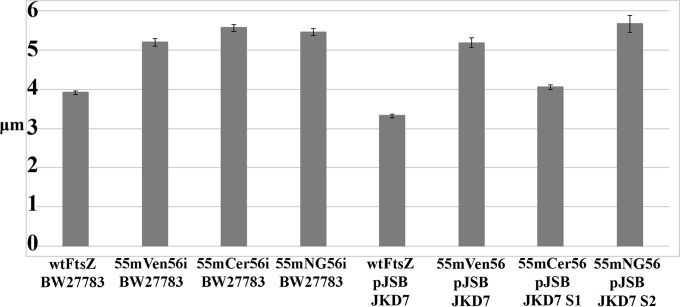
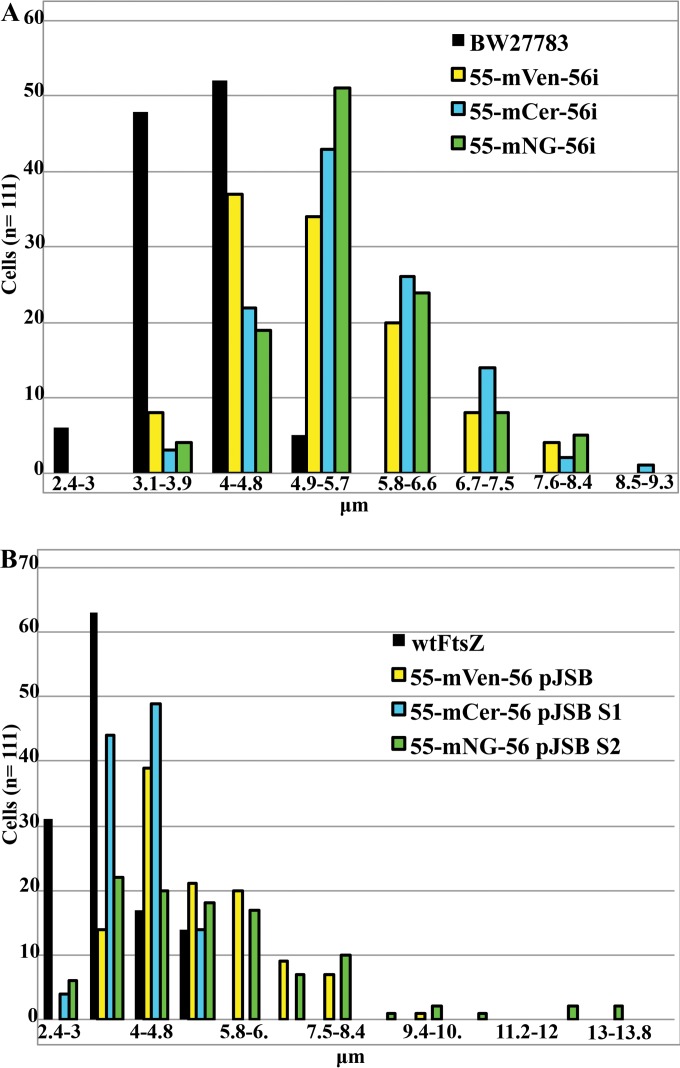
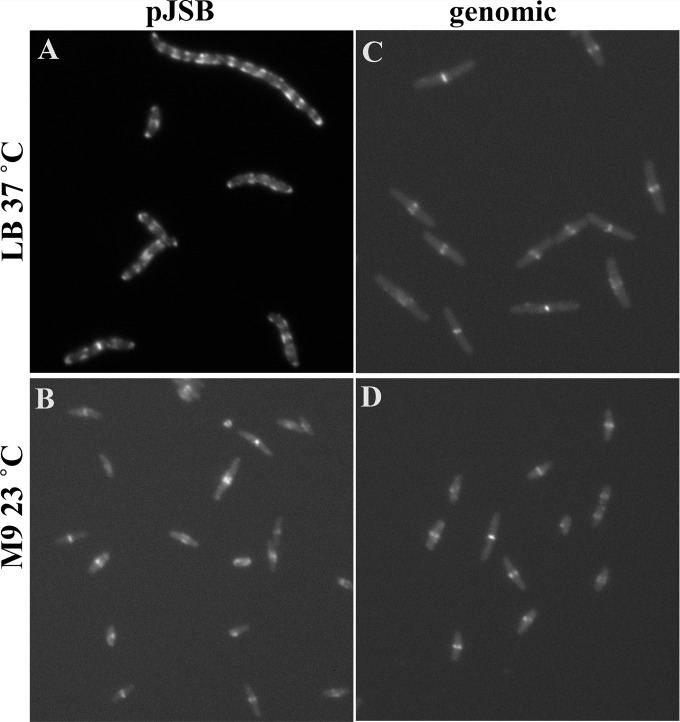
There was measurable variation in the lengths of cells expressing different FtsZ-FPs from the pJSB2 plasmid as the sole source of FtsZ in both complementing and suppressor strains (Fig. 3). When FtsZ-FPs were expressed genomically, the average cell length was shorter than that with plasmid expression but somewhat longer than that with wtFtsZ for all constructs (Fig. 4A). When FtsZ-FPs were expressed from pJSB2, mCerulean gave cells only slightly longer than those with wtFtsZ, while mVenus and mNeonGreen gave longer cells (Fig. 4B). When cells were examined by fluorescence microscopy, mNeonGreen showed a striking difference in FtsZ localization during exponential growth between genomic and plasmid expression. Plasmid expression gave cells with multiple Z rings, most of them aberrant (Fig. 5A). In contrast, BW27783 cells expressing mNeonGreen genomically had mostly one Z ring per cell (Fig. 5C). Some were sharp and completely normal (Fig. 5C, upper left corner), but most were somewhat fuzzier, and some were slanted.
FIG 3.
Average cell lengths of select strains grown in LB at 37°C. The left four bars show BW27783 strains with the FtsZ-FP-encoding gene incorporated (“i”) into the genome. The right four bars show JKD7 strains with FtsZ expressed from the pJSB2 plasmid (with 0.2% arabinose). The FtsZ-FP constructs (with the FP inserted at Q55-G56) were mVenus, mCerulean, and mNeonGreen, with wtFtsZ as a control. mCerulean and mNeonGreen functioned in suppressor strains, indicated as S1 and S2. The number of cells measured for each strain was 111.
FIG 4.
Ranges of cell length for select strains. (A) BW27783 strains with genomically expressed FtsZ-FPs. (B) JKD7 strains expressing FtsZ from the pJSB2 plasmid (with 0.2% arabinose). Cells were grown in LB at 37°C. Note the different x axes for panels A and B.
FIG 5.
Cells expressing FtsZ-G55-mNeonGreen-Q56 during exponential growth. (A and B) JKD7 suppressor strain S2 (with FtsZ-G55-mNeonGreen-Q56 expressed from pJSB2) grown in LB plus 0.2% arabinose at 37°C (A) or in M9 medium plus 0.2% arabinose at 23°C (B). (C and D) BW27783 with genomically expressed FtsZ-G55-mNeonGreen-Q56i, grown in LB at 37°C (C) or in M9 medium at 23°C (D). Note that the light levels were adjusted for individual images to optimize the visibility of the Z ring and the cytoplasm. Panel A was originally much brighter than the others.
Better phenotypes with slow growth at 23°C.
Cells grown in LB at 37°C and supported by FtsZ-mNeonGreen expressed from pJSB were 5.66 μm long, compared to 3.38 μm for cells expressing wtFtsZ from pJSB2. When cells were examined by fluorescence microscopy, they had multiple Z rings and aberrant patches and spirals (Fig. 5A). We then tested FtsZ-mNeonGreen pJSB2 cells grown in M9 medium at 23°C, which increased the doubling time to 6 h. Under these slow-growth conditions, most cells had single, normal-looking Z rings and were 3.64 μm long, compared to 2.93 μm for wtFtsZ pJSB2 cells in M9 medium (Fig. 5B). Cells expressing FtsZ-mNeonGreen from the genome were also improved by growth in M9 medium at 23°C (Fig. 5C and D). We noted that mNeonGreen pJSB2 suppressor cells grown in LB at 37°C had significantly brighter Z rings than cells genomically expressing mNeonGreen that were grown in M9 medium at 23°C (Fig. 5), suggesting that the aberrant Z rings may have been caused by overexpression.
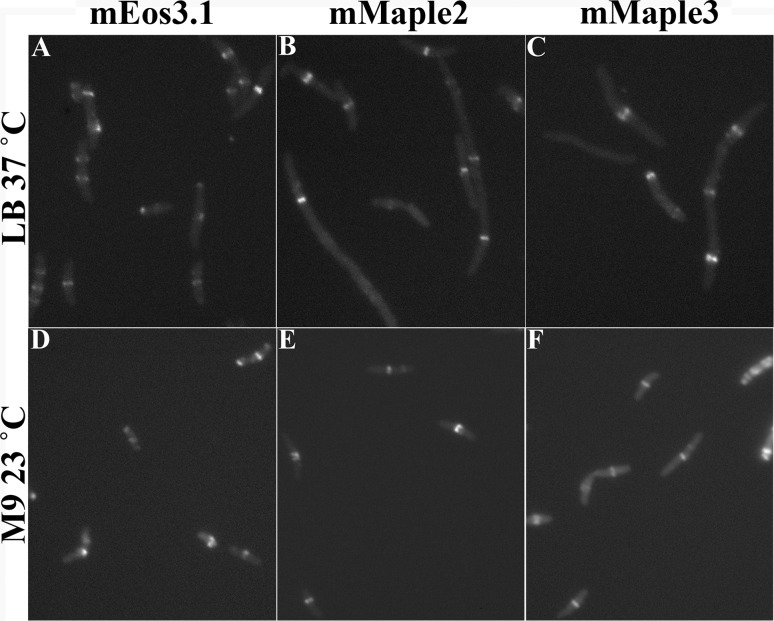
Cells expressing FtsZ-mEos3.1, FtsZ-mMaple2, and FtsZ-mMaple3 from pJSB2 were elongated, with multiple, aberrant Z rings (Fig. 6A to C), when grown in LB at 37°C. These were all greatly improved when cells were grown slowly in M9 medium at 23°C, as a large fraction of cells had a single Z ring (Fig. 6D to F). FtsZ-mEos3.1 cells in M9 medium were much brighter than those in LB at 37°C, probably because the doubling time was 8 h, which is longer than the short maturation time of mEos3.1.
FIG 6.
(A and D) JKD7 suppressor strain S6 (with FtsZ-G55-mEos3.1-Q56 expressed from pJSB2) grown in LB at 37°C (A) and in M9 medium at 23°C (D). (B and E) JKD7 suppressor strain S1 (with FtsZ-G55-mMaple2-Q56 expressed from pJSB2) grown in LB at 37°C (B) and in M9 medium at 23°C (E). (C and F) JKD7 suppressor strain S2 (with FtsZ-G55-mMaple3-Q56 expressed from pJSB2) grown in LB at 37°C (C) and in M9 medium at 23°C (F). Elongated cells with multiple Z rings were found for growth in LB at 37°C. However, the cells in M9 medium at 23°C were shorter, with single Z rings. Note that the light levels were adjusted for individual images to show the Z ring and the cytoplasm. Growth in LB at 23°C gave improved Z rings similar to those from growth in M9 at 23°C, suggesting that temperature was the important parameter.
The improved Z rings for cells grown in M9 medium at 23°C may have been due to the minimal medium or to the temperature. We therefore tested cells expressing FtsZ-mEos3.1 grown in LB at 23°C. These had normal-looking Z rings that were similar to those in M9 medium at 23°C (data not shown). This suggests that the defect of some FP inserts grown at 37°C is due primarily to thermosensitivity.
Peptide insertions within loops.
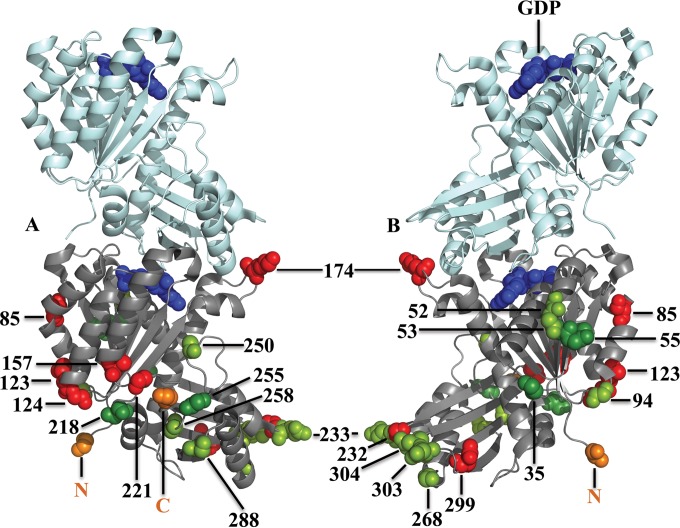
Proteins with the mVenus insert were nonfunctional for every tested site in the globular domain except for G55-Q56. Because FPs are relatively large protein inserts, we decided to test whether variants might function with a smaller insert, a 10-aa peptide, at some sites (Table 3; Fig. 7). As expected, FtsZ with the 10-aa insert at G55 provided complementation. In addition, FtsZ with the 10-aa insert at four additional sites, E35, A53, S218, and S255, gave full complementation. There were 10 additional sites where the 10-aa insertion yielded FtsZ that was functional after generation of suppressor strains and 8 tested sites where it gave a completely nonfunctional protein (Table 3). Three of these nonpermissive sites are on the left surface of FtsZ, in the N-terminal subdomain, and two are on the right surface of the C-terminal subdomain. Reducing the peptide from 10 aa to 3 aa allowed the protein with an insert at E232 to function in a suppressor strain, while the protein with a 3-aa insert at D299 remained completely nonfunctional.
TABLE 3.
Sites in the globular domain tested for complementation by insertion of a small peptide
| Insertion sitea | Location in tubulin core | Function as sole FtsZ source |
|---|---|---|
| E35-linker10-G36 | Back | C |
| T52-linker10-A53 | Back | S |
| A53-linker10-V54 | Back | C |
| G55-linker10-Q56 | Back | C |
| G71-linker10-A72 | Top | N |
| R85-linker10-D86 | Left | N |
| G94-linker10-A95 | Left bottom | S |
| L123-linker10-G124 | Left bottom | N |
| G124-linker10-I125 | Left bottom | N |
| V157-linker10-D158 | Front | N |
| R174-linker10-G175 | Right top | N |
| M206-linker10-N207 | Bottom (T7 loop) | N |
| S218-linker10-E219 | Front | C |
| G221-linker10-Y222 | Front | N |
| G232-linker10-E233 | Back right | N |
| G232-linker3-E233 | Back right | S |
| E233-linker10-D234 | Back right | S |
| E250-linker10-D251 | Front | S |
| S255-linker10-G256 | Front | C |
| R258-linker10-G259 | Front | S |
| F268-linker10-D269 | Back | S |
| D288-linker10-N289 | Front | S |
| D299-linker10-P300 | Back | N |
| D299-linker3-P300 | Back | N |
| N303-linker10-D304 | Back right | S |
| D304-linker10-E305 | Back right | S |
The linker10 peptide sequence was GSTLELEGST. The linker3 peptide was GST.
FIG 7.
Cartoon model showing two subunits of an FtsZ pf (PDB entry 3VOA [58]; Staphylococcus aureus FtsZ). The bottom subunit shows sites where the 10-aa GSTLELEGST peptide was inserted. (A) View of the front of FtsZ, from which the C terminus emanates (orange). (B) View of the back of FtsZ, from which the N terminus emanates (orange). Proteins with insertions at dark green sites provided complementation, those with insertions at light green sites generated suppressors, and those with insertions at red sites failed to function in vivo. GDP is shown in blue. The amino acids preceding inserts are shown as spheres, and the E. coli amino acid numbers are indicated.
In a previous study, we tested a range of point mutants for the ability to assemble pf sheets in DEAE-dextran and pf bundles in Ca2+. We had hoped that these in vitro assemblies might involve lateral bonds that were essential in the Z ring in vivo. None of the point mutants tested in that study affected the in vitro assemblies (46), although several of those mutants disrupted function in vivo (30). Hence, that study failed to identify candidates for lateral bonds. We turned to this assay to test some of the 10-aa insertions that failed to provide a functional protein in the present study. However, we were unable to express and purify some of these insertion mutants. Proteins with 10-aa inserts at L123 and G124 were highly expressed but insoluble, and a protein with the insert at R85 had very low expression and was also insoluble. A protein with the insert at D299 was expressed well and purified, but it did not polymerize in vitro. Thus, the failure of these insertion mutants to function in vivo may have been due to protein folding problems, not specifically to a block of lateral bonds.
A protein with the 10-aa insert at R174 was well expressed and could be purified. It formed one-stranded pfs under normal assembly conditions, pf sheets in DEAE-dextran, thin bundles in Ca2+, and large curved bundles in the crowding agent polyvinyl alcohol (Fig. 8). The sheets and bundles were identical to those assembled by wild-type FtsZ. Thus, the protein with the insert at R174 showed no defect in the lateral bonds that assemble these structures in vitro. We also tested two point mutants with mutations of R174, R174A and R174D, which were previously reported to fail in assembly of bundles in Ca2+ (47). We found that both mutants assembled normal pf bundles in Ca2+, sheets of pfs in DEAE-dextran, and large bundles in polyvinyl alcohol (similar to those shown in Fig. 8) (data not shown). Assemblies of these point mutants were done in buffer at pH 6.5 with 50 mM KCl, the same as in the work of Koppelman et al. (47).
FIG 8.
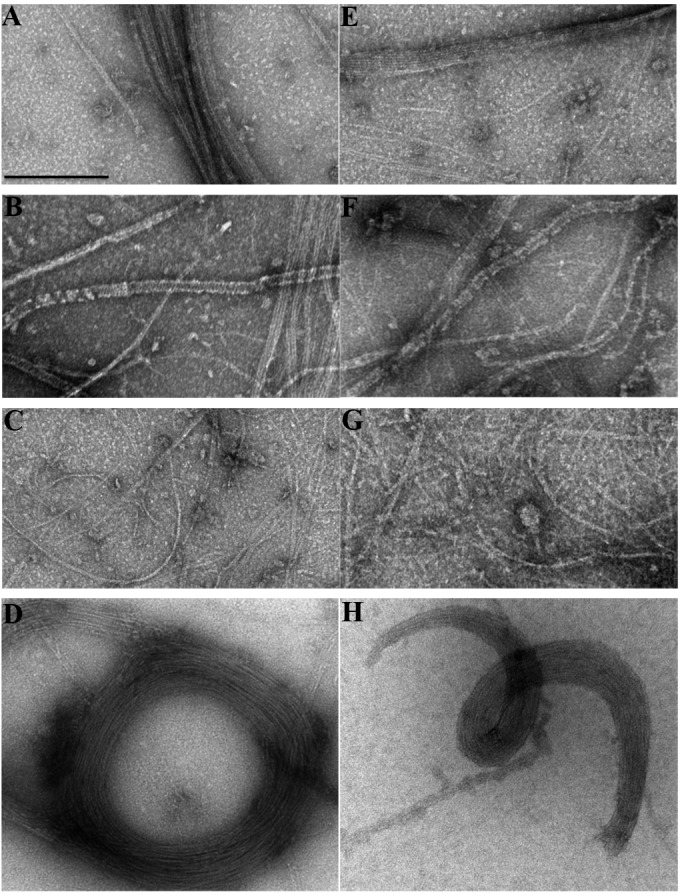
Negative-stain EM images of polymerization with the 10-aa insert at R174. (A to D) Wild-type FtsZ. (E to H) FtsZ with the 10-aa insertion after R174. FtsZ was present at 1 mg/ml. (A and E) DEAE-dextran (0.5 mg/ml) and 5 mM EDTA in HMK100 buffer. (B and F) DEAE-dextran (0.5 mg/ml) in HMK100 buffer. (C and G) Ca2+ (10 mM) in HMK100 buffer. (D and H) Ten percent polyvinyl alcohol in HMK100 buffer. Bar, 200 nm.
mVenus insertions in the linker.
A recent study showed that the 50-aa C-terminal linker (aa 317 to 367) functions as an intrinsically disordered peptide (IDP) and can be replaced by unrelated IDPs within a length range of 43 to 95 aa (9). We originally expected that insertion of mVenus almost anywhere in the linker would be tolerated. However, a previous study showed that proteins with a dVenus insertion at V326 or T348 needed the generation of suppressor strains in order to function, while a protein with an insert at Q333 nearly provided complementation (8, 12). In the present study, we tested proteins with mVenus inserts at the beginning (after aa G316) and the end (after aa A366) of the linker. Neither provided complementation, but both resulted in formation of suppressor strains (Table 4).
TABLE 4.
Testing of proteins with FP insertions in the linkers of FtsZ for ability to function as the sole FtsZ sourcea
| FP insertion site | Complementation |
|---|---|
| mVenus-M1 | S |
| G316-mVenus-M317 | S |
| G316-3mVenus229-M317 | N |
| G316-mVenus-K367 | N |
| G316-mVenusAdd20-K367 | N |
| G316-mVenusAdd39-K367 | N |
| Q333-mVenus-P334 | S |
| A366-mVenus-K367 | S |
S, function in suppressor strains; N, neither complementation nor function in suppressor strains. Unless indicated otherwise, FP constructs contained the full protein sequence and additional 5-aa linkers at the N (GSTLE) and C (LEGST) termini. This is specifically indicated for the mVenus constructs, as we also tested variations in the linkers: 3mVenus229 was missing the first 2 and last 10 aa of the mVenus sequence.
We considered the possibility that the 2-nm spacing between the N and C termini of the structured YFP plus the ∼16 unstructured aa at the two ends interferes with function by increasing the length of the linker beyond its ideal range. We therefore tested the truncated construct G316-3mVenus229-M317. Surprisingly, this was worse than the full-length insert. The protein with G316-mVenus-M317 generated suppressor strains, while that with G316-3mVenus229-M317 was completely nonfunctional. Leaving the mVenus insert at G316 and replacing the remaining linker with adducin segments of 20 or 39 aa (G316-mVenus-Add20-367 and G316-mVenus-Add39-367) also resulted in a nonfunctional FtsZ protein. Previous work showed that the complete linker can be replaced by several unrelated disordered peptides from adducin (9), but for reasons that are not understood, inserting the globular mVenus construct interferes with function.
DN effects of FP inserts.
Previous work showed that some FtsZ mutants and deletion constructs exhibited dominant negative (DN) effects when expressed in the presence of wtFtsZ expressed from the genomic locus. These included any construct with an intact and normal N-terminal subdomain but a missing or damaged C-terminal subdomain (8, 29). In addition, constructs with a deletion of the C-terminal FtsA-binding peptide showed DN effects. The DN effects were weak, requiring expression of the mutant FtsZ protein at levels 3 to 5 times higher than that of wtFtsZ for a partial effect, as indicated by a reduced colony size. Complete growth arrest, indicated by no colonies, was seen only at the highest expression level, with 0.005% arabinose, and only for the most toxic constructs. In contrast to these effects, constructs with a missing or defective N-terminal subdomain had no detectable DN effects. The results for some of our insert constructs are summarized in Table 1.
As a control, we tested one previously characterized truncation mutant, FtsZ1-195, which contains the N-terminal globular subdomain, including half of helix H7. This gave small colonies at 0.0005% arabinose; it still produced colonies at 0.005% arabinose, but they were smaller. This result was similar to the results of the previous study (8). Dimeric Venus and Dreiklang were the only G55-insert-Q56 constructs that showed DN effects, producing smaller colonies beginning at 0.0015% arabinose. D299-mVenus-P300 and D304-mVenus-E305 displayed strong DN effects, like FtsZ1-195, with smaller colonies beginning at 0.0005% arabinose (Table 1). These effects may have been due to misfolding of the C-terminal subdomain. Various inserts at G316, the junction of the globular domain and the linker, gave minimal DN effects, with a small reduction in colony size only at the highest expression level. In the Ct tail linker, A366-mVenus-K367 displayed the strongest DN effect. Other inserts in the linker had minimal DN effects.
Most of the inserts were designed to put the FP in the lateral faces of the protofilament and to avoid the longitudinal interface within the protofilament. However, we engineered two “cap” mutants, G71-mVenus-A72 and M206-mVenus-N207, designed to poison assembly by placing the FP in the upper and lower interfaces, respectively. Consistent with previous work (29), the top cap (G71-mVenus-A72) showed no DN effect, while the bottom cap (M206-mVenus-N207) showed strong DN effects (Table 1). In fact, the M206-mVenus-N207 colony size became smaller with as little as 0.00015% arabinose, with no colonies seen at 0.0015% arabinose. A complete blockage of the bottom interface appears to have a stronger DN effect than complete removal of that domain.
DISCUSSION
FtsZ-FPs that are functional as the sole source of FtsZ.
An important advance in our study was the discovery of one site within FtsZ, G55-Q56, where inserts of several FPs gave proteins that were functional for cell division without the need for wild-type FtsZ. We tested a number of FPs at this site, and we discovered some general principles governing function. First, FPs with the original dimerization tendency, or even ones with residual weak dimerization, were not functional. The dimerization of FPs is too weak to bring monomers together, so the damaging effect is probably dimerization of pfs. This might result either from enhanced stabilization of naturally paired pfs or from an artificial pairing of pfs into a nonfunctional structure. Second, the best FPs were those based on Aequorea victoria GFP. FPs based on coral proteins gave variable results, although most of them were able to function in suppressor strains. Functionality was grossly defined as the ability of cells to divide and make colonies. The best FPs also produced cells that were only slightly elongated and had predominantly single, normal Z rings. Some FPs, although they could support division and colony formation, produced elongated cells with abnormal Z rings. The very best results were obtained with mVenus, mCerulean, and mNeonGreen incorporated at the genomic locus, where they are expressed under the normal genomic controls for FtsZ. These strains should be valuable for general observations of Z rings.
Apart from the loop at G55-Q56, the only sites that gave functional FtsZ-FPs were in the C-terminal linker. In our previous study, we could replace the linker with IDPs with completely unrelated sequences and a range of lengths (9). Similar results were obtained with the linkers of Bacillus and Caulobacter (48, 49). We expected that YFP could be inserted almost anywhere in the C-terminal linker, but most inserts could not function without a suppressor strain being generated. The globular FP domain may cause some steric interference that the IDP permits, but we do not understand the mechanism.
We selected mEos3.1 and mMaple3 as the best PAFPs at the G55-Q56 insertion site. However, cells expressing mEos3.1 or mMaple3 from pJSB were typically elongated, with multiple and abnormal Z rings, when grown in LB at 37°C. We therefore tested the effect of growing the cells more slowly, in M9 defined medium and at room temperature, as suggested in a recent PALM study using FtsZ-mEos2 (16). This gave a major improvement, with almost all cells showing normal length and a single, normal-appearing Z ring (Fig. 6). Growth in LB at room temperature gave a similar improvement in Z-ring morphology, suggesting that the primary factor is lower temperature. However, another advantage of slower growth is that it gives more time for the FPs to mature. The doubling time for FtsZ-mEos3.1 was 8 h in M9 medium, which is longer than the maturation half-time of 2 h (37) or 5 h (41) reported for mEos derivatives. Our FtsZ-mEos3.1 and FtsZ-mMaple3 constructs should be ideal for PALM; details favoring one or the other have yet to be determined.
Probing sites for steric blockage in a search for lateral contacts.
A second goal of our study was to identify faces of FtsZ where an inserted FP would permit function. This face could then be excluded from binding of any other essential protein, including lateral interactions of pfs. Inserting an FP or a 10-aa peptide should provide a substantial steric block of any interactions at the insertion site. Of the four faces, the back, which faces away from the membrane (50), could support the FP insert at G55-Q56, and the front could support the FP insert after G316, from which the linker and membrane tether emerge. Several proteins with 10-aa inserts on the front and back were also functional. We concluded that the front and back faces are not involved in lateral bonding of pfs or in essential binding of other proteins.
The failure of an FtsZ-FP to function, either directly for complementation or to generate a suppressor strain in which it can function, could be due to blocking of an essential protein binding interaction, but it could also be due to failure of FtsZ to fold properly. Complete misfolding was apparently not the case for most inserts, because they had diffuse fluorescence rather than precipitating into inclusion bodies. Also, all of the failed inserts in the C-terminal subdomain that were examined showed DN effects, suggesting that the proteins were soluble and capable of interacting with the Z ring. The DN effects were similar to those previously observed for mutant FtsZ with a complete N-terminal subdomain but a missing or defective C-terminal subdomain (8, 29). The failure of these inserts may be due to subtle folding defects in the C-terminal subdomain.
These defects might be caused by the complexity of folding mVenus. We therefore tested a simpler insert, a 10-aa peptide with no folded structure. Especially when inserted into a loop, this might have a minimal effect on the folding of the parent protein. Indeed, we found several additional sites on the front and back faces which had not supported insertion of mVenus but did support inserts of the smaller, 10-aa peptide. Although it is smaller than a globular FP, the 10-aa peptide is large enough that it should block any protein-protein interaction at its site of insertion.
We then used this 10-aa peptide for a more focused search for lateral interactions. Lateral contacts of pfs, which are still not certain to exist, should be limited to the left and right faces. We looked especially at sites that protrude the farthest on the left and right. Our observations and interpretations are detailed below.
(i) R85-D86.
The R85-D86 bend in helix H3 is the most protrusive loop on the left face. The protein with the 10-aa insert here failed to function, consistent with this being a site of lateral contact. However, we also failed to express a soluble protein in the pET system, so this insertion variant may have folding problems. A D86K point mutant caused pfs to pair in vitro, suggesting a role for this site in lateral bonding (46). However, the D86K mutant was functional in vivo (30). Overall, the evidence is ambiguous regarding whether the R85-D86 loop might mediate physiologically important lateral bonds.
(ii) G94-A95.
The G94-A95 loop also protrudes on the left side, below R85-D86 and toward the back. The protein with the 10-aa insert at this site was functional, so we excluded this as an attractive candidate for lateral contact. We note, however, that an E93R point mutant caused a substantial increase in pf bundling in vitro, suggesting that this site is capable of mediating lateral interactions (51).
(iii) L123 to I125.
The L123-to-I125 loop is adjacent to and slightly in front of G94-A95. Two proteins with 10-aa inserts at this loop were nonfunctional, so this loop remains a candidate for lateral contact. However, we failed to express a protein in the pET system, suggesting that the proteins may have folding problems unrelated to lateral bonding.
(iv) R174-G175.
The R174-G175 site is on a large loop projecting to the upper right. The protein with the 10-aa insert at this site was nonfunctional in vivo, consistent with this being an essential lateral bonding site. The insert did not block any of our in vitro tests for lateral association (sheet and bundle formation in DEAE-dextran, Ca2+, and polyvinyl alcohol). It is possible that a site for lateral bonding in vivo is different from those mediating these in vitro pf associations.
(v) G232-E233-D234.
The G232-E233-D234 loop protrudes from the right side at the bottom. Although the 10-aa insert at G232-E233 inactivated FtsZ function in vivo, a 3-aa insert at this site gave a functional protein. The 10-aa insert at the adjacent E233-D234 region gave a functional protein. Therefore, this loop is not an attractive candidate for lateral interaction.
(vi) N303-E304.
The N303-E304 loop is adjacent to G232-D234 but more toward the back. Two proteins with 10-aa inserts there were functional in vivo, so this is not attractive as a lateral contact site.
Li et al. reported a crystal structure of Mycobacterium tuberculosis FtsZ that showed an antiparallel pair of spiraling pfs (52). The pfs interacted through a salt bridge between R229 and D301, which are equivalent to S231 and D304 in E. coli. Our proteins with 10-aa inserts at or near these sites functioned for cell division. This suggests that this site may be specific to M. tuberculosis FtsZ or to this crystal but is not an essential lateral contact site conserved across species.
Another site that has been implicated in pf bundling in vitro is the short “C-terminal variable” peptide (10). The peptide NRNKR from B. subtilis caused bundling in low-salt buffers, while the KQAD peptide from E. coli did not. We did not explore this site in the present study.
These results do not affirm or reject the possibility of lateral association of FtsZ pfs, but they place constraints on some of the most attractive candidates. It is still debated whether pfs associate in ribbons, with lateral contacts (53, 54), or are more widely scattered, with few contacts (17, 55). Our new FP insert constructs giving 100% functional FtsZ-FPs may lead to improved superresolution light microscopy and perhaps resolve this question.
MATERIALS AND METHODS
Creation of ftsZ mutants.
To clone FtsZ-FPs, we first amplified the FP of interest. For example, the primers used to amplify Venus (a fast-folding YFP [56]) were as follows: sense primer, 5′-phosphorylated ATGGTGAGCAAGGGC (aa sequence, MVSKG; N terminus of Venus); and antisense primer, 3′-phosphorylated CTTGTACAGCTCGTCC (aa sequence, DELYK; C terminus of Venus). We also made an insert by truncating 2 aa from the N terminus by using the sense primer AGCAAGGGCGAGGAGC (aa sequence, SKGEE) and 10 aa from the C terminus by using the antisense primer CCCGGCGGCGGTCACGAACTCCAGCA (aa sequence, LEFVTAAG). Next, we linearized the plasmid containing ftsZ at the site of insertion. For example, for insertion between G55 and Q56, we used the sense primer 5′CTCGAGGGATCCACCcagacgattcaaatcgg (aa sequence, LEGSTqtiqi) and the antisense primer 3′CTCGAGGGTGGATCCtccaaccgctgttttac (aa sequence, ktavgGSTLE) (uppercase letters show the gene sequences coding for the linker sequences connecting FtsZ to Venus, which were added to all inserts [unless otherwise specified]). These linkers each have both a BamHI site and an XhoI site. The purified PCR products (FP and linearized FtsZ-linkers) were ligated using blunt-end cloning. Because there is a 50% chance that Venus will ligate correctly, we screened colonies first to confirm fluorescence and subsequently for FtsZ localization by expressing the protein by use of 0.2% arabinose. Each construct was confirmed by sequencing the segment of ftsZ around the insert. The FtsZ portion of FtsZ-mNeonGreen in pJSB2 was sequenced entirely, and it did not contain mutations.
In order to create the 10-aa insertion, we took the PCR product of the linearized plasmid containing ftsZ with linkers at the site of insertion (before ligation of the FP) and used T4 polynucleotide kinase to phosphorylate the 3′ ends. The PCR product was then ligated together to circularize the plasmid.
In order to convert the 10-aa insertion to a 3-aa insertion, BamHI restriction digestion was performed, followed by religation. Prior to this, we removed a unique BamHI restriction site in the pJSB2 plasmid.
Monomerizing YFP.
Venus and Dreiklang were monomerized to mVenus and mDreiklang by A206K point mutagenesis (31), using the sense primer 5′-TGAGCTACCAGTCCaagCTGAGCAAAGACC-3′ and the antisense primer 5′-GGTCTTTGCTCAGcttGGACTGGTAGCTCA-3′.
Complementation and suppressor generation test.
E. coli strain JKD7-1/pKD3 (28) was used as a host to perform complementation and suppressor strain assays with cells plated at 42°C with 0.2% arabinose as previously described (29, 30). Complementing strains grew colonies overnight after plating of as few as 100 cells. To generate suppressor strains, 106 cells were plated and grown for 2 to 3 days to form colonies. Suppressor strains were confirmed if they demonstrated no growth in both the absence of arabinose and the presence of ampicillin.
Microscopy.
Samples were viewed by differential interference contrast and fluorescence microscopy on a Zeiss Axiophot microscope with a Plan-Neofluar 100×, 1.3-numerical-aperture (NA) oil-immersion lens. Filter cubes optimized for CFP, YFP, GFP, and RFP were used for fluorescence microscopy. Images were acquired with a Coolsnap HQ charge-coupled device (CCD) camera (Roper Scientific) and processed with Adobe Photoshop.
Cell length measurements.
Single colonies were picked and grown in liquid LB or M9 medium, as specified, to a log-phase optical density at 600 nm (OD600) of 0.45 to 0.55. One milliliter of the culture was centrifuged for 1 min at 5,000 rpm. The pellet was resuspended in 100 μl of 10 mM HEPES, pH 7.5, 150 mM NaCl, 5 mM glucose, and 5 mM MgCl2. Cells were transferred (5 μl) to a fresh agarose pad [40 mM 3-(N-morpholino)propanesulfonic acid, pH 7 (7.3), 0.2% glycerol, 0.1% agarose, 0.2% arabinose] and immediately imaged. Cells were imaged with an exposure time of 100 ms for differential interference contrast and 3 s for fluorescence microscopy. ImageJ was used to identify and count cells with single Z rings. Cells showing an invagination by differential interference contrast microscopy were included only if there was a Z ring at the site of invagination.
FtsZ-FP strains were diluted 1:200 from overnight cultures and grown in liquid LB or M9 medium, as specified, to a log-phase OD600 of 0.1 to 0.32. M9 buffer included M9 salts, 1 mM MgSO4·7H2O, 0.00005% vitamin B1, 0.2% glucose, 0.2% arabinose, and antibiotic. After testing of various carbon sources, glucose served as the best carbon source under these minimal medium conditions. Although glucose alone suppresses pJSB expression, glucose plus arabinose gave optimal growth and expression of FtsZ-FP.
Genomic incorporation.
G55-mVenus-Q56, G55-mCerulean-Q56, and G55-mNeonGreen-Q56 were cloned into the plasmid pMAK705 to replace wild-type ftsZ in E. coli strain BW27783 (57), using the procedure of Hamilton et al. (42). Each FtsZ-FP-encoding gene was amplified using primers 5′-agagaCGATCGatgtttgaaccaatggaacttacc-3′ and 5′-tctctAAGCTTttaatcagcttgcttacgcaggaa-3′ and inserted between PvuI and HindIII sites (restriction sequences are shown in uppercase letters) in the plasmid pMAK705. Genomic gene replacement was verified by sequencing, and the loss of pMAK705 was confirmed by chloramphenicol sensitivity testing.
DN assay.
E. coli strain BW27783 (57) was used as a host for the DN assay. Cells were plated at 30°C on 0.2% glucose or 0.00015 to 0.005% arabinose to produce 1× to 5× expression of mutant FtsZ compared to genomic wtFtsZ, as previously described (8, 29, 30). One hundred cells were plated and then observed after overnight growth for a change in colony size or number compared to cells grown on glucose. Cells expressing wtFtsZ from pJSB2 showed no change in colony size over this range of arabinose concentrations.
Electron microscopy.
Proteins were expressed using the pET11b vector as described by Gardner et al. (9). All assembly was conducted in HMK100 buffer (50 mM HEPES, pH 7.5, 100 mM KCl, 5 mM magnesium acetate). Uranyl acetate was used as a negative stain. For Ca2+ assembly, reactions lasted for 5 min at 37°C; all other assembly reactions were performed for 2 min at 23°C after GTP was added.
ACKNOWLEDGMENTS
We thank Loren Looger, Janelia Farms, for plasmids encoding the mEos proteins and Xiaowei Zhuang, Harvard, for the mMaple constructs.
This work was supported by NIH grant R01 GM66014 to Harold P. Erickson.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Osawa M, Anderson DE, Erickson HP. 2008. Reconstitution of contractile FtsZ rings in liposomes. Science 320:792–794. doi: 10.1126/science.1154520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Erickson HP, Anderson DE, Osawa M. 2010. FtsZ in bacterial cytokinesis: cytoskeleton and force generator all in one. Microbiol Mol Biol Rev 74:504–528. doi: 10.1128/MMBR.00021-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coltharp C, Buss J, Plumer TM, Xiao J. 2016. Defining the rate-limiting processes of bacterial cytokinesis. Proc Natl Acad Sci U S A 113:E1044-53. doi: 10.1073/pnas.1514296113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Erickson H. 2016. F1000Prime dissenting opinion on [Coltharp C et al., Proc Natl Acad Sci U S A 2016, 113(8):E1044-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiao J. 2016. Recommendations, dissents and comments for [Coltharp C et al., Proc Natl Acad Sci U S A 2016, 113(8):E1044-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Housman M, Milam SE, Moore D, Osawa M, Erickson HP. 2016. FtsZ protofilament curvature is the opposite of tubulin rings. Biochemistry 55:4085–4091. doi: 10.1021/acs.biochem.6b00479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oliva MA, Cordell SC, Lowe J. 2004. Structural insights into FtsZ protofilament formation. Nat Struct Mol Biol 11:1243–1250. doi: 10.1038/nsmb855. [DOI] [PubMed] [Google Scholar]
- 8.Osawa M, Erickson HP. 2005. Probing the domain structure of FtsZ by random truncation and insertion of GFP. Microbiology 151:4033–4043. doi: 10.1099/mic.0.28219-0. [DOI] [PubMed] [Google Scholar]
- 9.Gardner KA, Moore DA, Erickson HP. 2013. The C-terminal linker of Escherichia coli FtsZ functions as an intrinsically disordered peptide. Mol Microbiol 89:264–275. doi: 10.1111/mmi.12279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buske PJ, Levin PA. 2012. Extreme C terminus of bacterial cytoskeletal protein FtsZ plays fundamental role in assembly independent of modulatory proteins. J Biol Chem 287:10945–10957. doi: 10.1074/jbc.M111.330324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma X, Ehrhardt DW, Margolin W. 1996. Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. Proc Natl Acad Sci U S A 93:12998–13003. doi: 10.1073/pnas.93.23.12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Osawa M, Erickson HP. 2006. FtsZ from divergent foreign bacteria can function for cell division in Escherichia coli. J Bacteriol 188:7132–7140. doi: 10.1128/JB.00647-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Strauss MP, Liew AT, Turnbull L, Whitchurch CB, Monahan LG, Harry EJ. 2012. 3D-SIM super resolution microscopy reveals a bead-like arrangement for FtsZ and the division machinery: implications for triggering cytokinesis. PLoS Biol 10:e1001389. doi: 10.1371/journal.pbio.1001389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rowlett VW, Margolin W. 2014. 3D-SIM super-resolution of FtsZ and its membrane tethers in Escherichia coli cells. Biophys J 107:L17–L20. doi: 10.1016/j.bpj.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jennings PC, Cox GC, Monahan LG, Harry EJ. 2011. Super-resolution imaging of the bacterial cytokinetic protein FtsZ. Micron 42:336–341. doi: 10.1016/j.micron.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 16.Buss J, Coltharp C, Huang T, Pohlmeyer C, Wang SC, Hatem C, Xiao J. 2013. In vivo organization of the FtsZ-ring by ZapA and ZapB revealed by quantitative super-resolution microscopy. Mol Microbiol 89:1099–1120. doi: 10.1111/mmi.12331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu G, Huang T, Buss J, Coltharp C, Hensel Z, Xiao J. 2010. In vivo structure of the E. coli FtsZ-ring revealed by photoactivated localization microscopy (PALM). PLoS One 5:e12682. doi: 10.1371/journal.pone.0012682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holden SJ, Pengo T, Meibom KL, Fernandez Fernandez C, Collier J, Manley S. 2014. High throughput 3D super-resolution microscopy reveals Caulobacter crescentus in vivo Z-ring organization. Proc Natl Acad Sci U S A 111:4566–4571. doi: 10.1073/pnas.1313368111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacq M, Adam V, Bourgeois D, Moriscot C, Di Guilmi AM, Vernet T, Morlot C. 2015. Remodeling of the Z-ring nanostructure during the Streptococcus pneumoniae cell cycle revealed by photoactivated localization microscopy. mBio 6:e01108-15. doi: 10.1128/mBio.01108-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levin PA, Kurtser IG, Grossman AD. 1999. Identification and characterization of a negative regulator of FtsZ ring formation in Bacillus subtilis. Proc Natl Acad Sci U S A 96:9642–9647. doi: 10.1073/pnas.96.17.9642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fleurie A, Manuse S, Zhao C, Campo N, Cluzel C, Lavergne JP, Freton C, Combet C, Guiral S, Soufi B, Macek B, Kuru E, VanNieuwenhze MS, Brun YV, Di Guilmi AM, Claverys JP, Galinier A, Grangeasse C. 2014. Interplay of the serine/threonine-kinase StkP and the paralogs DivIVA and GpsB in pneumococcal cell elongation and division. PLoS Genet 10:e1004275. doi: 10.1371/journal.pgen.1004275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ehrmann M, Boyd D, Beckwith J. 1990. Genetic analysis of membrane protein topology by a sandwich gene fusion approach. Proc Natl Acad Sci U S A 87:7574–7578. doi: 10.1073/pnas.87.19.7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Biondi RM, Baehler PJ, Reymond CD, Veron M. 1998. Random insertion of GFP into the cAMP-dependent protein kinase regulatory subunit from Dictyostelium discoideum. Nucleic Acids Res 26:4946–4952. doi: 10.1093/nar/26.21.4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kratz PA, Bottcher B, Nassal M. 1999. Native display of complete foreign protein domains on the surface of hepatitis B virus capsids. Proc Natl Acad Sci U S A 96:1915–1920. doi: 10.1073/pnas.96.5.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siegel MS, Isacoff EY. 1997. A genetically encoded optical probe of membrane voltage. Neuron 19:735–741. doi: 10.1016/S0896-6273(00)80955-1. [DOI] [PubMed] [Google Scholar]
- 26.Bendezú FO, Hale CA, Bernhardt TG, de Boer PA. 2009. RodZ (YfgA) is required for proper assembly of the MreB actin cytoskeleton and cell shape in E. coli. EMBO J 28:193–204. doi: 10.1038/emboj.2008.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oliva MA, Trambaiolo D, Lowe J. 2007. Structural insights into the conformational variability of FtsZ. J Mol Biol 373:1229–1242. doi: 10.1016/j.jmb.2007.08.056. [DOI] [PubMed] [Google Scholar]
- 28.Dai K, Lutkenhaus J. 1991. ftsZ is an essential cell division gene in Escherichia coli. J Bacteriol 173:3500–3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Redick SD, Stricker J, Briscoe G, Erickson HP. 2005. Mutants of FtsZ targeting the protofilament interface: effects on cell division and GTPase activity. J Bacteriol 187:2727–2736. doi: 10.1128/JB.187.8.2727-2736.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stricker J, Erickson HP. 2003. In vivo characterization of Escherichia coli ftsZ mutants: effects on Z-ring structure and function. J Bacteriol 185:4796–4805. doi: 10.1128/JB.185.16.4796-4805.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zacharias DA, Violin JD, Newton AC, Tsien RY. 2002. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science 296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- 32.Zhang J, Campbell RE, Ting AY, Tsien RY. 2002. Creating new fluorescent probes for cell biology. Nat Rev Mol Cell Biol 3:906–918. doi: 10.1038/nrm976. [DOI] [PubMed] [Google Scholar]
- 33.Li X, Zhang G, Ngo N, Zhao X, Kain SR, Huang CC. 1997. Deletions of the Aequorea victoria green fluorescent protein define the minimal domain required for fluorescence. J Biol Chem 272:28545–28549. doi: 10.1074/jbc.272.45.28545. [DOI] [PubMed] [Google Scholar]
- 34.Shimozono S, Hosoi H, Mizuno H, Fukano T, Tahara T, Miyawaki A. 2006. Concatenation of cyan and yellow fluorescent proteins for efficient resonance energy transfer. Biochemistry 45:6267–6271. doi: 10.1021/bi060093i. [DOI] [PubMed] [Google Scholar]
- 35.Brakemann T, Stiel AC, Weber G, Andresen M, Testa I, Grotjohann T, Leutenegger M, Plessmann U, Urlaub H, Eggeling C, Wahl MC, Hell SW, Jakobs S. 2011. A reversibly photoswitchable GFP-like protein with fluorescence excitation decoupled from switching. Nat Biotechnol 29:942–947. doi: 10.1038/nbt.1952. [DOI] [PubMed] [Google Scholar]
- 36.Rizzo MA, Springer GH, Granada B, Piston DW. 2004. An improved cyan fluorescent protein variant useful for FRET. Nat Biotechnol 22:445–449. doi: 10.1038/nbt945. [DOI] [PubMed] [Google Scholar]
- 37.McKinney SA, Murphy CS, Hazelwood KL, Davidson MW, Looger LL. 2009. A bright and photostable photoconvertible fluorescent protein. Nat Methods 6:131–133. doi: 10.1038/nmeth.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paez-Segala MG, Sun MG, Shtengel G, Viswanathan S, Baird MA, Macklin JJ, Patel R, Allen JR, Howe ES, Piszczek G, Hess HF, Davidson MW, Wang Y, Looger LL. 2015. Fixation-resistant photoactivatable fluorescent proteins for CLEM. Nat Methods 12:215–218. doi: 10.1038/nmeth.3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang M, Chang H, Zhang Y, Yu J, Wu L, Ji W, Chen J, Liu B, Lu J, Liu Y, Zhang J, Xu P, Xu T. 2012. Rational design of true monomeric and bright photoactivatable fluorescent proteins. Nat Methods 9:727–729. doi: 10.1038/nmeth.2021. [DOI] [PubMed] [Google Scholar]
- 40.McEvoy AL, Hoi H, Bates M, Platonova E, Cranfill PJ, Baird MA, Davidson MW, Ewers H, Liphardt J, Campbell RE. 2012. mMaple: a photoconvertible fluorescent protein for use in multiple imaging modalities. PLoS One 7:e51314. doi: 10.1371/journal.pone.0051314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang S, Moffitt JR, Dempsey GT, Xie XS, Zhuang X. 2014. Characterization and development of photoactivatable fluorescent proteins for single-molecule-based superresolution imaging. Proc Natl Acad Sci U S A 111:8452–8457. doi: 10.1073/pnas.1406593111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hamilton CM, Aldea M, Washburn BK, Babitzke P, Kushner SR. 1989. New method for generating deletions and gene replacements in Escherichia coli. J Bacteriol 171:4617–4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shaner NC, Lambert GG, Chammas A, Ni Y, Cranfill PJ, Baird MA, Sell BR, Allen JR, Day RN, Israelsson M, Davidson MW, Wang J. 2013. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat Methods 10:407–409. doi: 10.1038/nmeth.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol 22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 45.Habuchi S, Tsutsui H, Kochaniak AB, Miyawaki A, van Oijen AM. 2008. mKikGR, a monomeric photoswitchable fluorescent protein. PLoS One 3:e3944. doi: 10.1371/journal.pone.0003944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu C, Stricker J, Erickson HP. 2001. Site-specific mutations of FtsZ—effects on GTPase and in vitro assembly. BMC Microbiol 1:7. doi: 10.1186/1471-2180-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koppelman CM, Aarsman ME, Postmus J, Pas E, Muijsers AO, Scheffers DJ, Nanninga N, Den Blaauwen T. 2004. R174 of Escherichia coli FtsZ is involved in membrane interaction and protofilament bundling, and is essential for cell division. Mol Microbiol 51:645–657. [DOI] [PubMed] [Google Scholar]
- 48.Buske PJ, Levin PA. 2013. A flexible C-terminal linker is required for proper FtsZ assembly in vitro and cytokinetic ring formation in vivo. Mol Microbiol 89:249–263. doi: 10.1111/mmi.12272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sundararajan K, Miguel A, Desmarais SM, Meier EL, Casey Huang K, Goley ED. 2015. The bacterial tubulin FtsZ requires its intrinsically disordered linker to direct robust cell wall construction. Nat Commun 6:7281. doi: 10.1038/ncomms8281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Osawa M, Erickson HP. 2011. Inside-out Z rings—constriction with and without GTP hydrolysis. Mol Microbiol 81:571–579. doi: 10.1111/j.1365-2958.2011.07716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jaiswal R, Patel RY, Asthana J, Jindal B, Balaji PV, Panda D. 2010. E93R substitution of Escherichia coli FtsZ induces bundling of protofilaments, reduces GTPase activity, and impairs bacterial cytokinesis. J Biol Chem 285:31796–31805. doi: 10.1074/jbc.M110.138719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li Y, Hsin J, Zhao L, Cheng Y, Shang W, Huang KC, Wang HW, Ye S. 2013. FtsZ protofilaments use a hinge-opening mechanism for constrictive force generation. Science 341:392–395. doi: 10.1126/science.1239248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Milam SL, Osawa M, Erickson HP. 2012. Negative-stain electron microscopy of inside-out FtsZ rings reconstituted on artificial membrane tubules show ribbons of protofilaments. Biophys J 103:59–68. doi: 10.1016/j.bpj.2012.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Szwedziak P, Wang Q, Bharat TA, Tsim M, Lowe J. 2014. Architecture of the ring formed by the tubulin homologue FtsZ in bacterial cell division. eLife 3:e04601. doi: 10.7554/eLife.04601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li Z, Trimble MJ, Brun YV, Jensen GJ. 2007. The structure of FtsZ filaments in vivo suggests a force-generating role in cell division. EMBO J 26:4694–4708. doi: 10.1038/sj.emboj.7601895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. 2002. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol 20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 57.Khlebnikov A, Datsenko KA, Skaug T, Wanner BL, Keasling JD. 2001. Homogeneous expression of the P(BAD) promoter in Escherichia coli by constitutive expression of the low-affinity high-capacity AraE transporter. Microbiology 147:3241–3247. doi: 10.1099/00221287-147-12-3241. [DOI] [PubMed] [Google Scholar]
- 58.Matsui T, Yamane J, Mogi N, Yamaguchi H, Takemoto H, Yao M, Tanaka I. 2012. Structural reorganization of the bacterial cell-division protein FtsZ from Staphylococcus aureus. Acta Crystallogr D Biol Crystallogr 68:1175–1188. doi: 10.1107/S0907444912022640. [DOI] [PubMed] [Google Scholar]