ABSTRACT
Antimicrobial resistance through extended-spectrum beta-lactamases (ESBLs) and transferable (plasmid-encoded) cephamycinases (pAmpCs) represents an increasing problem in human and veterinary medicine. The presence of ESBL-/pAmpC-producing commensal enterobacteria in farm animals, such as broiler chickens, is considered one possible source of food contamination and could therefore also be relevant for human colonization. Studies on transmission routes along the broiler production chain showed that 1-day-old hatchlings are already affected. In this study, ESBL-/pAmpC-positive broiler parent flocks and their corresponding eggs, as well as various environmental and air samples from the hatchery, were analyzed. The eggs were investigated concerning ESBL-/pAmpC-producing enterobacteria on the outer eggshell surface (before/after disinfection), the inner eggshell surface, and the egg content. Isolates were analyzed concerning their species, their phylogroup in the case of Escherichia coli strains, the respective resistance genes, and the phenotypical antibiotic resistance. Of the tested eggs, 0.9% (n = 560) were contaminated on their outer shell surface. Further analyses using pulsed-field gel electrophoresis showed a relationship of these strains to those isolated from the corresponding parent flocks, which demonstrates a pseudo-vertical transfer of ESBL-/pAmpC-producing enterobacteria into the hatchery. Resistant enterobacteria were also found in environmental samples from the hatchery, such as dust or surfaces which could pose as a possible contamination source for the hatchlings. All 1-day-old chicks tested negative directly after hatching. The results show a possible entry of ESBL-/pAmpC-producing enterobacteria from the parent flocks into the hatchery; however, the impact of the hatchery on colonization of the hatchlings seems to be low.
IMPORTANCE ESBL-/pAmpC-producing enterobacteria occur frequently in broiler-fattening farms. Recent studies investigated the prevalence and possible transmission route of these bacteria in the broiler production chain. It seemed very likely that the hatcheries play an important role in transmission and/or contamination events. There are only few data on transmission investigations from a grandparent or parent flock to their offspring. However, reliable data on direct or indirect vertical transmission events in the hatchery are not available. Therefore, we conducted our study and intensively investigated the broiler hatching eggs from ESBL-/pAmpC-positive broiler parent flocks as well as the hatchlings and the environment of the hatchery.
KEYWORDS: antibiotic resistance, Enterobacteriaceae, broiler chicken, hatchery, ESBL, pAmpC, AmpC
INTRODUCTION
Antimicrobial resistance is a challenging problem in public health. Transferable resistances to extended-spectrum cephalosporins (ESCs) have a rising impact on treatment strategies in both human and veterinary medicine (1, 2). Especially, extended-spectrum beta-lactamases (ESBLs) and plasmid-encoded cephamycinases (pAmpCs) protect pathogens against a variety of beta-lactam antibiotics, including third- and fourth-generation antibiotics. The most widespread ESBL genes in Germany are CTX-M-1, CTX-M-15, and CTX-M-14, which occur in humans as well as in animals (3, 4). A frequently detected pAmpC beta-lactamase is the CIT-type CMY-2 enzyme (5, 6).
Pathogenic ESBL-/pAmpC-producing enterobacteria can cause severe problems; however, commensal enterobacteria harboring ESBL-/pAmpC resistance genes were also detected in companion animals as well as farm animals, including broiler chickens (7–14). Especially, those farm animals can act as reservoirs for resistant bacteria and introduce the ESBL-/pAmpC-producing enterobacteria into the food production process (4, 15–17). Various studies also demonstrated a high prevalence of ESBL/pAmpC-producing enterobacteria in broiler chicken farms and the respective environment in Germany (7, 8, 18–22). It turned out that the 1-day-old (parent) broiler chicks already seemed to be colonized by these resistant bacteria when arriving at the farms (19, 23).
However, there are different important levels within the broiler production chain that can in general contribute to the transfer of resistant bacteria. It starts with the grandparents of the broiler chicken and proceeds to the hatcheries, down to the broiler-fattening farms, and, subsequently, to the slaughterhouse and food production process. Previous studies showed possible transmissions of pathogenic as well as commensal resistant enterobacteria between the levels of broiler production (23–26). The transmission or spread of highly similar plasmids harboring the respective resistance genes was also discussed (27). All these data suggest that hatcheries responsible for either the grandparent chicks or the parent chicks seemed to be involved in possible transmission routes. A direct transmission from the grandparent or the parent chicken to their offspring through the eggs could not yet be confirmed. The objective of this study was therefore to analyze all potential transmission routes of ESBL-/pAmpC-producing enterobacteria within the first levels of the broiler production chain. For that, eggs from ESBL-/pAmpC-positive parent flocks were tracked through the various stations within the hatchery, and the corresponding hatchlings were finally analyzed. At every sampling, different environmental and air samples were also considered.
RESULTS
ESBL-/pAmpC-producing enterobacteria.
ESBL-/pAmpC-producing enterobacteria were found in samples from all four sampling time points. Out of the 36 samples from seven parent flocks (time point one), 24 samples (66.7%) were positive for ESBL-/pAmpC-producing enterobacteria after preenrichment in LB medium (Table 1). Without preenrichment, ESBL-/pAmpC-producing enterobacteria were detected in only 41.7% of the specimens. The microbial counts of the resistant bacteria varied between 1.67E + 01 and 7.17E + 05 CFU per boot swab or g of feces (geometric mean, 6.91E + 04 CFU).
TABLE 1.
Occurrence of ESBL-/pAmpC-producing enterobacteria and of total enterobacteria in samples of broiler parent flocks and in the hatchery
| Sampling | No. of positive samples/total no. of samples, prevalence (%)a |
||||||
|---|---|---|---|---|---|---|---|
| Flock A | Flock B | Flock C | Flock D | Flock E | Flock F | Flock G | |
| Parent flock | |||||||
| Boot swab | |||||||
| ESBL-/pAmpC-producing enterobacteria | 1/1 | 1/1 | 1/1 | 1/1 | 3/3 | 0/3 | 1/4 |
| Total enterobacteria | 1/1 | 1/1 | 1/1 | 1/1 | 3/3 | 3/3 | 4/4 |
| Feces | |||||||
| ESBL-/pAmpC-producing enterobacteria | 3/3 | 3/3 | 3/3 | 2/3 | 3/3 | 1/3 | 1/4 |
| Total enterobacteria | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 4/4 |
| Before disinfection of eggs (hatchery) | |||||||
| Egg surface | |||||||
| ESBL-/pAmpC-producing enterobacteria | 0/40 | 4/40, 10 | 0/40 | 0/40 | 0/40 | 1/40, 2.5 | 0/40 |
| Total enterobacteria | 12/40, 30 | 24/40, 60 | 12/40, 30 | 7/40, 17.5 | 16/40, 40 | 29/40, 72.5 | 21/40, 52.5 |
| Egg inner surface | |||||||
| ESBL-/pAmpC-producing enterobacteria | 0/40 | 0/40 | 0/40 | 0/40 | 0/40 | 0/40 | 0/40 |
| Total enterobacteria | 0/40 | 5/40, 12.5 | 5/40, 12.5 | 2/40, 5 | 5/40, 12.5 | 0/40 | 2/40, 5 |
| Egg content | |||||||
| ESBL-/pAmpC-producing enterobacteria | 0/40 | 0/40 | 0/40 | 0/40 | 0/40 | 0/40 | 0/40 |
| Total enterobacteria | 0/40 | 0/40 | 0/40 | 0/40 | 0/40 | 0/40 | 0/40 |
| Environmental samples | |||||||
| ESBL-/pAmpC-producing enterobacteria | 0/3 | 0/5 | 0/4 | 0/6 | 0/7 | 0/3 | 0/6 |
| Total enterobacteria | 1/3, 33.3 | 0/5 | 2/4, 50 | 0/6 | 3/7, 42.9 | 0/3 | 0/6 |
| Air samples | |||||||
| ESBL-/pAmpC-producing enterobacteria | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 | 0/2 |
| Total enterobacteria | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 | 0/2 |
| After disinfection of eggs (hatchery) | |||||||
| Egg surface | |||||||
| ESBL-/pAmpC-producing enterobacteria | 0/40 | 1/40, 2.5 | 0/40 | 0/40 | 0/40 | 0/40 | 0/40 |
| Total enterobacteria | 0/40 | 4/40, 10 | 1/40, 2.5 | 1/40, 2.5 | 0/40 | 1/40, 2.5 | 3/40, 7.5 |
| Environmental samples | |||||||
| ESBL-/pAmpC-producing enterobacteria | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 | 0/3 | 0/5 |
| Total enterobacteria | 0/4 | 0/4 | 1/4, 25 | 0/4 | 0/4 | 0/3 | 1/5, 20 |
| Air samples | |||||||
| ESBL-/pAmpC-producing enterobacteria | 0/4 | 0/2 | 0/2 | 0/2 | 0/2 | 0/2 | 0/2 |
| Total enterobacteria | 0/4 | 0/2 | 0/2 | 0/2 | 0/2 | 0/2 | 0/2 |
| Hatching of chicks (hatchery) | |||||||
| Cloacal swabs | |||||||
| ESBL-/pAmpC-producing enterobacteria | 0/40 | 0/40 | 0/40 | 0/40 | 0/40 | 0/40 | 0/40 |
| Total enterobacteria | 0/40 | 0/40 | 2/40, 5 | 0/40 | 0/40 | 0/40 | 1/40, 2.5 |
| Environmental samples | |||||||
| ESBL-/pAmpC-producing enterobacteria | 0/8 | 3/9, 33.3 | 0/8 | 0/11 | 0/9 | 0/9 | 1/11, 9.1 |
| Total enterobacteria | 0/8 | 5/9, 55.6 | 3/8, 37.5 | 10/11, 90.9 | 7/9, 77.8 | 5/9, 55.6 | 9/11, 81.8 |
For the ESBL-/pAmpC-producing enterobacteria data, numbers of ESBL-/pAmpC-positive samples and total numbers of samples of each sample type are stated. For the total enterobacteria data, the prevalences of samples positive for ESBL-/pAmpC-producing enterobacteria and for total enterobacteria are given.
From the hatchery in total, 200 specimens from eggs and chicks as well as 22 to 27 environmental and air samples from each of the flocks (n = 1,571) were analyzed. Before disinfection, ESBL-/pAmpC-producing enterobacteria were found on the outer surface of five eggs (1.8%), with four eggs belonging to flock B and one egg belonging to flock F (Table 1). The resistant bacteria were detected by the enrichment method only. None of the samples from the egg content and the inner surface or from the air and the environment were positive before routine egg disinfection. After disinfection, one egg surface (0.4%) belonging to flock B was positive for ESBL-/pAmpC-producing enterobacteria using the preenrichment method (Table 1). Again, all environmental and air samples were negative at this time. At the hatching of the chicks, we detected ESBL-/pAmpC-producing enterobacteria in four out of 65 environmental samples (6.2%) (Table 1). The positive samples were dust collected inside the hatcher, a crushed egg shell sample, and an environmental swab collected from flock B and a swab taken from the station for the automatic separation of chicks and eggshells of flock G. All 280 cloacal swabs from the hatchlings were negative concerning the resistant bacteria.
Total enterobacteria.
In all boot swabs (n = 14) and feces samples (n = 22) from the parent flocks, enterobacteria were detected on MacConkey agar no. 3 (Oxoid, Wesel, Germany) without cefotaxime (MC−) agar plates after enrichment (Table 1). In addition, enumeration of total enterobacteria was done for the feces samples. We counted up to 1.01E + 08 enterobacteria/g of feces. The geometric mean (1.54E + 07 CFU/g of feces) was higher than for the resistant enterobacteria and resulted in a mean proportion of 0.6% ESBL-/pAmpC-producing enterobacteria of the total enterobacteria.
At the hatchery, all 1,571 samples were also analyzed concerning total enterobacteria. After the preenrichment, 200 samples (12.7%) were positive on MC− agar plates (Table 1). Of these samples, 73% (n = 146) were collected during the arrival of the eggs. Outer surfaces of the eggs (43.2%) from all seven flocks as well as inner eggshell surfaces (6.8%) and environmental swabs (21.4%) were positive concerning enterobacteria. The egg contents and the air samples were all negative. For the 120 egg surfaces with enterobacterial growth on MC− agar plates, enumeration was possible for 42 eggs (35.0%). The geometric mean was calculated as 8.71E + 04 enterobacteria/egg surface (minimum, 2.67E + 01 enterobacteria/egg surface; maximum, 3.47E + 06 enterobacteria/egg surface). After the disinfection of eggs, we still found 10 out of 280 outer egg surfaces (3.6%) to be positive for total enterobacteria (Table 1). For two of them, the total amount could be counted as 8.36E + 07 CFU and 7.33E + 02 CFU. In addition, enterobacteria could be detected in two feather samples from the hatchery (flocks C and G). All collected air samples after disinfection of the eggs were negative. After hatching of the chicks, 39 of 65 environmental samples (60.0%), including hatchlings dust and crushed eggshells, were positive on MC− agar plates after preenrichment (Table 1). At the same time point, only three cloacal swabs (1.1%) from the hatchlings were positive for enterobacteria, two from flock C and one from flock G. For one cloacal swab from flock C, enumeration on MC− agar plates led to a total of 9.48E + 05 enterobacteria.
Characterization of beta-lactamase genes and phylotyping.
Molecular analyses showed that all seven parent flocks were colonized with up to four different types of ESBL-/pAmpC-producing enterobacteria, as shown in Table 2.
TABLE 2.
ESBL/pAmpC resistance genes and species plus phylogroups of enterobacteria isolated from samples of the parent flocks and the hatchery
| Flock | Sampling | Sample(s) | Species | Phylogroup | Gene(s) |
|---|---|---|---|---|---|
| A | Parent flock | Boot swab, pooled feces | E. coli | A | TEM-1a/TEM-52 |
| B | Parent flock | Boot swab, pooled feces | E. fergusonii | Negativeb | TEM-52 |
| E. coli | A/C | TEM-1a/CTX-M1 | |||
| A | TEM-1a/CTX-M1 | ||||
| A | CTX-M1 | ||||
| B1c | CTX-M1 | ||||
| Before disinfection | Outer egg shell surface | E. coli | B1c | CTX-M1 | |
| After disinfection | Outer egg shell surface | E. fergusonii | Negativeb | TEM-52 | |
| Hatching | Hatchlings dust | E. coli | E | TEM-1a/CMY-2 | |
| Crushed egg shell, Environmental sample | K. pneumoniae | Negative | SHV-1a/SHV-2 | ||
| C | Parent flock | Boot swab, pooled feces | E. fergusonii | Negative | TEM-1a/CMY-2 |
| Negative | TEM-52 | ||||
| E. coli | A/C | CMY-2 | |||
| B1 | TEM-1a/CMY-2 | ||||
| F | CMY-2 | ||||
| D | Parent flock | Boot swab, pooled feces | E. coli | A | TEM-1a/CMY-2 |
| A | CMY-2 | ||||
| F | CTX-M1 | ||||
| F | CMY-2 | ||||
| E | Parent flock | Boot swab, pooled feces | E. coli | F | CTX-M1 |
| F | Parent flock | Boot swab, pooled feces | E. coli | A/C | TEM-1a/CTX-M15 |
| Before disinfection | Outer egg shell surface | C. freundii | CMY | ||
| G | Parent flock | Boot swab, pooled feces | E. coli | A/C | TEM-1a/TEM-52 |
| B1 | TEM-52 | ||||
| B1 | CMY-2 | ||||
| Hatching | Environmental sample | E. coli | A | TEM-1a/TEM-52 |
Isolates of the four positive eggs of flock B were all determined to be Escherichia coli of the phylogroup B1 harboring a CTX-M-1 gene. These strains show molecular characteristics identical to those of strains of the respective parent flock (E. coli, phylogroup B1, CTX-M-1 resistance gene [Table 2]). Therefore, randomly chosen isolates from pooled feces and the boot swab of the parent flock B as well as from the four egg surfaces were further analyzed by pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST). The fifth ESBL-/pAmpC-positive outer eggshell before disinfection was determined from flock F. From this specimen, a Citrobacter freundii isolate which was positive for a CIT-type CMY gene was identified.
After disinfection of the eggs at the hatchery, one egg was still positive for ESBL-/pAmpC-producing enterobacteria on the outer surface. This egg originated also from flock B, and phylogroup-negative Escherichia fergusonii strains harboring the ESBL gene TEM-52 could be isolated. Again, comparable isolates were found in the respective parent flock B (Table 2), and therefore, PFGE and MLST analyses were applied also for those strains.
During hatching of the chicks of flock B, we found the dust of the hatching breeder, a crushed egg shell sample, and an environmental swab from the station for the automatic separation of chicks and eggshells to be positive for resistant enterobacteria. E. coli of phylogroup E/D harboring the broad-spectrum beta-lactamase (BSBL) gene TEM-1 and a CIT-type pAmpC gene (CMY-2) was isolated from the hatchling dust. Both other environmental samples were positive for Klebsiella pneumoniae harboring the resistance genes SHV-1 (BSBL) and SHV-2. From a swab from the station for the automatic separation of chicks and eggshells of flock G, E. coli strains of phylogroup A harboring the resistance genes TEM-1 (BSBL) and TEM-52 were isolated.
Antimicrobial resistance testing.
The applied disk diffusion tests showed variation in the disk diffusion diameters according to the ESBL/pAmpC resistance genes verified by sequencing. Theses variations were independent from the respective phylogroups of the isolates. Strains with resistance against cefotaxime (CTX) showed a CTX-M-1 gene, whereas strains harboring a CTX-M-15 or TEM-52 gene showed resistance to CTX and ceftazidime (CAZ). The diameters of the inhibition zone of the CTX-M-15 isolates were up to 5 mm smaller for both antibiotics compared to those of the TEM-52 strains. All isolates with a sequenced ESBL gene showed an increase in the inhibition zone diameter of CTX in the presence of clavulanic acid of up to 20 mm. Isolates that are resistant against CTX, CAZ, and cefoxitin (FOX) and show an increase in the inhibition zone diameter of CTX in the presence of clavulanic acid of up to 5 mm on average harbor the CMY-2 gene. Isolates harboring both a BSBL TEM-1 gene and a TEM-52 gene or an SHV-1 (BSBL) and an SHV-2 gene showed only an intermediate response against CTX and CAZ.
PFGE and MLST analyses of flock B.
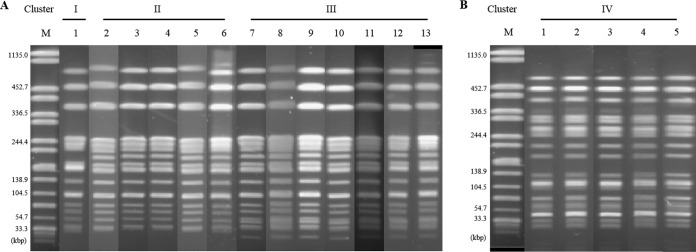
From flock B, we isolated ESBL-producing Escherichia strains with comparable genotypes (phylogroup plus resistance gene) from specimens from the parent flock as well as from the hatchery. To determine the phylogenetic relationships of these strains, we did PFGE analyses of randomly chosen isolates from all specimens (Fig. 1).
FIG 1.
PFGE pattern of ESBL-positive isolates from flock B. (A) Comparison of Escherichia coli isolates from the parent flock and the outer surface of four eggs at the time of arrival at the hatchery. Isolates were of phylogroup B1 and harbor a CTX-M1 gene. Lane M, Salmonella Braenderup H9812; lanes 1 and 12, feces sample 2, parent flock; lanes 2 and 9, surface egg 4; lanes 3 and 7, feces sample 3, parent flock; lanes 4 and 13, boot swab, parent flock; lanes 5 and 10, surface egg 3; lane 6, surface egg 1; lane 8, feces sample 1, parent flock; lane 11, surface egg 2. (B) Comparison of phylogroup-negative Escherichia fergusonii isolates which were positive for the TEM-52 gene. Samples were taken from the parent flock and at the hatchery after disinfection of the eggs. Lane M, Salmonella Braenderup H9812; lane 1, feces sample 3, parent flock; lane 2, feces sample 2, parent flock; lane 3, feces sample 1, parent flock; lane 4, boot swap, parent flock; lane 5, surface egg 5.
Evaluation of the PFGE gels showed four different band patterns, of which clusters I to III comprise the tested B1/CTX-M-1 isolates (Fig. 1A). The PFGE pattern of cluster I was found only once, in the boot swab from the parent flock. Cluster II included isolates from one pooled feces sample and all four egg surfaces before disinfection. Cluster III comprises identical band patterns from strains from the parent flock (boot swab and pooled feces) as well as from two of the eggs before disinfection. The band patterns of clusters I to III differ by only one band (I to II) or two bands (I to III). Identical band patterns in the PFGE gel imply a direct phylogenetic relationship. Therefore, we also checked the MLST type as a control for one isolate of each of the clusters. It turned out that the isolates were assigned to the same MLST type, sequence type 1665 (ST1665).
The PFGE cluster IV was restricted to the phylogroup-negative E. fergusonii isolates from all specimens collected from the parent flock and the outer surface of the disinfected egg (Fig. 1B). The tested isolates showed identical band patterns and were therefore determined to be phylogenetically related. For the phylogroup-negative strain, the MLST type could not be determined.
DISCUSSION
It is already known that commensal ESBL-/pAmpC-producing E. coli strains occur in healthy chickens on broiler farms (7, 8, 28), and even 1-day-old chicks seemed to be affected (19). Recently published studies described the introduction and the circulation of ESBL-/pAmpC-producing E. coli strains in broiler farms and along the whole broiler production process (23, 25, 27). They showed that there is a clonal spread through grandparent flocks as well as parent flocks and broiler farms. However, there were missing links in the longitudinal follow up, especially in the hatcheries. Therefore, we conducted our study to investigate the transmission routes of ESBL-/pAmpC-producing enterobacteria from the parent flocks to the hatchery and, subsequently, the hatched chicks.
Parent flocks.
The detected resistance genes as well as the respective prevalences were in concordance with previously published studies on broiler chickens in Germany (3, 17, 19). We found ESBL-/pAmpC-producing E. coli strains of different phylogroups comparable to those in a recent study on the genetic diversity of E. coli in poultry production (29). In addition, we isolated E. fergusonii strains harboring resistance genes against beta-lactam antibiotics from two parent flocks. Since a multidrug-resistant E. fergusonii strain was also found in chickens (30), studies on ESBL/pAmpC producers in poultry and veterinary public health agencies should also consider these bacteria.
For all phenotypically resistant enterobacteria except Enterobacter spp., a respective ESBL or pAmpC gene could be identified. Enterobacter spp. are known to show intrinsic resistance to beta-lactam antibiotics due to alterations of outer membrane proteins (31).
Interestingly, the results of the disk diffusion diameters showed variations according to the respective gene type, which have not, to our knowledge, been described before in detail.
Transmission to the hatchery via egg surfaces.
From each of the seven parent flocks, we tracked the corresponding eggs to the hatchery and analyzed the outer surfaces of 40 eggs of the same batch before and after disinfection, respectively. In contrast to Mezhoud et al. (32), we used a modified shell rinse method (33) instead of crushing the whole eggshell to distinguish between the bacteria originating from the outer and inner shell surfaces. Using the shell rinse method, we could demonstrate the contamination of five eggs with ESBL-/pAmpC-producing enterobacteria prior to their disinfection (flock B, E. coli B1 plus CTX-M-1; flock G, Citrobacter freundii plus CMY). In a recent study on egg contamination in Belgium, the Netherlands, and France, ESBL producers could be isolated from broiler chicken eggs as well (32). Using comparative analyses, we could show that the ESBL strains found on egg surfaces of flock B harboring a CTX-M-1 are highly related to those isolated from their parent flock (Fig. 1). This clearly demonstrates that the introduction of ESBL-producing E. coli strains occurs directly from the parent flock into the hatchery. Furthermore, we could show that up to 3.47E + 06 total enterobacteria were present on the outer surfaces of the eggs which were transported to the hatchery. This finding matches previously published data (34). The prevalence of total enterobacteria with 42.9% positive eggs demonstrates, in principle, a high input of bacteria into the hatchery.
The same batch of eggs was again sampled after their disinfection by formaldehyde fumigation, which is routinely done in hatcheries for egg sanitation (34). On one egg surface, a still-viable E. fergusonii isolate harboring a TEM-52 gene was found. Again, PFGE analyses verified the phylogenetically relationship to the respective isolates from its parent flock B. This emphasizes the hypothesis of a direct transmission via the eggs. In total, 280 egg surfaces were analyzed after disinfection, of which 3.6% were positive for enterobacterial growth on MC− agar plates. Therefore, even after the disinfection of the eggs, they could serve as a contamination source; however, it seems to have a rather low impact.
Transmission to hatchery via the environment.
Before disinfection, all environmental and air samples from the hatchery were negative concerning ESBL-/pAmpC-producing enterobacteria. However, in three flocks, we found up to 50% of the environmental samples to be positive for non-ESBL/pAmpC enterobacteria. It seems that resistant strains in the environment of the hatchery where the eggs arrive are absent or their load is very low; therefore, they are under the detection limit of our sampling method.
After disinfection of the eggs, only two feather samples showed enterobacterial growth on MC− agar plates. As feathers are introduced into the hatchery usually stuck to the eggshells via feces, they have an impact on bacterial transmission into the hatchery similar to that of the eggs themselves. This could be avoided by removing the feathers in the parent flock farms. Also, none of the air samples taken in the particular rooms before and after disinfection showed enterobacterial growth. Taken together, a relevant transmission of ESBL-/pAmpC-producing enterobacteria via air or contaminated surfaces seems to be unlikely. A study on this transmission route was not done before.
Transmission to hatchlings via eggs.
One possible transmission route of ESBL-/pAmpC-producing enterobacteria to hatchlings we investigated was the direct vertical transfer through reproductive organs of the hens. This was shown for layering hens in cases of experimental infections with Salmonella enterica serovar Enteritidis (35) but, to our knowledge, not for bacteria of the genus Escherichia. Therefore, we investigated the total egg content (egg white and albumin) of 40 eggs from each parent flock after their arrival in the hatchery. None of the tested eggs were positive for any enterobacterial growth; therefore, we would exclude this transmission route for ESBL-/pAmpC-producing enterobacteria.
Early contamination of eggs can also occur after the lay through penetration of the eggshells. Eggshell and eggshell membranes are natural defense barriers, but penetration through and multiplication within the egg were shown for salmonellae in various studies (35–39). To our knowledge, this was also not yet demonstrated for E. coli. Penetration of pathogens through the eggshell occurs more likely in cases of incomplete cuticle or with variations in pH, temperature, humidity, or vapor (35, 38). In the investigated hatchery, the conditions at our samplings were constant and, therefore, preventive for the penetration of bacteria into the eggs. However, from 6.8% of the inner surfaces of the eggs, enterobacteria could be isolated which were not resistant to a beta-lactam antibiotic. This demonstrates a potential growth of enterobacteria on the inner surface of an egg and is comparable to the results of another study (37). Nevertheless, further validation of the method needs to be done. There is no suitable method to analyze the bacterial growth on the outer and inner surfaces of the same egg so far. We intensively tested and optimized our methods for the investigation of the outer and inner egg surfaces before starting the samplings to clearly show a penetration of the pathogens through the eggshell and avoid contamination. However, this might happen because of tiny breaks in the eggshell or cuticle occurring during the filling of the eggshells with the hot liquid MC− agar (56°C).
At the last sampling time point, we investigated the gut microbiota of the recently hatched chicks using cloacal swabs. In our study, none of the 280 recently hatched chicks were positive for ESBL-/pAmpC-producing enterobacteria. In fact, only three chicks (1.1%) were already colonized with enterobacteria.
Taken together, our results show that a strict vertical transmission of ESBL-/pAmpC-producing enterobacteria from broiler parent flocks to their hatchlings through the eggs could not be verified. We also demonstrated that a difference of 1 or 2 days in the sampling of hatchlings can change the outcome of a study. The colonization process of the hatchlings with the gut microbiota occurs within a few days and needs, therefore, to be further analyzed.
Transmission to hatchlings via the environment.
Studies reported that 1-day-old broiler chicks are already and the grandparent chicks are colonized with ESBL-/pAmpC-producing E. coli (19, 40); they therefore contribute to the spread of these bacteria at the farm level. As discussed before, it is more likely that the chicks get colonized by the uptake of resistant bacteria from the environment of the hatchery. This pseudo-vertical transfer of ESBL-/pAmpC-producing enterobacteria might occur in the hatcheries or during the process of discarding eggshells, chicks' inspection, vaccination, counting, and loading them into the transportation boxes. We found four ESBL-/pAmpC-positive samples in the hatchery after hatching of the chicks in hatchling dust, crushed egg shells, and two swabs from the environment of the hatchery connected to the process of separation of eggshells from the chicks. This supports the hypothesis of a potential pseudo-vertical transfer from the hatchery environment to the hatchlings.
To our knowledge, the outer eggshell surface (before and after disinfection) as well as the inner shell surface and the egg content of broiler hatching eggs were not analyzed before in a published study. Microbiological analyses and the comparison of molecular data from isolates of parent flocks, eggs, and environmental samples showed a low level of transmission of ESBL-/pAmpC-producing enterobacteria from the parent flocks to the hatchery. We found resistant bacteria in the environment of the hatchery after hatching of chicks, but a strict vertical transfer for the ESBL-/pAmpC-producing enterobacteria could not be shown.
MATERIALS AND METHODS
Samplings.
In our study, seven different broiler parent flocks, their corresponding eggs at the hatchery, their hatchlings, and various environmental samples were investigated concerning the occurrence of ESBL-/pAmpC-producing enterobacteria in Germany in the years 2014 and 2015.
(i) Parent flocks.
Sixteen parent flocks of an integrated broiler production were initially investigated using at least three pooled feces and one boot swab. Out of these, only ESBL-/pAmpC-positive flocks were then included in the study. Seven positive parent flocks were selected, which were of different ages: 29 weeks (flock A), 57 weeks (flock B), 43 weeks (flock C), 50 weeks (flocks D and F), 51 weeks (flock E), and 58 weeks (flock G). Analyses of the parent flocks were done approximately 1 week before starting the samplings at the hatchery. The eggs of all studied parent flocks were transferred to the same hatchery.
(ii) Before egg disinfection.
Samplings in the hatchery were performed at three different time points. The first sampling was done during the arrival of the eggs at the hatchery. Environmental samples were taken from the wall and the ground or the drain using sterile swabs (dry cultural swab with flexible polystyrene [PS] stick; Nerbe Plus GmbH, Winsen [Luhe], Germany), moistened with phosphate-buffered saline (PBS), before the eggs arrived. The sampled spots were approximately 100 cm2 in size. In addition, two air samples were taken using two different sampling techniques analyzing 1,000 liters of air. The first instrument was the impactor Biotest Hycon RCS Plus air sampler (Biotest AG, Dreieich, Germany) used with an airflow of 50 liters/min. For the detection of resistant and nonresistant enterobacteria, Hycon blank strips (Millipore, Darmstadt, Germany) were filled with MacConkey agar no. 3 (Oxoid, Wesel, Germany) containing 1 mg/liter cefotaxime (AppliChem, Darmstadt, Germany) (MC+) and MacConkey agar no. 3 without cefotaxime (MC−), respectively. As a second air sampling instrument, the automated impinger Coriolis μ (Bertin Technologies) was used, and air was collected in 10 ml of PBS with an airflow of 250 liters/min.
After the arrival of the eggs at the hatchery, two air samples were again taken, as well as environmental swabs from the transport trolleys of the eggs and the truck (except for flock A). From each of the parent flocks, eggs were collected for analyzing the outer surface of the shell, the egg contents, and the inner shell surface. Therefore, 40 eggs were directly put into sterile Whirl-Pak stand-up bags (540 ml volume) for analyses of the outer surface using the shell rinse method according to Musgrove et al. (33).
For analyses of the egg contents and the inner shell surface, an additional 40 eggs were collected.
(iii) After egg disinfection.
The second sampling at the hatchery was performed after routine disinfection of the eggs by formaldehyde fumigation, which was usually done twice. Again, 40 eggs from the same batch were collected for outer surface analyses only. The collecting procedure was the same as described before. Two air samples were taken, as well as three environmental swabs from the wall, the ground, and the incubator racks. If possible, feathers or insects, like flies, were collected from the respective room in the hatchery directly into sterile sample bags.
(iv) Hatching.
The third sampling at the hatchery was done after the hatching of the chicks originating from the previously sampled parent flock. Therefore, 40 cloacal swabs of the hatchlings (dry cultural swab with aluminum stick; Nerbe Plus GmbH, Winsen [Luhe], Germany), 8 to 10 g of hatchling dust directly collected from the inside the hatchers, and two pooled eggshell samples each containing five crushed eggshells were collected into sterile Whirl-Pak stand-up bags. Up to five swabs from the environment of the hatchery were taken during the process of discarding eggshells and chicks' inspection, vaccination, counting, and loading into transport boxes.
Laboratory analyses. (i) Pooled feces and boot swabs.
Boot swabs and 20 g of feces were inoculated in stomacher bags in 200 ml and 180 ml of Luria-Bertani (LB) medium (Carl Roth, Karlsruhe, Germany), respectively, and were treated by stomaching for 2 min at 200 rpm (stomacher 400 circulator; Seward Limited, West Sussex, United Kingdom). After that, aliquots of the sample suspensions were taken for enumeration of ESBL-/pAmpC-producing enterobacteria on MC+ agar plates and an enumeration of all enterobacteria in feces samples on MC− agar plates. Therefore, 100 μl of an appropriate dilution was plated out onto three plates of the respective agar. Colonies were counted according to their morphology after aerobic incubation at 37°C for 24 h. For preenrichment, the stomacher bags with the LB sample mixtures were incubated aerobically 24 h at 37°C. After that, 10 μl of the incubated LB mixtures was streaked out with an inoculation loop on MC+ and MC− agar plates. Agar plates were aerobically incubated at 37°C for 24 h for qualitative detection of ESBL-/pAmpC-producing enterobacteria as well as for analysis of the total enterobacterial composition. Species identification was done using matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS; MALDI Microflex LT and Biotyper database, Bruker Daltonics, Bremen, Germany). Up to 10 suspicious ESBL-/pAmpC-producing enterobacteria of each sample with different colony morphology (two isolates each morphology) on MC+ agar plates were isolated and stored for further susceptibility testing and molecular analyses.
(ii) Outer surface of eggs.
Analyses of the outer surface of the eggs before and after disinfection were done using a slightly modified shell rinse method (33). After adding 10 ml of LB medium to the Whirl-Pak stand-up bags, eggs were shaken for 10 min at 20°C and 200 rpm in a shaking incubator. Retained samples were taken for enumeration of ESBL-/pAmpC-producing enterobacteria on MC+ agar plates and of all enterobacteria on MC− agar plates, as described above. Bags containing eggs and LB medium were incubated aerobically for 24 h at 37°C without shaking, and 10 μl of the incubated LB mixtures was streaked out on MC+ and MC− agar plates, respectively. Colony counting and isolate identification were done as described above. Isolates from MC− agar plates were cross-checked for their growth on MC+ agar plates.
(iii) Inner surface of eggs and egg contents.
Analyses of the inner surface and the egg content were done for the same egg. Therefore, the top of the egg was dipped for 2 s into 70% ethanol for disinfection, with a subsequent exposure time of at least 30 s and then opened sterile with the egg slicer Cregg (Brainstream, Oerlinghausen, Germany), which cuts the top of the egg and opens the egg at a diameter of approximately 35 mm. Egg yolk and albumin were put into a sterile 50-ml beaker with a screw cap (Sarstedt, Nümbrecht, Germany), gently mixed together with 20 ml of LB medium (10 ml for flock A), and incubated 24 h at 37°C. The empty eggshell was dried under a hood for at least 2 h and then filled with liquid MC− agar (56°C). The eggs were incubated at 37°C up to 48 h, and the shell was removed from the egg-shaped agar using sterile scalpels to analyze possible growth of enterobacteria originating from the inner surface of the eggs. If possible, colonies were picked. Additionally, the egg-shaped MacConkey agar was put into a Whirl-Pak stand-up bag, crushed and mixed manually with 30 ml of LB medium, and incubated for 24 h at 37°C. Enrichments of the egg content as well as the inner surface were streaked out on MC+ and MC− agar plates and incubated 24 h at 37°C. Plates were further analyzed concerning the growth of (resistant) enterobacteria, and isolation of colonies was done as previously mentioned.
(iv) Cloacal swabs.
Swabs were cut off into a tube containing 1.5 ml of sterile PBS and were gently vortexed. Seven hundred fifty microliters of the sample was stored at 4°C as a retained sample. Seven hundred fifty microliters, including the swab, was used for a cultural enrichment in 9 ml of LB medium and a subsequent analysis for ESBL-/pAmpC-producing enterobacteria, as described before. In case of a positive enrichment of ESBL-/pAmpC-producing enterobacteria, the retained sample was used for enumeration of the (resistant) bacteria.
(v) Environmental samples.
Environmental swabs were investigated according to the cloacal swab protocol using 1 ml of PBS for enrichment in LB medium. The pooled crushed eggshells as well as the dust samples were processed as follows: 30 ml of LB medium was added to the Whirl-Pak stand-up bag, and the mixture was rigorously shaken by hand and incubated for 24 h following the enrichment procedure. The same procedure was applied for collected flies and feathers using 10 ml of LB medium. Environmental samples from flock A were not analyzed on MC− agar plates concerning the growth of total enterobacteria.
(vi) Air samples.
The strips of the RCS Plus air sampler were aerobically incubated for 24 h at 37°C after air sampling. Colonies were counted according to their morphology and the respective species identified via MALDI-TOF MS. Up to 10 suspicious ESBL-/pAmpC-producing enterobacteria with two isolates of each colony morphology were isolated from the MC+ agar strip for further susceptibility testing and molecular analyses. Regarding the Coriolis, 3 ml of the PBS was added to 27 ml of LB medium and incubated aerobically at 37°C for 24 h for analyses of the ESBL-/pAmpC-producing enterobacteria as well as for analysis of the total enterobacterial composition.
(vii) Antimicrobial susceptibility testing.
For all suspicious isolates, disk diffusion tests with 30 μg of cefotaxime, 30 μg of cefotaxime plus 10 μg of clavulanic acid, 30 μg of ceftazidime, and 30 μg of cefoxitin (Liofilchem s.r.l., Roseto degli Abruzzi, Italy) were applied. Isolates were grown on MC+ agar plates, and a suspension in 0.85% NaCl solution with a McFarland standard of 0.5 was prepared from this culture. The following procedure was done according to the EUCAST guidelines (41). Disk diameters were analyzed using the EUCAST breakpoint tables (42). All enterobacteria suspicious for resistance concerning the tested beta-lactams were further investigated.
(viii) Real-time PCR and sequencing.
Subsequently to the antimicrobial susceptibility testing, isolates were further analyzed using real-time PCR for the detection of the predominant beta-lactamase genes blaCTX-M, blaSHV, blaTEM, as well as CIT-type pAmpC genes (43). Therefore, DNA was extracted by a simple boiling method, and the supernatant was used for all PCR analyses (43). Positive controls and no-template controls were used as published previously (43). Detected ESBL/pAmpC genes for at least one isolate of each sample were verified by sequencing, each with the primers shown in Table 3. The novel primers SHV(−28)-F, SHV(+1000)-R, CMY2(−80)-F, CMY2(+1242)-R were designed with OligoPerfect designer (Thermo Fisher Scientific, Inc., St. Leon Roth, Germany) using sequences with GenBank accession no. X53433 (SHV) and FR719923 (CIT-type pAmpCs) as templates. For both PCRs, 25 μl of the respective reaction mixture contained 12.5 μl of DreamTaq Green PCR Mastermix (Thermo Fisher Scientific, Inc.), 1 μl of each primer (10 μM), and 7.5 μl of PCR water. The PCR conditions for both genes were as follows: 5 min of initial denaturation at 94°C, 35 cycles of 30 s of denaturation at 94°C, 30 s of primer binding at 57°C, 1 min of elongation at 72°C, and a final elongation step at 72°C for 5 min. Nucleotide sequences were analyzed with BioNumerics version 6.6 (Applied Maths, Sint-Martens-Latem, Belgium) and compared with reference sequences from GenBank to determine the assigned gene number according to www.lahey.org/studies/.
TABLE 3.
Sequencing primers used in the study for determining the respective beta-lactamase genes
| Primer | Sequence (5′ → 3′) | Product size (bp) | Reference |
|---|---|---|---|
| TEM-F | GCGGAACCCCTATTTG | 963 | 27 |
| TEM-R | ACCAATGCTTAATCAGTGAG | 27 | |
| CTX-M1-SEQ_F | CCCATGGTTAAAAAATCACTGC | 944 | 33 |
| CTX-M1-SEQ_R | CAGCGCTTTTGCCGTCTAAG | 33 | |
| SHV(−28)-F | GGCCCTCACTCAAGGATGTA | 1,028 | This study |
| SHV(+1000)-R | CCACGTTTATGGCGTTACCT | This study | |
| CMY2(−80)-F | CAACACGGTGCAAATCAAAC | 1,322 | This study |
| CMY2(+1242)-R | CATGGGATTTTCCTTGCTGT | This study |
(ix) Phylogenetic typing.
Isolates which were determined to be Escherichia coli or Escherichia fergusonii by MALDI-TOF MS were further analyzed for their phylogenetic group, as described previously (44). The multiplex PCR was performed using primers as published previously, and PCR conditions were adjusted as follows: 3 min of initial denaturation at 94°C, 33 cycles of a 30-s denaturation at 94°C, primer binding for 30 s at 57°C, and a 1-min elongation at 72°C, with a final elongation step of 5 min at 72°C. The total reaction mixture of 15 μl contained 0.15 μl of each primer (0.3 μl of TspE4C2.1b and TspE4C2.2b), 7.5 μl of DreamTaq Green PCR Mastermix, and 5 μl of PCR water. Isolates showing band patterns in the agarose gel electrophoresis which were not specific for just one phylogroup were assigned as a combined phylogroup, for example, A/C.
(x) Pulsed-field gel electrophoresis.
Isolates from parent flock B and from the corresponding egg surfaces of the same phylogroup harboring an identical ESBL gene were investigated by pulsed-field gel electrophoresis (PFGE) to further analyze their phylogenetic relationship. At least one isolate of each sample from the parent flock (feces and boot swab) and the egg surfaces at arrival at the hatchery and after disinfection of the eggs was analyzed. Isolates were grown aerobically in LB medium for 24 h at 37°C. From that, a bacterial suspension in PBS (Oxoid, Wesel, Germany) was made with an optical density at 600 nm (OD600) of about 0.33. Plugs were prepared by mixing 250 μl of bacterial suspension and 375 μl of 1.5% LE GP agarose (Biozym Scientific GmbH, Hessisch Oldendorf, Germany), incubated at 56°C for 24 h in cell lysis buffer (500 mM EDTA [pH 9.5], 1% sarcosine, 0.9 mg of proteinase K/ml), and washed six times in Tris-EDTA buffer. Whole-genomic DNA within agarose plug slices was enzymatically restricted for 4 h at 37°C using 15 U of XbaI (Thermo Fisher Scientific, Inc., St. Leon Roth, Germany) for each slice. Running conditions for the gel electrophoresis were used as published by Schaufler et al. (45). Salmonella enterica serovar Braenderup H9812 was used as a size standard, according to Hunter et al. (46).
(xi) Multilocus sequence typing.
From each PFGE cluster, one isolate was analyzed with multilocus sequence typing (MLST) to determine their sequence type (ST). Primers were used as published previously (47). Sequences were analyzed with BioNumerics version 6.6, and the respective STs were assigned according to http://mlst.warwick.ac.uk/mlst/dbs/Ecoli.
ACKNOWLEDGMENTS
We thank all farmers and the hatchery staff for their kind cooperation. Many thanks to Nicole Roschanski for scientific advice and Maja Thieck for excellent technical assistance in the laboratory.
This project was funded by the Federal Ministry of Education and Research (grant 01KI1313C) and is part of the RESET research consortium (http://www.reset-verbund.de/index.htm).
REFERENCES
- 1.Pfeifer Y, Cullik A, Witte W. 2010. Resistance to cephalosporins and carbapenems in Gram-negative bacterial pathogens. Int J Med Microbiol 300:371–379. doi: 10.1016/j.ijmm.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 2.Pitout JDD, Laupland KB. 2008. Extended-spectrum β-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect Dis 8:159–166. doi: 10.1016/S1473-3099(08)70041-0. [DOI] [PubMed] [Google Scholar]
- 3.Valentin L, Sharp H, Hille K, Seibt U, Fischer J, Pfeifer Y, Michael GB, Nickel S, Schmiedel J, Falgenhauer L, Friese A, Bauerfeind R, Roesler U, Imirzalioglu C, Chakraborty T, Helmuth R, Valenza G, Werner G, Schwarz S, Guerra B, Appel B, Kreienbrock L, Kasbohrer A. 2014. Subgrouping of ESBL-producing Escherichia coli from animal and human sources: an approach to quantify the distribution of ESBL types between different reservoirs. Int J Med Microbiol 304:805–816. doi: 10.1016/j.ijmm.2014.07.015. [DOI] [PubMed] [Google Scholar]
- 4.Ewers C, Bethe A, Semmler T, Guenther S, Wieler LH. 2012. Extended-spectrum beta-lactamase-producing and AmpC-producing Escherichia coli from livestock and companion animals, and their putative impact on public health: a global perspective. Clin Microbiol Infect 18:646–655. doi: 10.1111/j.1469-0691.2012.03850.x. [DOI] [PubMed] [Google Scholar]
- 5.Bauernfeind A, Chong Y, Lee K. 1998. Plasmid-encoded AmpC beta-lactamases: how far have we gone 10 years after the discovery? Yonsei Med J 39:520–525. doi: 10.3349/ymj.1998.39.6.520. [DOI] [PubMed] [Google Scholar]
- 6.Rodríguez I, Barownick W, Helmuth R, Mendoza MC, Rodicio MR, Schroeter A, Guerra B. 2009. Extended-spectrum {beta}-lactamases and AmpC {beta}-lactamases in ceftiofur-resistant Salmonella enterica isolates from food and livestock obtained in Germany during 2003–07. J Antimicrob Chemother 64:301–309. doi: 10.1093/jac/dkp195. [DOI] [PubMed] [Google Scholar]
- 7.Dierikx C, van der Goot J, Fabri T, van Essen-Zandbergen A, Smith H, Mevius D. 2013. Extended-spectrum-beta-lactamase- and AmpC-beta-lactamase-producing Escherichia coli in Dutch broilers and broiler farmers. J Antimicrob Chemother 68:60–67. doi: 10.1093/jac/dks349. [DOI] [PubMed] [Google Scholar]
- 8.Briñas L, Moreno MA, Zarazaga M, Porrero C, Saenz Y, Garcia M, Dominguez L, Torres C. 2003. Detection of CMY-2, CTX-M-14, and SHV-12 β-lactamases in Escherichia coli fecal-sample isolates from healthy chickens. Antimicrob Agents Chemother 47:2056–2058. doi: 10.1128/AAC.47.6.2056-2058.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blanc V, Mesa R, Saco M, Lavilla S, Prats G, Miro E, Navarro F, Cortes P, Llagostera M. 2006. ESBL- and plasmidic class C beta-lactamase-producing E. coli strains isolated from poultry, pig and rabbit farms. Vet Microbiol 118:299–304. doi: 10.1016/j.vetmic.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 10.Dierikx CM, van Duijkeren E, Schoormans AHW, van Essen-Zandbergen A, Veldman K, Kant A, Huijsdens XW, van der Zwaluw K, Wagenaar JA, Mevius DJ. 2012. Occurrence and characteristics of extended-spectrum-beta-lactamase- and AmpC-producing clinical isolates derived from companion animals and horses. J Antimicrob Chemother 67:1368–1374. doi: 10.1093/jac/dks049. [DOI] [PubMed] [Google Scholar]
- 11.Kaesbohrer A, Schroeter A, Tenhagen B-A, Alt K, Guerra B, Appel B. 2012. Emerging antimicrobial resistance in commensal Escherichia coli with public health relevance. Zoonoses Public Health 59(Suppl 2):S158–S165. doi: 10.1111/j.1863-2378.2011.01451.x. [DOI] [PubMed] [Google Scholar]
- 12.Schmiedel J, Falgenhauer L, Domann E, Bauerfeind R, Prenger-Berninghoff E, Imirzalioglu C, Chakraborty T. 2014. Multiresistant extended-spectrum beta-lactamase-producing Enterobacteriaceae from humans, companion animals and horses in central Hesse, Germany. BMC Microbiol 14:187. doi: 10.1186/1471-2180-14-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valenza G, Nickel S, Pfeifer Y, Eller C, Krupa E, Lehner-Reindl V, Holler C. 2014. Extended-spectrum-beta-lactamase-producing Escherichia coli as intestinal colonizers in the German community. Antimicrob Agents Chemother 58:1228–1230. doi: 10.1128/AAC.01993-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu G, Day MJ, Mafura MT, Nunez-Garcia J, Fenner JJ, Sharma M, van Essen-Zandbergen A, Rodriguez I, Dierikx C, Kadlec K, Schink A-K, Chattaway M, Wain J, Helmuth R, Guerra B, Schwarz S, Threlfall J, Woodward MJ, Woodford N, Coldham N, Mevius D. 2013. Comparative analysis of ESBL-positive Escherichia coli isolates from animals and humans from the UK, The Netherlands and Germany. PLoS One 8:e75392. doi: 10.1371/journal.pone.0075392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Michael GB, Freitag C, Wendlandt S, Eidam C, Fessler AT, Lopes GV, Kadlec K, Schwarz S. 2015. Emerging issues in antimicrobial resistance of bacteria from food-producing animals. Future Microbiol 10:427–443. doi: 10.2217/fmb.14.93. [DOI] [PubMed] [Google Scholar]
- 16.Börjesson S, Ny S, Egervarn M, Bergstrom J, Rosengren A, Englund S, Lofmark S, Byfors S. 2016. Limited dissemination of extended-spectrum beta-lactamase- and plasmid-encoded AmpC-producing Escherichia coli from food and farm animals, Sweden. Emerg Infect Dis 22:634–640. doi: 10.3201/eid2204.151142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reich F, Atanassova V, Klein G. 2013. Extended-spectrum beta-lactamase- and AmpC-producing enterobacteria in healthy broiler chickens, Germany. Emerg Infect Dis 19:1253–1259. doi: 10.3201/eid1908.120879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dierikx C, van Essen-Zandbergen A, Veldman K, Smith H, Mevius D. 2010. Increased detection of extended spectrum beta-lactamase producing Salmonella enterica and Escherichia coli isolates from poultry. Vet Microbiol 145:273–278. doi: 10.1016/j.vetmic.2010.03.019. [DOI] [PubMed] [Google Scholar]
- 19.Laube H, Friese A, von Salviati C, Guerra B, Käsbohrer A, Kreienbrock L, Roesler U. 2013. Longitudinal monitoring of extended-spectrum-beta-lactamase/AmpC-producing Escherichia coli at German broiler chicken fattening farms. Appl Environ Microbiol 79:4815–4820. doi: 10.1128/AEM.00856-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blaak H, van Hoek AH, Hamidjaja RA, van der Plaats RQ, Kerkhof-de Heer L, de Roda Husman AM, Schets FM. 2015. Distribution, numbers, and diversity of ESBL-producing E. coli in the poultry farm environment. PLoS One 10:e0135402. doi: 10.1371/journal.pone.0135402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwaiger K, Bauer J, Holzel CS. 2013. Selection and persistence of antimicrobial-resistant Escherichia coli including extended-spectrum beta-lactamase producers in different poultry flocks on one chicken farm. Microb Drug Resist 19:498–506. doi: 10.1089/mdr.2012.0257. [DOI] [PubMed] [Google Scholar]
- 22.Laube H, Friese A, von Salviati C, Guerra B, Rösler U. 2014. Transmission of ESBL/AmpC-producing Escherichia coli from broiler chicken farms to surrounding areas. Vet Microbiol 172:519–527. doi: 10.1016/j.vetmic.2014.06.008. [DOI] [PubMed] [Google Scholar]
- 23.Dierikx CM, van der Goot JA, Smith HE, Kant A, Mevius DJ. 2013. Presence of ESBL/AmpC-producing Escherichia coli in the broiler production pyramid: a descriptive study. PLoS One 8:e79005. doi: 10.1371/journal.pone.0079005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giovanardi D, Campagnari E, Ruffoni LS, Pesente P, Ortali G, Furlattini V. 2005. Avian pathogenic Escherichia coli transmission from broiler breeders to their progeny in an integrated poultry production chain. Avian Pathol 34:313–318. doi: 10.1080/03079450500179046. [DOI] [PubMed] [Google Scholar]
- 25.Nilsson O, Borjesson S, Landen A, Bengtsson B. 2014. Vertical transmission of Escherichia coli carrying plasmid-mediated AmpC (pAmpC) through the broiler production pyramid. J Antimicrob Chemother 69:1497–1500. doi: 10.1093/jac/dku030. [DOI] [PubMed] [Google Scholar]
- 26.Petersen A, Christensen JP, Kuhnert P, Bisgaard M, Olsen JE. 2006. Vertical transmission of a fluoroquinolone-resistant Escherichia coli within an integrated broiler operation. Vet Microbiol 116:120–128. doi: 10.1016/j.vetmic.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 27.Zurfluh K, Wang J, Klumpp J, Nüesch-Inderbinen M, Fanning S, Stephan R. 2014. Vertical transmission of highly similar blaCTX-M-1-harbouring IncI1 plasmids in Escherichia coli with different MLST types in the poultry production pyramid. Front Microbiol 5:519. doi: 10.3389/fmicb.2014.00519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smet A, Martel A, Persoons D, Dewulf J, Heyndrickx M, Catry B, Herman L, Haesebrouck F, Butaye P. 2008. Diversity of extended-spectrum beta-lactamases and class C beta-lactamases among cloacal Escherichia coli isolates in Belgian broiler farms. Antimicrob Agents Chemother 52:1238–1243. doi: 10.1128/AAC.01285-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pasquali F, Lucchi A, Braggio S, Giovanardi D, Franchini A, Stonfer M, Manfreda G. 2015. Genetic diversity of Escherichia coli isolates of animal and environmental origins from an integrated poultry production chain. Vet Microbiol 178:230–237. doi: 10.1016/j.vetmic.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 30.Forgetta V, Rempel H, Malouin F, Vaillancourt R, Topp JRE, Dewar K, Diarra MS. 2012. Pathogenic and multidrug-resistant Escherichia fergusonii from broiler chicken. Poult Sci 91:512–525. doi: 10.3382/ps.2011-01738. [DOI] [PubMed] [Google Scholar]
- 31.Hopkins JM, Towner KJ. 1990. Enhanced resistance to cefotaxime and imipenem associated with outer membrane protein alterations in Enterobacter aerogenes. J Antimicrob Chemother 25:49–55. doi: 10.1093/jac/25.1.49. [DOI] [PubMed] [Google Scholar]
- 32.Mezhoud H, Chantziaras I, Iguer-Ouada M, Moula N, Garmyn A, Martel A, Touati A, Smet A, Haesebrouck F, Boyen F. 2016. Presence of antimicrobial resistance in coliform bacteria from hatching broiler eggs with emphasis on ESBL/AmpC-producing bacteria. Avian Pathol 45:1–30. doi: 10.1080/03079457.2016.1167837. [DOI] [PubMed] [Google Scholar]
- 33.Musgrove MT, Jones DR, Northcutt JK, Harrison MA, Cox NA, Ingram KD, Hinton AJ Jr. 2005. Recovery of Salmonella from commercial shell eggs by shell rinse and shell crush methodologies. Poult Sci 84:1955–1958. doi: 10.1093/ps/84.12.1955. [DOI] [PubMed] [Google Scholar]
- 34.Cadirci S. 2009. Disinfection of hatching eggs by formaldehyde fumigation–a review. Arch Geflügelk 73:116–123. [Google Scholar]
- 35.Cox NA, Berrang ME, Cason JA. 2000. Salmonella penetration of egg shells and proliferation in broiler hatching eggs–a review. Poult Sci 79:1571–1574. doi: 10.1093/ps/79.11.1571. [DOI] [PubMed] [Google Scholar]
- 36.Messens W, Grijspeerdt K, Herman L. 2005. Eggshell characteristics and penetration by Salmonella enterica serovar Enteritidis through the production period of a layer flock. Br Poult Sci 46:694–700. doi: 10.1080/00071660500395582. [DOI] [PubMed] [Google Scholar]
- 37.De Reu K, Grijspeerdt K, Messens W, Heyndrickx M, Uyttendaele M, Debevere J, Herman L. 2006. Eggshell factors influencing eggshell penetration and whole egg contamination by different bacteria, including Salmonella enteritidis. Int J Food Microbiol 112:253–260. doi: 10.1016/j.ijfoodmicro.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 38.Wellman-Labadie O, Picman J, Hincke MT. 2008. Antimicrobial activity of cuticle and outer eggshell protein extracts from three species of domestic birds. Br Poult Sci 49:133–143. doi: 10.1080/00071660802001722. [DOI] [PubMed] [Google Scholar]
- 39.Rathgeber BM, McCarron P, Budgell KL. 2013. Salmonella penetration through eggshells of chickens of different genetic backgrounds. Poult Sci 92:2457–2462. doi: 10.3382/ps.2013-03139. [DOI] [PubMed] [Google Scholar]
- 40.Mevius DJK, Koene MGJ, Witt B, van Pelt W, Bondt W. 2009. Monitoring of antimicrobial resistance and antibiotic usage in animals in the Netherlands in 2008. Central Veterinary Institute of Wageningen University and Research Centre, Wageningen, the Netherlands. [Google Scholar]
- 41.Matuschek E, Brown DFJ, Kahlmeter G. 2014. Development of the EUCAST disk diffusion antimicrobial susceptibility testing method and its implementation in routine microbiology laboratories. Clin Microbiol Infect 20:66. doi: 10.1111/1469-0691.12570. [DOI] [PubMed] [Google Scholar]
- 42.European Committee on Antimicrobial Susceptibility Testing. 2016. Breakpoint tables for interpretation of MICs and zone diameters. Version 6.0. European Committee on Antimicrobial Susceptibility Testing, Växjö, Sweden: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_6.0_Breakpoint_table.pdf. [Google Scholar]
- 43.Roschanski N, Fischer J, Guerra B, Roesler U. 2014. Development of a multiplex real-time PCR for the rapid detection of the predominant beta-lactamase genes CTX-M, SHV, TEM and CIT-type AmpCs in Enterobacteriaceae. PLoS One 9:e100956. doi: 10.1371/journal.pone.0100956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clermont O, Christenson JK, Denamur E, Gordon DM. 2013. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ Microbiol Rep 5:58–65. doi: 10.1111/1758-2229.12019. [DOI] [PubMed] [Google Scholar]
- 45.Schaufler K, Semmler T, Wieler LH, Wöhrmann M, Baddam R, Ahmed N, Müller K, Kola A, Fruth A, Ewers C, Guenther S. 2016. Clonal spread and interspecies transmission of clinically relevant ESBL-producing Escherichia coli of ST410–another successful pandemic clone? FEMS Microbiol Ecol 92:fiv155. doi: 10.1093/femsec/fiv155. [DOI] [PubMed] [Google Scholar]
- 46.Hunter SB, Vauterin P, Lambert-Fair MA, van Duyne MS, Kubota K, Graves L, Wrigley D, Barrett T, Ribot E. 2005. Establishment of a universal size standard strain for use with the PulseNet standardized pulsed-field gel electrophoresis protocols: converting the national databases to the new size standard. J Clin Microbiol 43:1045–1050. doi: 10.1128/JCM.43.3.1045-1050.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MCJ, Ochman H, Achtman M. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol 60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]