ABSTRACT
NAH7 and pWW0 from gammaproteobacterial Pseudomonas putida strains are IncP-9 conjugative plasmids that carry the genes for degradation of naphthalene and toluene, respectively. Although such genes on these plasmids are well-characterized, experimental investigation of their conjugation systems remains at a primitive level. To clarify these conjugation systems, in this study, we investigated the NAH7-encoded conjugation system by (i) analyzing the origin of its conjugative transfer (oriT)-containing region and its relaxase, which specifically nicks within the oriT region for initiation of transfer, and (ii) comparing the conjugation systems between NAH7 and pWW0. The NAH7 oriT (oriTN) region was located within a 430-bp fragment, and the strand-specific nicking (nic) site and its upstream sequences that were important for efficient conjugation in the oriTN region were identified. Unlike many other relaxases, the NAH7 relaxase exhibited unique features in its ability to catalyze, in a conjugation-independent manner, the site-specific intramolecular recombination between two copies of the oriTN region, between two copies of the pWW0 oriT (oriTW) region (which is clearly different from the oriTN region), and between the oriTN and oriTW regions. The pWW0 relaxase, which is also clearly different from the NAH7 relaxase, was strongly suggested to have the ability to conjugatively and efficiently mobilize the oriTN-containing plasmid. Such a plasmid was, in the presence of the NAH7Δnic derivative, conjugatively transferable to alphaproteobacterial and betaproteobacterial strains in which the NAH7 replication machinery is nonfunctional, indicating that the NAH7 conjugation system has a broader host range than its replication system.
IMPORTANCE Various studies have strongly suggested an important contribution of conjugative transfer of catabolic plasmids to the rapid and wide dissemination of the plasmid-loaded degradation genes to microbial populations. Degradation genes on such plasmids are often loaded on transposons, which can be inserted into the genomes of the recipient bacterial strains where the transferred plasmids cannot replicate. The aim was to advance detailed molecular knowledge of the determinants of host range for plasmids. This aim is expected to be easily and comprehensively achieved using an experimental strategy in which the oriT region is connected with a plasmid that has a broad host range of replication. Using such a strategy in this study, we showed that (i) the NAH7 oriT-relaxase system has unique properties that are significantly different from other well-studied systems and (ii) the host range of the NAH7 conjugation system is broader than previously thought.
KEYWORDS: Pseudomonas, conjugation, oriT, plasmid, relaxase
INTRODUCTION
Conjugative transfer of plasmids that carry various genetic traits contributes greatly to the rapid adaptation and evolution of host bacteria (1). The conjugative transfer of plasmids in Gram-negative bacteria consists of DNA transfer and replication (Dtr) and mating pair formation (Mpf) systems that are connected by the function of a coupling protein (CP) (2). The Dtr-specified relaxase catalyzes site-specific and strand-specific cleavage at the nic site within the origin of transfer (oriT) region and covalently binds to the 5′ end of the cleaved single-stranded DNA (ssDNA). Rolling-circle-type replication of the plasmid is initiated from the 3′ end of the cleaved ssDNA, and the relaxase-ssDNA complex that is recruited to the Mpf system by CP is transferred into the recipient cell. The transferred relaxase next cleaves and ligates the two copies of the nic site in the transferred ssDNA molecule, and its complementary strand is synthesized (3). All of the self-transferable and mobilizable plasmids carry their oriT regions as the essential cis-acting sequences. Each of these regions usually possesses, in addition to the nic site, direct and/or inverted repeats, where relaxase and (an)other auxiliary protein(s) bind, and there is usually strict constraint between the oriT region and its cognate conjugative transfer machinery for efficient conjugation (4, 5). In addition to their essential role in conjugative transfer, the relaxases encoded by several plasmids have also been shown to be capable of catalyzing site-specific recombination reactions between two identical copies of the oriT region (i.e., intramolecular resolution between two directly repeated copies of the oriT region and/or intermolecular integration at the two copies of the oriT region, each on a different molecule) (6–9). Although the oriT-relaxase systems have been experimentally identified and well characterized in several plasmids, such systems in many other plasmids have only been predicted by their in silico analysis. This is also the case with plasmids that encode enzymes for the degradation of various recalcitrant chemical compounds.
Complete microbial degradation of naphthalene, a representative recalcitrant aromatic compound, is mediated by a variety of bacterial strains belonging to diverse taxa, and the most extensively analyzed example is degradation by the gammaproteobacterial strain Pseudomonas putida G7 (10). The degradation pathway in this strain is encoded by an 82-kb self-transferable plasmid, NAH7, which belongs to an IncP-9 incompatibility group. The biochemical and genetic properties of its naphthalene degradation have been well clarified (11), and our analysis has also revealed fundamental and unique properties of NAH7 with respect to its replication and conjugative transfer (12, 13). This plasmid is transferable by conjugation between Pseudomonas and Escherichia coli strains and can replicate in both genera (14). The complete sequence of NAH7 revealed that (i) its Mpf system is encoded by the mpf operon, (ii) the traC and traB genes in the traABC operon are postulated to encode relaxase and CP, respectively (Fig. 1a) (15), and (iii) the mpfC and traC products are classified as the MPFT and MOBF families, respectively (3). The third traDEF operon on NAH7 is located 430 bp downstream of the traABC operon so that the two operons are oriented in a head-to-head configuration (Fig. 1a). Although none of the individual gene products in the traDEF operon are essential for conjugative transfer (12), the traF product is considered to be a host-range modifier since the NAH7 traF mutant was self-transferable from P. putida to P. putida and E. coli and from E. coli to E. coli but not from E. coli to P. putida (13). We further showed that a P. putida derivative with a defective chromosomal gene (i.e., ptsO for the nitrogen-related phosphotransferase [PTSNTR] system) can conjugatively receive the NAH7 traF mutant from E. coli. However, the detailed mechanism(s) governing the phenomenon of the host-range specificity of NAH7 remains unclear. To obtain additional information on the conjugation machinery of NAH7 to better understand its host-range specificity, we attempted in this study to (i) identify and characterize the oriT region and relaxase of NAH7 and (ii) show the host range of its conjugation system using various proteobacterial strains as the recipients.
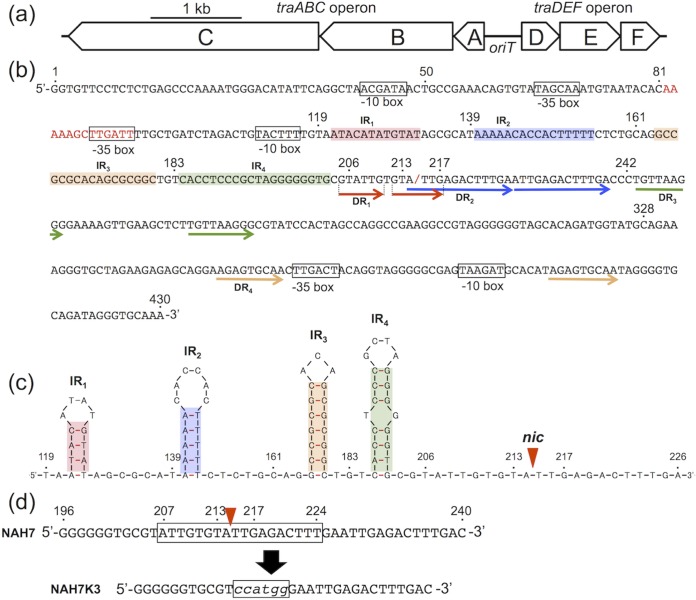
FIG 1.
NAH7 oriT region. (a) oriT and its flanking regions. The 430-bp oriT (oriTN) region is located between the traA and traD genes, and the mpf operon is located downstream of the traABC operon. (b) The 430-bp oriTN sequence. Its first and last nucleotides are those located just 1 base upstream of the start codons of the traA and traD genes, respectively, and relevant nucleotide positions (see Fig. 4) are indicated above the sequence. A putative IHF-binding sequence is indicated in red letters, and the nic site is indicated by a red slash. Boxes show the predicted promoters for the traABC and traDEF operons. Direct repeats are indicated by arrows below the sequence, and the nucleotides able to form the inverted repeats (IRs) with hairpin loop structures are shadowed in different colors. (c) Predicted secondary structure of the internal oriTN region between base positions 119 and 226. The nic site is indicated by a red triangle. (d) Introduction of mutation into the nic-containing sequence in the oriTN region. The boxed sequence was replaced by the KpnI recognition sequence (see Materials and Methods for details).
RESULTS
Identification of the oriT region on NAH7.
The toluene-catabolic plasmid pWW0 is another well-studied IncP-9 self-transferable plasmid (16). Its conjugative transfer-related genes belong to the MOBF-MPFT family (3) and exhibit the same organization as NAH7, as well as high (approximately 74%) amino acid sequence similarity with NAH7 (13). Lambertsen et al. (17) described briefly that a 471-bp fragment containing the traA-traD intergenic region on pWW0 has its oriT function. On the basis of such information, we predicted that the oriT region on NAH7 is located within a 430-bp fragment between its traA and traD genes (i.e., bp positions from 19758 to 20187 on the NAH7 map [Fig. 1a]) (15). This fragment was cloned into a nonmobilizable and broad-host-range vector plasmid, pNIT6012, to obtain pNIT101. pNIT101 had no transferability by conjugation from P. putida KT2440 to P. putida KT2440G (Table 1). The tetracycline resistance (Tcr) marker on pNIT101, but not that on pNIT6012, was mobilized by conjugation from P. putida G7(NAH7K2) to P. putida KT2440G at a frequency of 8.0 × 10−5 per donor cell (Table 1). Twenty-three percent of the Tcr transconjugants were sensitive to kanamycin (Km), and subsequent agarose gel electrophoresis analysis of the cleared lysates from such transconjugants revealed the conjugative mobilization of pNIT101. These results indicated that the 430-bp fragment, i.e., the NAH7 oriT (oriTN) region, indeed has the oriT activity.
TABLE 1.
Conjugative transfer and mobilization of plasmidsa
| Host | Plasmid(s) | Transfer frequency |
Mobilization frequency |
||
|---|---|---|---|---|---|
| Kmr transconjugants | Tcs clones | Tcr transconjugants | Kms clones | ||
| G7 | NAH7K2 + pNIT6012 | 1.2 × 10−3 | 48/48 | <1.2 × 10−8 | N.T.b |
| G7 | NAH7K2 + pNIT101 | 6.0 × 10−5 | 37/48 | 8.0 × 10−5 | 11/48 |
| G7 | NAH7K2 + pNIT201 | 1.4 × 10−3 | 40/48 | 1.3 × 10−3 | 8/48 |
| G7 | NAH7K3 + pNIT6012 | <1.1 × 10−8 | N.T. | <1.1 × 10−8 | N.T. |
| G7 | NAH7K3 + pNIT101 | <5.1 × 10−8 | N.T. | 5.3 × 10−4 | 48/48 |
| G7 | NAH7K4 + pNIT6012 | <4.4 × 10−8 | N.T. | <4.4 × 10−8 | N.T. |
| G7 | NAH7K4 + pNIT101 | <4.2 × 10−8 | N.T. | <4.2 × 10−8 | N.T. |
| G7dCLC | NAH7K4 + pNIT6012 | 7.6 × 10−3 | N.T. | <8.0 × 10−8 | N.T. |
| G7dCLC | NAH7K4 + pNIT101 | 3.7 × 10−4 | 40/48 | 4.8 × 10−4 | 13/48 |
| KT2440 | pNIT6012 | <1.4 × 10−8 | N.T. | <1.4 × 10−8 | N.T. |
| KT2440 | pNIT101 | <4.8 × 10−8 | N.T. | <4.8 × 10−8 | N.T. |
| KT2440 | pMT1405 + pNIT6012 | 1.1 × 10−3 | 48/48 | 2.8 × 10−5c | 12/48c |
| KT2440 | pMT1405 + pNIT101 | 1.2 × 10−3 | 3/48 | 5.3 × 10−2 | 14/48 |
| KT2440 | pMT1405 + pNIT201 | 4.1 × 10−4 | 13/48 | 3.9 × 10−2 | 20/48 |
All the recipients used were KT2440G. The NAH7 derivatives and pMT1405 carry the Kmr gene, and the pNIT series of plasmids the Tcr gene. The transfer and mobilization frequencies, which are the mean values from at least three independent experiments, are expressed by dividing the numbers of Kmr and Tcr transconjugants, respectively, by the number of donor cells. The number of Tcs clones among 48 Kmr transconjugants and that of the Kms clones among the 48 Tcr transconjugants are shown in the fourth and sixth columns, respectively.
N.T., not tested.
Agarose gel electrophoresis analysis of the cleared lysates prepared from the Tcr transconjugants revealed no detection of the plasmids less than 20 kb in size, indicating no conjugative mobilization of the intact 8.2-kb form of pNIT6012 itself. The mechanism(s) for the formation of the Tcr transconjugants is unknown.
Requirement of NAH7 traC product for conjugative transfer and site-specific recombination.
R388 is a very well characterized and broad-host-range IncW plasmid (2), and its TrwC protein is a representative of the MOBF family relaxases, which contain N-terminal relaxase and C-terminal helicase domains (see Fig. S2 in the supplemental material). The R388 relaxase has been reported to be essential for conjugative transfer and to catalyze the site-specific recombination between the directly repeated copies of its cognate oriT region on the same molecule, and the crossover site coincides with the nic site (6). On the basis of their phylogenetic relationship, the NAH7 and pWW0 TraC proteins have been classified as members of the MOBF family relaxases (3). The NAH7 TraC protein exhibits 81% amino acid sequence identity with that of pWW0 and only 48% identity with the R388 TrwC protein (Fig. S2). To date, however, there have been no experimental results that support the relaxase activities of the NAH7 and pWW0 TraC proteins.
To clarify whether the NAH7 traC gene is indeed essential for its conjugative transfer and mobilization, this gene on NAH7 was deleted to construct NAH7K4. No transfer of the Kmr marker on NAH7K4 was observed by conjugation from the P. putida G7 background to P. putida KT2440G (Table 1). NAH7K4 regained its conjugative transferability when the donor chromosome had an insert of the traC gene [in strain G7dCLC(NAH7K4)]. pNIT101 was nonmobilizable from G7(NAH7K4) but mobilizable from G7dCLC (Table 1). These results indicated the essential role of the traC gene in conjugative transfer and mobilization. When the R388 trwC gene was introduced into G7(NAH7K4), NAH7K4 did not exhibit conjugative transferability (data not shown).
To investigate whether the NAH7-encoded conjugative transfer-related protein(s) also has site-specific recombinase activity, we constructed a pNIT6012 derivative, pNIT301, in which the sacB and gentamicin resistance (Gmr) genes are flanked by directly oriented copies of the oriTN region. pNIT301 was introduced into an E. coli recA strain, EC100, and its derivative harboring NAH7K2. The resulting strains were cultivated overnight in Tc-containing and Tc- and Km-containing one-third LB broth, respectively, and these two cultures were plated on one-third LB agar supplemented with Tc and sucrose. EC100(NAH7K2)(pNIT301) generated Tc- and sucrose-resistant colonies at a 60-fold higher frequency than EC100(pNIT301) (Table 2). Most of the sucrose-resistant derivatives from the first strain conferred the Gms phenotype on the host cells, and analysis of the pNIT6012-based plasmids in these Gms derivatives indicated the intramolecular site-specific recombination between the two copies of the oriTN region on pNIT301 by the plausible involvement of an NAH7-loaded conjugative transfer-related gene(s). To examine whether the NAH7 traC gene product was involved in site-specific recombination, we constructed pUC18traC so that the NAH7-derived traC transcription was under the control of the lac promoter and was inducible by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG). pUC18traC or pUC18 was introduced into EC100(pNIT301), and the resulting strain was, after cultivation overnight in Tc-, ampicillin (Ap)-, and IPTG-containing one-third LB broth, plated on one-third LB agar supplemented with Tc and sucrose. EC100(pNIT301)(pUC18traC) generated the Tc- and sucrose-resistant colonies at a 70-fold higher frequency than EC100(pNIT301)(pUC18) (Table 2). Our subsequent analysis of the plasmids from the resulting colonies with the Gms phenotype also revealed the expected recombination event, showing the crucial role of the NAH7 traC product in site-specific recombination.
TABLE 2.
Site-specific recombination between two copies of the oriT regions from NAH7 and pWW0a
| Plasmid | Recombination frequency (Gms/Tcr and sucrose resistant)b |
||||
|---|---|---|---|---|---|
| None | NAH7K2 | pMT1405 | pUC18 | pUC18traC | |
| pNIT301 | 9.2 × 10−6 (48/48) | 5.9 × 10−4 (46/48) | 3.7 × 10−5 (48/48) | 4.7 × 10−6 (48/48) | 3.3 × 10−4 (48/48) |
| pNIT302 | 3.3 × 10−6 (48/48) | 3.1 × 10−4 (48/48) | 3.9 × 10−4 (48/48) | 4.6 × 10−8 (48/48) | 2.2 × 10−6 (47/48) |
| pNIT303 | 5.1 × 10−6 (48/48) | 1.2 × 10−3 (48/48) | 4.5 × 10−3 (48/48) | 7.3 × 10−6 (48/48) | 1.3 × 10−3 (48/48) |
| pNIT304 | 5.4 × 10−5 (48/48) | 1.4 × 10−3 (48/48) | 6.8 × 10−3 (48/48) | 8.7 × 10−5 (48/48) | 4.4 × 10−3 (48/48) |
The E. coli EC100 derivative that carried a traC-containing plasmid and one of the pNIT series of plasmids with two copies of oriT regions was cultivated in Tc- and Km-containing or Tc- and Ap-containing one-third LB and plated on one-third LB agar plates supplemented with only Tc and Tc plus 10% sucrose.
The recombination frequency was calculated by dividing the number of Tc- and sucrose-resistant colonies by that of Tcr colonies. The mean value obtained from at least three independent experiments is shown. The number of Gms clones per 48 Tc- and sucrose-resistant colonies is indicated in parentheses.
Identification of the nic site on NAH7.
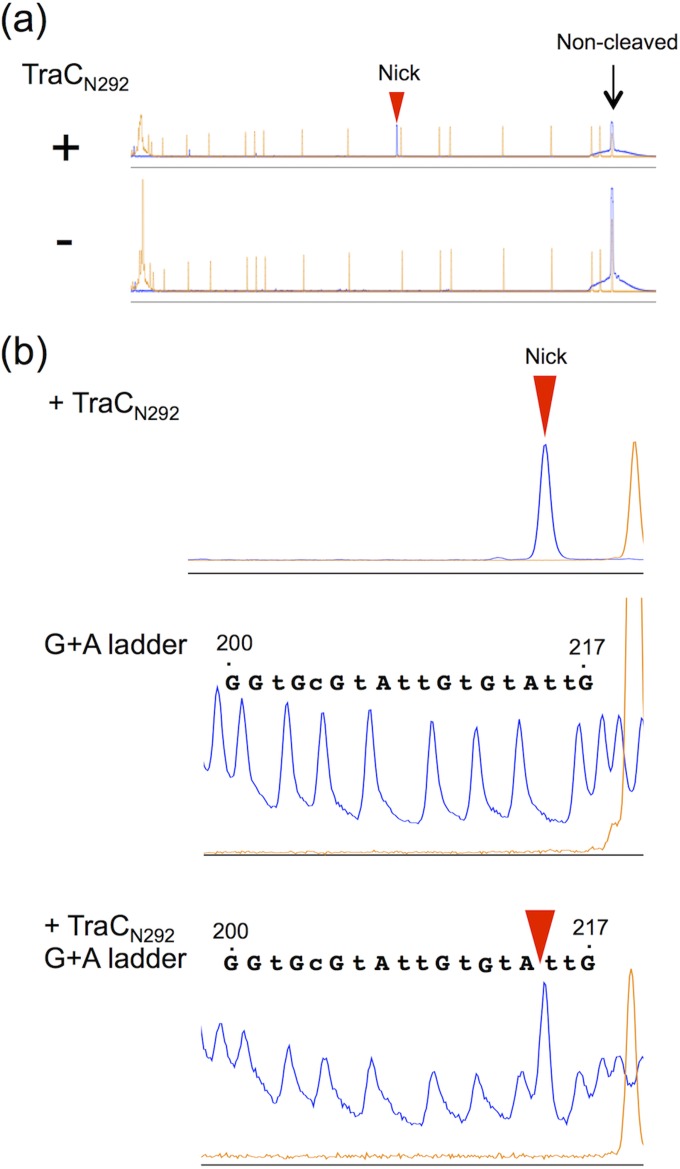
Since the NAH7 TraC protein was considered to function as a relaxase, in vitro experiments were performed to determine the NAH7 nic site. Our repeated attempts to purify the full-length and His-tagged form of the TraC protein were unsuccessful. Therefore, the His-tagged TraC derivative containing the N-terminal relaxase domain with a length of 292 amino acid residues (TraCN292) was purified (see Fig. S1 in the supplemental material) and used for its in vitro activity to specifically nick the oriT-containing double-stranded DNA (dsDNA) fragment, in which the 5′ end of only one strand (see Fig. 1b for the sequence of the top strand) was fluorescently labeled with 6-carboxyfluorescein (FAM). Incubation of TraCN292 with the top-strand-labeled dsDNA fragment and subsequent analysis using a Sanger sequencer led to the detection of a cleaved and FAM-labeled ssDNA fragment that was smaller than the noncleaved fragment (Fig. 2a). Use of the G+A sequencing ladder (see Materials and Methods for details) showed that the 3′ end of the cleaved ssDNA fragment corresponds to base position 214 in the oriTN region (Fig. 1b and 2b). Such a cleaved and FAM-labeled ssDNA fragment was not detected in the absence of TraCN292, and incubation of TraCN292 with the bottom-strand-labeled dsDNA fragment gave rise to no clear signal for the cleaved ssDNA fragment (data not shown). These results showed that the NAH7 nic site is located at a position between 214 and 215 (Fig. 1b), which is 11 bases downstream from an inverted repeat (IR4) (Fig. 1b and c). This experimentally determined nic site was 1 base downstream from the predicted nic site by comparative sequence analysis (Fig. 3a).
FIG 2.
Identification of NAH7 nic site by in vitro assay. The oriT-containing dsDNA fragment whose 5′ end of the top strand was labeled with FAM was incubated with or without the TraCN292 protein under the in vitro nicking assay conditions (see the Materials and Methods). The resulting DNA products were mixed with the GeneScan 500 LIZ size standard and then electrophoresed using a capillary sequencer (ABI 3130xl) with or without the G+A sequencing ladder. (a) Fluorescence intensity patterns of DNA fragments after treatment with and without TraCN292. x axis, fragment size (left to right, smaller to larger); y axis, arbitrary fluorescence intensity. The peak that was specifically generated by the TraCN292 treatment is indicated by a red triangle. Orange peaks, GeneScan 500 LIZ size standard. (b) Enlarged fluorescence patterns of three samples: the TraCN292-treated sample, the G+A sequencing ladder sample, and a mixture of both samples. Since piperidine catalyzes strand breakage at the 5′ end of the formic acid-modified purine residue of the DNA fragment, the nucleotide identified by the G+A sequencing ladder analysis is depicted at the position 1 base downstream of each fluorescence signal peak.
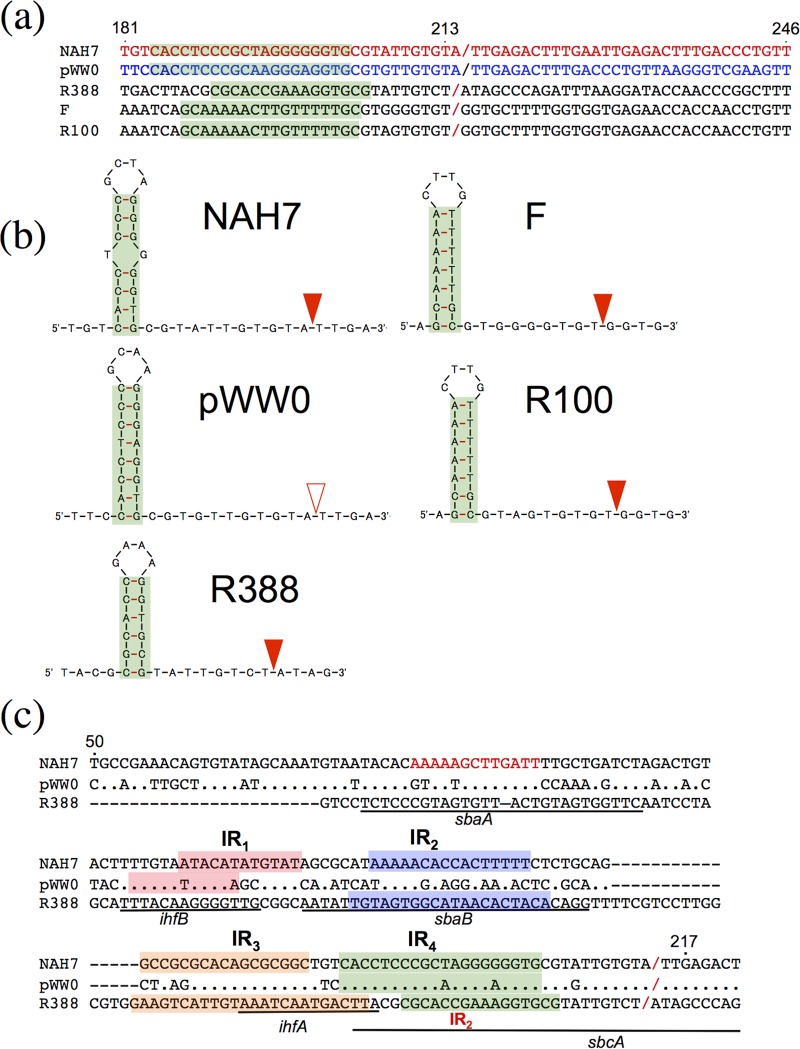
FIG 3.
Comparison of nic-containing sequences. (a) Similarity of the nic-containing sequences among five plasmids whose relaxases belong to the MOBF family. The experimentally determined and predicted nic sites are shown by red and blue slashes, respectively (22). (b) Predicted secondary structures that are located just upstream of the nic sites of the five plasmids. The experimentally determined and putative nic sites are shown by filled and open triangles, respectively. (c) Comparison of the nic- and IR-containing oriT sequences among NAH7, pWW0, and R388. Numerals above the NAH7 sequence are those depicted in Fig. 1b. Dot, the nucleotide is identical to that of NAH7; hyphen, no nucleotide. The first nucleotide in the R388 sequence is located 1 base upstream of its trwA gene. The shadowed sequences are IRs. The red letters of the NAH sequence are a putative IHF-binding site, and the underlined portions of the R388 sequence are as follows: ihfA and ihfB, IHF-binding sites; sbaA and sbaB, TrwA-binding sites; and sbcA, TrwC-binding site (41).
Mobilization ability of an NAH7 derivative lacking its nic site.
To investigate whether (i) the nic site of NAH7 is indeed essential in cis for its conjugative transfer and (ii) NAH7 has an additional nic site at one or more regions on its replicon, we constructed an NAH7 derivative, NAH7K3, in which the nic-containing 18-bp sequence was replaced by a KpnI recognition sequence (Fig. 1d). P. putida G7(NAH7K3) did not exhibit the conjugative transferability of NAH7K3 to P. putida KT2440G (Table 1). However, NAH7K3 still had the ability to mobilize pNIT101 to the recipient strain at a frequency of 5.3 × 10−4. These results showed that the nic site is located only within the 18-bp sequence and that NAH7K3 can still express all of the trans-acting products that are required for the conjugative transfer.
Importance of the sequence upstream of the NAH7 nic site for mobilization.
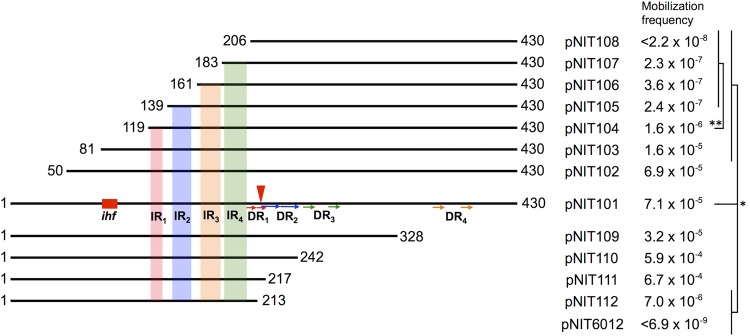
The oriT regions in many conjugative plasmids usually contain multiple copies of inverted repeats (IRs) and direct repeats (DRs), and such repeats have been indicated to be important for efficient conjugative transfer/mobilization (5). There are also several repeats in the oriTN region, including IR1 to IR4 downstream and DR1 to DR4 upstream of the nic site (Fig. 1b and c). To determine which part(s) in the oriTN region is important for mobilization, various fragments in the oriTN region starting from either of the traA- or traD-proximal ends were cloned into pNIT6012, and the resulting plasmids, pNIT102 to pNIT112, were examined for their conjugative mobilization in the presence of NAH7K3 (Fig. 4). The deletion of the leftmost 49-bp sequence (pNIT102) exhibited no change in mobilization frequency. Additional left-hand deletions up to bp 183 led to stepwise (5-fold to 320-fold) reductions in the mobilization frequencies, which were statistically ordered as follows: pNIT101 ≈ pNIT102 > pNIT103 ≈ pNIT104 > pNIT105 to pNIT107. The deletion of the IR1-containing 20-bp sequence from pNIT104 led to a 10-fold reduction in mobilization frequency, and the deletion of the IR4-containing 23-bp sequence from pNIT107 resulted in no mobilization of the resulting plasmid, pNIT108. Conversely, the right-hand deletions up to bp 218 (pNIT111) yielded no drastic reductions in the mobilization frequencies. A 4-bp sequence at the oriTN-derived 3′ end on pNIT111 is missing in pNIT112. Although pNIT112 lacks the nic site (Fig. 1b), it was mobilized at a frequency 10-fold lower than pNIT101. The results using the deletion derivatives of the oriTN region suggested the importance of the nic upstream fragment in the oriTN region, especially the IR4-containing sequence, for efficient mobilization.
FIG 4.
Deletion derivatives of the oriTN region and their mobilization. The numerals at both ends of each fragment are the nucleotide positions in the 430-bp oriTN region. Four predicted IRs with hairpin loop structures (see Fig. 1c) are indicated by different colored boxes, DRs are indicated by arrows, and the putative IHF-binding site is depicted as a red box. The frequencies of mobilization of pNIT101 to pNIT114 from G7(NAH7K3) to KT2400Gm are expressed by the numbers of the Tcr transconjugants per donor cell. Each frequency is the mean value obtained from at least three independent experiments. Statistical analysis was performed using the t test: statistical significance (P < 0.05) in comparison with pNIT101 (*) and with pNIT104 (**).
Functional interchangeability of conjugative mobilization systems between NAH7 and pWW0.
The 431-bp pWW0 oriT (oriTW) region, which is located between its traA and traD genes, exhibits 63% nucleotide identity with the oriTN region (Fig. 5b), and NAH7 and pWW0 carry similar conjugation-related genes (see above). To clarify whether the nonconjugative plasmid carrying the oriTW region is mobilized by the NAH7 conjugation machinery, the oriTW region was cloned into pNIT6012 to obtain pNIT201. The Tcr marker on pNIT201 was successfully mobilized from P. putida G7(NAH7K2) to KT2440G at a frequency of 1.3 × 10−3 (Table 1). Subsequent plasmid profile analysis of the Tcr transconjugants showing the Kms phenotype confirmed the conjugative mobilization of pNIT201, revealing the ability of the NAH7 conjugation machinery to mobilize the oriTW-loaded plasmid into the recipient strain. To investigate whether the pWW0 conjugation machinery can mobilize the oriTN-containing plasmid, pMT1405, a pWW0 derivative with an insertion of the Kmr gene (18), was introduced into KT2440(pNIT101) and KT2440(pNIT6012). pMT1405 mobilized the pNIT101-encoded Tcr marker at a high frequency (Table 1), and analysis of such Tcr transconjugants that did not receive the Kmr marker indicated the mobilization of pNIT101. (Note that the low-frequency mobilization of the pNIT6012-loaded Tcr marker by pMT1045 was not due to the mobilization of the intact form of pNIT6012 itself [Table 1, footnote c].) These reciprocal conjugation experiments showed the functional interchangeability of the oriT regions and the plasmid-encoded conjugation-related proteins between NAH7 and pWW0.
FIG 5.
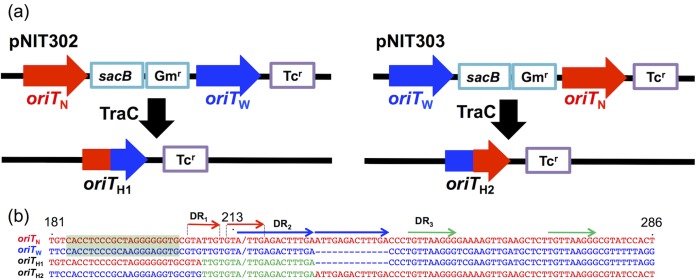
Site-specific recombination between oriTN and oriTW regions to form the hybrid oriT region. (a) Schematic structures of the sacB-Gmr gene cassette-flanking regions on pNIT302 and pNIT303. Their intramolecular recombination between the oriTN and oriTW regions in the presence of NAH7K2, pMT1405, or pUCtraC generated the hybrid oriT regions, oriTH1 and oriTH2, respectively. sacB, levansucrase gene as a couterselective suicidal marker; Gmr and Tcr, gentamicin and tetracycline resistance genes, respectively. (b) The sequences of the internal parts of oriTN, oriTW, oriTH1, and oriTH2 regions. The numerals above the oriTN sequence are their nucleotide positions that are depicted in Fig. 1b. The oriTN- and oriTW-derived sequences are shown in red and blue, respectively, and the sequence containing the crossover site in the oriTH1 and oriTH2 regions is shown in green. The nic site in the oriTN region is indicated by a red slash, and the predicted nic sites in the three other oriT regions are indicated by a blue or green slash.
We next examined the ability of the NAH7- and pWW0-encoded gene product(s) to catalyze the site-specific recombination between two copies of the oriTW region and between the oriTN and oriTW regions. For this purpose, we used pNIT304 and the two pNIT301 derivatives, pNIT302 and pNIT303, in each of which one of the two oriTN regions was replaced by an oriTW region (Fig. 5a). The Tc- and sucrose-resistant colonies were obtained more frequently from the EC100(pNIT304), EC100(pNIT302), and EC100(pNIT303) derivatives carrying NAH7K2 or pMT1405 than from EC100(pNIT304), EC100(pNIT302), and EC100(pNIT303), respectively (Table 2). Most of the Tcr plasmid derivatives from the NAH7K2- or pMT1405-carrying colonies lost the Gmr marker and were suggested to be generated by the deletion between the two oriT regions. Further sequence analysis of such independently obtained plasmids showed that the deletion plasmids from pNIT302 and pNIT303 have identical 19-bp sequences (5′-TTGTGTA/TTGAGACTTTGA-3′; the slash indicates the NAH7 nic site) that are shared by the oriTN and oriTW regions (Fig. 5), indicating the presence of a recombination site within the common 19-bp sequence. The same results were obtained when similar experiments were performed by using pUC18traC instead of NAH7K2 or pMT1405 (Table 2), showing the involvement of the NAH7 relaxase in the recombination.
Host range of the NAH7 conjugation system.
Our previous study (12) showed that NAH7 has conjugative transfer and replication abilities in the gammaproteobacterial (e.g., Pseudomonas and E. coli) strains but lacks one or both of these abilities in the alphaproteobacterial (e.g., Sphingobium) and betaproteobacterial (e.g., Burkholderia) strains. To investigate whether NAH7 exhibits conjugative transferability to strains belonging to these three classes, mobilization of pNIT101 and its parental plasmid, pNIT6012, to 11 (three Pseudomonas strains, three Burkholderia strains, one Ralstonia strain, three Sphingobium strains, and one Sinorhizobium strain) strains in the presence of NAH7K3 was investigated (Table 3). While pNIT101 and pNIT6012 can be introduced into all of these strains by transformation, only the former plasmid was mobilizable from G7(NAH7K3) to all of the recipient strains except for Sphingobium sp. TKS::Gm. These results show that the NAH7 conjugation system has a broader host range than its replication system.
TABLE 3.
Mobilization of oriTN-containing plasmid to various bacterial strainsa
| Recipient strain | Mobilization frequencyb |
|
|---|---|---|
| pNIT6012 | pNIT101 | |
| P. putida KT2440::Gm | <1.1 × 10−8 | 3.6 × 10−3 |
| P. fluorescens Pf-5G | <4.0 × 10−8 | 4.8 × 10−4 |
| P. aeruginosa KGG | <3.5 × 10−8 | 1.4 × 10−4 |
| B. multivorans ATCC 17616 | <3.5 × 10−8 | 5.8 × 10−4 |
| B. vietnamiensis G4::Gm | <3.6 × 10−8 | 1.5 × 10−4 |
| B. cenocepacia J2315::Gm | <3.6 × 10−8 | 7.6 × 10−5 |
| R. solanacearum RS1085::Gm | <3.8 × 10−8 | 3.8 × 10−6 |
| Sphingobium japonicum UT26::Gm | <2.9 × 10−8 | 1.0 × 10−6 |
| Sphingobium sp. MI1205::Gm | <3.3 × 10−8 | 2.4 × 10−5 |
| Sphingobium sp. TKS::Gm | <4.3 × 10−9 | <4.4 × 10−9 |
| Sinorhizobium meliloti 1021 | <5.6 × 10−8 | 1.4 × 10−3 |
Mobilization of pNIT6012 and pNIT101 from P. putida G7(NAH7K3) to the recipient strain was investigated by selecting the Tcr Gmr or Tcr Smr transconjugants.
Mobilization frequency is expressed by the number of transconjugants per donor cell, and the mean value was obtained from at least three independent experiments.
DISCUSSION
In this study, the functional oriT region of NAH7 was localized within a 430-bp sequence between traA and traD, and an in vitro nicking assay by use of TraCN292 led to the identification of the nic site at a position between 214 and 215 (Fig. 1b). The nic-flanking oriT regions in many conjugatively transferable and mobilizable plasmids usually carry several copies of IRs, and each nic site in these oriT regions has been shown to be located 8 to 10 bases downstream of one IR (Fig. 3b) (19). One such IR in the R388 oriT region (IR2, which is the most proximal to the nic site) is a binding site of TrwC and essential for DNA cleavage at the nic site (20). The oriTN region carries four IRs in the fragment upstream of the nic site so that IR4 (the most proximal IR to the nic site) is separated from it by 11 bases (Fig. 1 and 3a). The importance of the IR4- and IR1-containing sequences for efficient mobilization was revealed by our analysis of progressive deletants in the fragment upstream of the nic site (Fig. 4). The absence of mobilization of pNIT108, a deletant that lacks the IR4-containing upstream sequence but possesses the nic site, may be due to the inability of NAH7 relaxase to bind IR4 and/or cleave DNA at the nic site, assuming that the interaction between TrwC and IR2 of R388 is applicable to that between TraC and IR4 of NAH7. Removal of the IR1-containing sequence from pNIT104 (to generate pNIT105) resulted in a severalfold decrease in mobilization frequency. The presence of the IR1-containing sequence may contribute to the enhancement of the mobilization frequency by its binding to the unidentified accessory protein(s) for nicking. TrwA is such an accessory protein in the R388 conjugation system and can bind to the sequences upstream of the nic site and enhance the TrwC-mediated nicking activity (Fig. 3c) (21). trwA on R388 is the first gene in the trwABC operon, and it is reasonable to consider that the NAH7 TraA protein plays a role similar to that of TrwA, although TraA and TrwA differ significantly in their amino acid sequences and the oriTN region lacks sequences similar to the TrwA-binding sequences (sbaA and sbaB) in the R388 oriT (oriTR388) region. Further analysis will clarify the role of TraA in the conjugation of NAH7. A putative integration host factor (IHF)-binding site that is located upstream of IR1 in the oriTN region appeared to have a positive effect on mobilization (Fig. 4). The role of this site is unknown at present, but it is probably different from the two oriTR388-carrying IHF-binding sites since the latter two sites are much closer to the nic site in the oriTR388 region (Fig. 3c). In contrast to the fragment upstream of the nic site in the oriTN region, its DR-containing downstream fragment was not crucial for mobilization efficiency. pNIT112 lacks the intact nic site (between positions 214 and 215 in Fig. 1b) so that the wild-type sequence of ATTGAG (positions 214 to 219) is replaced by GCTAGC (NheI site), which is followed by the pNIT6012 sequence (Fig. 4). However, pNIT112 was mobilized, albeit at a frequency 10-fold lower than pNIT101 (Fig. 4). Detailed mutational analysis of the R388 nic-containing sequence has shown that the replacement of nic-covering dinucleotides by other dinucleotides does not result in the functional loss of the conjugative mobilization of the resulting oriTR388 derivatives (22, 23). The situation with R388 may also be applicable in the case of NAH7; the mutated nic site on pNIT112 may be cleaved and 5′ ligated with the NAH7 relaxase with an efficiency lower than that of the wild-type nic site, thus leading to the low-frequency mobilization of pNIT112.
Each of the relaxases involved in the Dtr system exhibits conjugation-dependent and intramolecular site-specific recombination between the two copies of the cognate nic site on the ssDNA molecule that is transferred to the recipient cell (24). Some but not all relaxases have been reported to additionally exhibit the intramolecular recombination activity in a conjugation-independent manner, and the R388 relaxase is the most representative example (25). This activity was also associated with the NAH7 relaxase (Fig. 5). This property of the NAH7 relaxase allowed us in this study to show that the NAH7 relaxase and most probably the pWW0 relaxase can catalyze intramolecular site-specific recombination between two identical copies of the oriTN or oriTW region and between the oriTN and oriTW regions. The crossover site between the heterogeneous oriT regions is located within the 19-base sequence that is shared by the two regions and contains the nic site in the case of the oriTN region. Based on these results, it is very likely that the nic site on the oriTW region is situated at a position that is identical to that on the oriTN region (Fig. 3). Moreover, pWW0 was able to mediate the efficient mobilization of pNIT101 (Table 1), showing the effectiveness of the pWW0 conjugation system for the mobilization of the oriTN-containing plasmid. The conjugation machinery usually shows high specificity toward its cognate oriT region (26–29); for example, the conjugation systems encoded by two Enterococcus plasmids, pAD1 and pAM373, do not mobilize the plasmids carrying the oriT regions from pAM373 and pAD1, respectively, although most of the conjugation-related genes and the oriT regions between the two plasmids show >95% identity. This type of high specificity is not observed between NAH7 and pWW0 due to the functional interchangeability of their relaxase-oriT systems irrespective of the 81% and 63% identities of the relaxases and the oriT sequences, respectively (Tables 1 and 2; see also Fig. S2b in the supplemental material). The high similarity of nic- and IR4-containing sequences between the oriTN and oriTW regions may enable both relaxases to function at the noncognate oriT regions. The high similarity of the IR1 sequence and considerable difference in the IR2 sequence between the two oriT regions (Fig. 3c) appears to be consistent with the importance of the IR1 but not the IR2 sequence of the oriTN region for its efficient mobilization (Fig. 4) and for the functional interchangeability of the two relaxase-oriT systems. The IR1 counterpart on pWW0 may also be important for its efficient conjugation. The oriTN and oriTR388 regions differ considerably not only in the overall nucleotide sequences (except for the identical 16-bp sequence immediately upstream of the nic sites) and the numbers and positions of IRs and putative IHF-binding sites but also in the amino acid sequences of relaxases (Fig. 3c; Fig. S2). Such differences must have led to the absence of mobilization of pNIT101 by R388.
We and other groups have previously shown that the narrow-host-range, self-transferable, and Pseudomonas-derived plasmids, such as those belonging to the IncP-9 and IncP-7 families, often carry the transposons that are responsible for the degradation of recalcitrant compounds (e.g., aromatic compounds) and resistance to antibiotics and heavy metal ions (10). When these plasmids are transferred by conjugation to the bacterial cells in which the replication systems of plasmids are nonfunctional, the transferred plasmids can allow the eventual insertion of the transposons in the stable replicons (e.g., chromosome) in the recipient cells with concomitant loss of the other parts of plasmids. Therefore, the broad-host-range property of plasmid-encoding conjugation systems can greatly contribute to the horizontal transfer of various genetic traits. Plasmids RP4, R388, and pKM101, which belong to the IncP-1, IncW, and IncN groups, respectively, are well-characterized and so-called broad-host-range plasmids because both their replication and conjugation systems can function in nearly all proteobacterial strains (30). The host ranges of these conjugation systems have been further shown to be broader than those of the replication systems; the former systems can deliver DNA into the bacterial recipient strains where the latter systems are not functional (31). Our results in this study similarly showed that the NAH7 conjugation system has a broader host range than its replication system; the conjugation system has an ability to conjugatively mobilize DNA into the alphaproteobacterial and betaproteobacterial strains, in which the replication system is most probably nonfunctional (Table 3). Shintani et al. (32) reported a preliminary result in which a betaproteobacterial Delftia strain from a soil community was able to receive NAH7 by conjugation from P. putida but did not allow the plasmid to replicate. This report also supports our finding that various genera in the betaproteobacterial class can work as recipients for the NAH7 conjugation system. Unlike the other alphaproteobacterial strains examined, Sphingomonas sp. TKS is unique in its inability to receive pNIT101 by the NAH7 conjugation system (Table 3). This property is apparently not observed in the conjugation systems encoded by other incompatibility plasmids since our preliminary experiments indicated the successful conjugative transfer of RP4 to TSK (data not shown). Either one or more of the following steps may be defective: the NAH7-encoding mating-pair formation between the donor and recipient cells, conjugative transfer of the ssDNA form of pNIT101, and its conversion to the dsDNA form in TSK. At present, it remains unclear which step(s) is defective and what mechanism(s) gives rise to the unique property of TSK. Isolation and characterization of the NAH7 and/or TSK mutant(s) that results in the successful conjugative mobilization of pNIT101 to TSK or its mutant(s) will provide some clues to uncover the plasmid and/or chromosomal factors that govern the host range of the conjugation system. Our previous finding that the traF product of NAH7 works as its host-range modifier (12) was obtained using a traF mutant of NAH7. Our identification of the NAH7 oriT region and relaxase in this study will be valuable for our future investigation of the role of the traF product in host-range modification by using the mobilization system of the plasmid carrying only the oriT region.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 4. E. coli strains were usually grown at 37°C in Luria-Bertani broth (33), and the other strains were grown at 30°C in one-third LB (0.33% tryptone, 0.16% yeast extract, 0.5% NaCl) broth. The E. coli cells carrying the NAH7 and pWW0 derivatives were cultivated at 30°C to avoid unstable maintenance at 37°C (14, 18). The solid medium was prepared with the addition of 1.5% agar. Antibiotics were added to the medium at the following concentrations: kanamycin (Km), 25 μg/ml; gentamicin (Gm), 30 μg/ml for Burkholderia cenocepacia, 10 μg/ml for E. coli, and 20 μg/ml for other strains; tetracycline (Tc), 150 μg/ml for B. cenocepacia, 50 μg/ml for Pseudomonas fluorescens, Pseudomonas aeruginosa, Burkholderia multivorans, and Burkholderia vietnamiensis, 20 μg/ml for E. coli, P. putida, and Sphingobium strains, 10 μg/ml for Sinorhizobium meliloti, and 2.5 μg/ml for Ralstonia solanacearum; streptomycin (Sm), 500 μg/ml for S. meliloti and 20 μg/ml for E. coli.
TABLE 4.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| Bacterial strains | ||
| E. coli | ||
| DH5α | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 Δ(lacZYA-argF) ϕ80lacZΔM15 | 33 |
| JM109 | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 Δ(lac-proAB)(F' traD36 proAB+ lacIqZΔM15) | 42 |
| BL21(DE3) | F− ompT hsdSB (rB− mB−) gal ompT, λ (DE3) | 43 |
| EC100 | mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80dlacZΔM15 ΔlacX74 recA1 endA1 araD139 Δ(ara- leu)7697 galU galK rpsL nupG | Epicentre, Inc. |
| P. putida | ||
| KT2440 | Wild-type strain | ATCC 470554 |
| KT2440G | KT2440::TnMod-OGm; Gmr | This study |
| G7 | Wild-type strain, NAH7 carrier | 44 |
| G7(NAH7K2) | G7 derivative harboring NAH7K2 instead of NAH7, formerly designated G7K2 | 45 |
| G7(NAH7K3) | G7 derivative harboring NAH7K3 instead of NAH7 | This study |
| G7(NAH7K4) | G7 derivative harboring NAH7K4 instead of NAH7 | This study |
| G7dCLC(NAH7K4) | G7 derivative carrying miniTn7T-LAC-traC in its chromosome, harboring NAH7K4 instead of NAH7 | This study |
| P. fluorescens | ||
| Pf-5G | Pf-5::TnMod-OGm; Gmr | 46 |
| P. aeruginosa | ||
| KG2512Gm | Km-sensitive and Gm-resistant derivative of PAO1 | 46 |
| B. multivorans | ||
| ATCC 17616 | Wild-type strain; Gmr | 47 |
| B. vietnamiensis | ||
| G4 | Wild-type strain | 48 |
| B. cenocepacia | ||
| J2315 | Wild-type strain | 49 |
| Ralstonia solanacearum | ||
| RS1085 | Wild-type strain | 50 |
| Sphingobium japonicum | ||
| UT26 | Wild-type strain | 51 |
| Sphingobium sp. | ||
| MI1205 | Wild-type strain | 52 |
| TKS | Wild-type strain | 53 |
| Sinorhizobium meliloti | ||
| 1021 | Wild-type strain; Smr | 54 |
| Plasmids | ||
| NAH7 | Tra+ Nah+, IncP-9 | 55 |
| NAH7K2 | NAH7ΔnahAc; Kmr | 45 |
| NAH7K3 | NAH7Δnic; Kmr | This study |
| NAH7K4 | NAH7ΔtraC; Kmr | This study |
| pMT1405 | pWW0 derivative carrying Kmr gene; Tol+ Xyl+ | 18 |
| pEX18Tc | pMB9 replicon; Tcr; sacB; suicide vector for gene replacement, carrying pUC18-derived multiple-cloning sites | 35 |
| pEX18Gm | pMB9 replicon; Gmr; sacB; suicide vector for gene replacement, carrying pUC18-derived multiple-cloning sites | 35 |
| pTnGm1 | pUC57 derivative carrying Gmr gene | Laboratory stock |
| pUC4K | pMB9 replicon; Kmr | 56 |
| pET22b(+) | Apr; C-Terminal His tag | TaKaRa |
| pET22b292TraC | pET22b(+) derivative for overexpression of traCN292 | This study |
| pUC18-mini-Tn7T-LACTraC | ColE1 replicon, Apr; Gmr, lacIq-Ptac::traC on mini-Tn7T | This study |
| pTNS2 | R6K replicon, Apr; source of TnsABCD transposase | 36 |
| pTnMod-OGm | pMB1 replicon, Tn5 inverted repeat; Gmr | 37 |
| pUC18 | Apr; E. coli vector | 42 |
| pUC18traC | pUC18 derivative carrying NAH7 traC gene | This study |
| pNIT6012 | pVS1 derivative; shuttle vector, Mob+ Tcr | 57 |
| pNIT101 to pNT112 | pNIT6012 derivatives carrying a 430-bp oriT region from NAH7 and its deletion fragments | This study |
| pNIT201 | pNIT6012 derivative carrying oriT region from pWW0 | This study |
| pNIT301 | pNIT6012 derivative carrying sacB and Gmr genes flanked by two copies of oriT NAH7 region | This study |
| pNIT302 and pNIT303 | pNIT6012 derivatives carrying sacB and Gmr genes flanked by oriT regions from NAH7 and pWW0 | This study |
| pNIT304 | pNIT6012 derivative carrying sacB and Gmr genes flanked by two copies of pWW0 oriT region | This study |
Nah+, utilization of naphthalene; Tol+ Xyl+, utilization of toluene and xylenes, respectively.
DNA manipulation.
Standard methods were used for the extraction of plasmid DNA, DNA digestion with restriction endonucleases, DNA ligation, and transformation of E. coli cells (33). Electrocompetent cells were prepared according to the 10-min method, and electroporation was carried out as described previously (34). An E. coli strain, DH5α or JM109, was used for the construction of plasmids. PCR was carried out using KOD-plus DNA polymerase (Toyobo, Osaka, Japan) or Ex Taq polymerase (TaKaRa, Ohtsu, Japan), and the primers used are listed in Table 5. Sequence determination was performed using an ABI Prism model 3130xl sequencer and an ABI Prism BigDye Terminator kit (Thermo Fisher Scientific, Waltham, MA).
TABLE 5.
Primers used in this study
| Primer name | Sequence (5′ to 3′)a | Purpose |
|---|---|---|
| XhoI_NAH7_oriT_F | GGGctcgagGGTGTTCCTCTCTGAGCCC | Cloning of oriTN on pNIT6012 |
| NheI_NAH7_oriT_R | GGGgctagcTTTGCACCCTATCTGCACCC | Cloning of oriTN on pNIT6012 |
| XhoI_NAH7_oriT_-50_F | GGGctcgagTGCCGAAACAGTGTATAGC | Cloning of derivative oriT on pNIT6012 |
| XhoI_NAH7_oriT_-81_F | GGGctcgagCAAAAAGCTTGATTTTGCTG | Cloning of derivative oriT on pNIT6012 |
| XhoI_NAH7_oriT_-119_F | GGGctcgagTAATACATATGTATAGCGC | Cloning of derivative oriT on pNIT6012 |
| XhoI_NAH7_oriT_-139_F | GGGctcgagTAAAAACACCACTTTTTCTCT | Cloning of derivative oriT on pNIT6012 |
| XhoI_NAH7_oriT_-161_F | GGGctcgagCAGGCCGCGCACAGCGCGGCT | Cloning of derivative oriT on pNIT6012 |
| XhoI_NAH7_oriT_-183_F | GGGctcgagGTCACCTCCCGCTAGGGGGGT | Cloning of derivative oriT on pNIT6012 |
| XhoI_NAH7_oriT_-206_F | GGGctcgagGTATTGTGTATTGAGACTTTG | Cloning of derivative oriT on pNIT6012 |
| NheI_NAH7_oriT_-328_R | GGGgctagcGCATACCATCTGTGCTACC | Cloning of derivative oriT on pNIT6012 |
| NheI_NAH7_oriT_-242_R | GGGgctagcGGGTCAAAGTCTCAATTCAA | Cloning of derivative oriT on pNIT6012 |
| NheI_NAH7_oriT_-217_R | GGGgctagcCAATACACAATACGCACC | Cloning of derivative oriT on pNIT6012 |
| NheI_NAH7_oriT_-213_R | GGGgctagcACACAATACGCACCCC | Cloning of derivative oriT on pNIT6012 |
| oriT_FAM_1b | [FAM]-GACAGGTGTTTATAGCCTGCC | Amplification of FAM-labeled oriT |
| oriT_FAM_2c | [FAM]-AATCAATGGCATCACCTGGCG | Amplification of FAM-labeled oriT |
| EcoRI_nic up_F | CGGgaattcGATGCTCCAACGTCGACCA | Deletion of nic site |
| KpnI_nic_up_R | CGGggtaccGAATTGAGACTTTGACCC | Deletion of nic site |
| KpnI_nic_down_F | CGGggtaccACGCACCCCCCTAGCG | Deletion of nic site |
| BamHI_nic_down_R | CGCggatccGCCATCCGGTTGAAGTCAT | Deletion of nic site |
| KpnI_traC_up_F | TAAggtaccAGAACTGGCACACCGTCAAG | Deletion of traC |
| BamI_traC_up_R | CGggatccGAACATGGCTACCTCGTTATGC | Deletion of traC |
| BamHI_traC_down_F | CGggatccAGGCGTGTAGATGATTTTTTG | Deletion of traC |
| XbaI_traC_down_R | GCtctagaCGAGGACATTATCGTAGGTC | Deletion of traC |
| NdeI_N292traC__F | GGGctcgagGCCGAACTCGATACCTAG | Cloning of traCN292 on pET22b(+) |
| XhoI_N292traC_R | GGAATTCcatatgTTCAACGTTACCTCTATCAA | Cloning of traCN292 on pET22b(+) |
| XhoI_pWW0_oriT_F | GGGctcgagGTTGTTCCCTCAAATTCCCCT | Cloning of oriTW on pNIT6012 |
| NheI_pWW0_oriT_R | GGGgctagcTGCACCCTACCTGCACCTAC | Cloning of oriTW on pNIT6012 |
| nic_con_F | GATCGTGTCGGCTTGCTGT | Confirmation of deletion of NAH7Δnic |
| nic_con_R | TTGCTCATCACTCGGATCG | Confirmation of deletion of NAH7Δnic |
| pNITGm_oriTN_F1 | TACCCGGGAGCTCGATTTGCACCCTATCTGCACC | Construction of pNIT301 and pNIT302 |
| pNITGm_oriTN_R1 | GGGGTGACGCCAAAGGGTGTTCCTCTCTGAGCC | Construction of pNIT301 and pNIT302 |
| pNITGm_oriTN_F2 | GATGTGTATAAGAGACAGTTTGCACCCTATCTGCACC | Construction of pNIT301 and pNIT303 |
| pNITGm_oriTN_R2 | TGGCAAAAGCTTCGAAGGTGTTCCTCTCTGAGCC | Construction of pNIT301 and pNIT303 |
| pNITGm_oriTW_F1 | TACCCGGGAGCTCGATGCACCCTACCTGCACCTAC | Construction of pNIT304 and pNIT303 |
| pNITGm_oriTW_R1 | GGGGTGACGCCAAAGGTTGTTCCCTCAAATTCC | Construction of pNIT304 and pNIT303 |
| pNITGm_oriTW_F2 | GATGTGTATAAGAGACAGTGCACCCTACCTGCACCTAC | Construction of pNIT304 and pNIT302 |
| pNITGm_oriTW_R2 | TGGCAAAAGCTTCGAAGTTGTTCCCTCAAATTCCCCT | Construction of pNIT304 and pNIT302 |
| pNITGm_oriTSacB_F | CTTTGGCGTCACCCCTTAC | Construction of pNIT301 to pNIT304 |
| pNITGm_oriTSacB_R | CAGATGTGTATAAGAGACAGGCGGCATCAGAGCAGATTG | Construction of pNIT301 to pNIT304 |
| Gm1_F | CCATTCAGGCTGCGCAACTGTTG | Construction of pNIT301 to pNIT304 |
| Gm1_R | GCAGCGAGTCAGTGAGCGAG | Construction of pNIT301 to pNIT304 |
| SpeI_traC_F | TAAactagtCCGGTCAAGCAGCAGCATAAC | Construction of pUCtraC and pTn7LactraC |
| XhoI_traC_R1 | GGGctcgagGCCTGGCCTATATTCTTC | Construction of pUC18traC |
| XhoI_traC_R2 | TAActcgagGGTAATGGTTTGGCAGGGG | Construction of pTn7LactraC |
Recognition sites for restriction enzymes are indicated by lowercase and italicized letters.
The nucleotide sequence able to anneal to the NAH7 traA gene is located at the positions from 20276 to 20256 on the NAH7 map.
The nucleotide sequence able to anneal to the strand of NAH7 traD gene is located at the positions from 19671 to 19691 on the NAH7 map.
Construction of plasmids and strains.
The 430-bp oriT region of NAH7 (oriTN) (Fig. 1) was PCR amplified using the total DNA of strain G7 as the template and the primer set XhoI_NAH7_oriT_F and NheI_NAH7_oriT_R, and the 431-bp oriT region of pWW0 (oriTW) was amplified using the total DNA of an E. coli strain carrying pMT1405 as the template and the primer set XhoI_pWW0_oriT_F and NheI_pWW0_oriT_R. After treatment with XhoI and NheI, the two amplicons were inserted at the corresponding sites of pNIT6012 to obtain pNIT101 and pNIT201, respectively. To construct the pNIT101 derivatives carrying a series of deletion variants in either the traA- or traD-proximal end of the oriTN region, parts of the oriTN region were PCR amplified with the appropriate primer sets and cloned into the NheI-XhoI sites of pNIT6012. The resulting plasmids were designated pNIT102 to pNIT112 (Fig. 4). pNIT301 and pNIT304 are the derivatives of pNIT6012, in which the sacB and Gm resistance (Gmr) genes are flanked by directly oriented copies of the oriTN and oriTW regions, respectively, and pNIT302 and pNIT303 are derivatives of pNIT301 in which one oriTN region is replaced by an oriTW region. These four plasmids were constructed as follows. The sacB gene as a counterselective suicidal marker (35) from pEX18Tc and the Gmr gene from pTnGm1 (Table 4) were PCR amplified using the primer set pNITGm_oriTSacB_F and pNITGm_oriTSacB_R and the set Gm1_F and Gm1_R, respectively, and the amplified Gmr gene amplicon was treated with PvuII. The oriTN region was PCR amplified using the primer set pNITGm_oriTN_F1 and pNITGm_oriTN_R1 and the set pNITGm_oriTN_F2 and pNITGm_oriTN_R2, and the oriTW region was PCR amplified using the primer set pNITGm_oriTW_F1 and pNITGm_oriTW_R1 and the set pNITGm_oriTW_F2 and pNITGm_oriTW_R2. These four amplicons were designated oriTN_1, oriTN_2, oriTW_1, and oriTW_2, respectively. A Gibson Assembly cloning kit (New England BioLabs, Beverly, MA) was used to insert the four amplicons, oriTN_1 or oriTW_1, the sacB and Gmr gene amplicons, and oriTN_2 or oriTW_2 in that order, into the EcoRI site of pNIT6012. The NAH7 traC gene was PCR amplified using the primer set SpeI_traC_F and XhoI_traC_R1, and the resulting amplicon was treated with SpeI and XhoI and inserted at the XbaI-SalI site of pUC18 to obtain pUC18traC. The NAH7 DNA fragment encoding the N-terminal 292 amino acid residues of the TraC protein (designated TraCN292) was PCR amplified using the total DNA of strain G7 as the template and the primer set NdeI_N292traC_F and XhoI_N292traC_R. After treatment with NdeI and XhoI, the amplicon was inserted at the corresponding sites of pET22b(+) to obtain pET22bN292TraC.
NAH7K3 is a derivative of NAH7K2 carrying a deletion mutation in its nic site, and this derivative was constructed using pEX18Tc. Approximately 1-kb regions located upstream and downstream of the nic site were amplified by PCR using the primer set EcoRI_nic_up_F and KpnI_nic_up_R and the set KpnI_nic_down_F and BamHI_nic_down_R, respectively. The two amplicons were treated with KpnI and EcoRI or BamHI and then inserted into the EcoRI and BamHI sites of pEX18Tc. The resulting plasmid was introduced into G7K2 by electroporation, and the Tcr transformants capable of growing on one-third LB agar were selected. The Tc-sensitive (Tcs) derivatives of such a transformant were next selected using one-third LB agar containing 10% sucrose. The expected double-crossover-mediated homologous recombination in the Tcs derivatives was confirmed by PCR.
To construct a traC deletion derivative of NAH7, approximately 1-kb regions located upstream and downstream of traC were amplified by PCR using the primer set KpnI_traC_up_F and BamHI_traC_up_R and the set BamHI_traC_down_F and XbaI_traC_down_R, respectively, and the two amplicons were treated with BamHI and KpnI or XbaI. These two restricted fragments and the BamHI-flanked Kmr gene fragment from pUC4K were cloned into the KpnI and XbaI sites of pEX18Gm (35). The resulting plasmid was introduced into G7 by electroporation, and the Kmr transformants able to grow on one-third LB agar containing 10% sucrose were selected. Among such transformants, Gms derivatives were chosen and the expected double-crossover-mediated homologous recombination in the Gms derivatives was confirmed by PCR. The resulting NAH7 derivative lacking its traC gene was designated NAH7K4.
To introduce the wild-type traC gene into the chromosome of G7(NAH7K4), the traC gene was first PCR amplified by the primer set SpeI_traC_F and XhoI_traC_R2. The amplicon was treated with SpeI and XhoI and inserted into the corresponding sites on pUC18-mini-Tn7T-LAC. The resulting plasmid and pTNS2 were used to cotransform G7(NAH7K4) by electroporation according to the method of Choi et al. (36), and the Gmr transformants that carried a single copy of the mini-Tn7T-LAC derivative at the chromosomal attTn7 site were selected. The Flp recombinase-mediated marker excision system was used to remove the Gmr gene from the chromosomal mini-Tn7T-LAC derivative to obtain G7dCLC(NAH7K4).
The genomes of six strains (B. vietnamiensis, B. cenocepacia, R. solanacearum, and three Sphingobium strains) were marked with the Gmr gene derived from a Tn5-based transposon on pTnMod-OGm (37). Electroporation of these strains by pTnMod-OGm to obtain the Gmr transposants led to the TnMod-OGm insertion in the recipient genomes.
Conjugative mating.
Donor and recipient cells cultivated in one-third LB broth with and without appropriate antibiotics, respectively, were harvested by centrifugation, washed with one-third LB broth, and resuspended in one-third LB broth. They were mixed at a ratio of 1 to 1, then spotted on a 0.45-μm membrane filter (Advantec Inc., Tokyo, Japan) that had been placed on a one-third LB agar plate. After incubation at 30°C for 24 h, the cells were suspended in one-third LB broth, diluted appropriately, and spread on selective agar plates. The conjugative transfer or mobilization frequency was expressed as the number of transconjugants per that of donor cells.
Resolution assay.
The intramolecular recombination between two oriT regions on the pNIT6012-based plasmids, pNIT301 to pNIT304, was investigated in an E. coli recA1 strain, EC100. One of the four plasmids was introduced into E. coli EC100, EC100(NAH7K2), EC100(pMT1405), EC100(pUC18traC), or EC100(pUC18) by transformation to select the Gmr transformants. Each transformant was cultivated overnight in one-third LB broth supplemented with Tc, and the culture was plated on one-third LB agar with Tc and one-third LB agar with Tc and 10% sucrose. The recombination frequency was expressed as the number of Tcr and sucrose-resistant colonies per that of Tcr colonies. The oriT-containing regions on the pNIT6012-based derivatives in the Tcr and sucrose-resistant colonies were analyzed by DNA sequencing.
Purification of His-tagged TraCN292 and its nicking activity.
The E. coli BL21(DE3) cells harboring pET22bN292TraC were cultured in LB at 37°C. When the optical density of the culture at 600 nm reached 1.0, IPTG was added at a final concentration of 0.5 mM. After additional incubation for 6 h at 30°C, the cells were disrupted using a CelLytic B reagent (Sigma-Aldrich, St. Louis, MO), and the IPTG-induced protein tagged with six histidine residues at the C terminus was purified using Talon metal affinity resins (TaKaRa) according to the manufacturer's recommendations. After purification, only a single protein band with a size of approximately 33 kDa was observed by SDS-polyacrylamide gel analysis (see Fig. S1 in the supplemental material).
To investigate the nicking activity of His-tagged TraCN292, it was mixed with the fluorophore-labeled DNA fragment, and the fluorescently labeled ssDNA fragment was detected by a capillary sequencer. For this purpose, the DNA fragment was PCR amplified using one primer whose 5′ end was labeled with 6-carboxyfluorescein (FAM) (Eurofins Genomics, Tokyo, Japan) and another unlabeled primer so that the 5′ end of one strand of amplified dsDNA fragment was labeled; the primer set oriT_FAM_1 and NheI_NAH7_oriT_R and the set XhoI_NAH7_oriT_F and oriT_FAM_2 (Table 5) were used to prepare the oriT-containing dsDNA fragments, and the 5′ ends of the top and bottom strands (Fig. 1b) of the resulting fragments, respectively, were labeled with FAM. A 25-ng FAM-labeled dsDNA fragment, 2.4 μM TraCN292, and 1 μg salmon sperm DNA were mixed in 20 mM Tris-HCl, 5 mM MgCl2, 5 mM NaCl, and 0.1 mM EDTA, pH 7.5. After incubation for 30 min at 30°C, the resulting DNA fragments were purified by a BigDye XTerminator purification kit (Thermo Fisher Scientific). The purified products were mixed with a GeneScan 500 LIZ size standard (Life Technologies) and separated using an ABI Prism model 3130xl sequencer. The method of Eckert (38) was used to treat the FAM-labeled dsDNA fragment with formic acid and piperidine to prepare a G+A sequencing ladder sample. The peak patterns of this sample and the TraCN292-treated PCR fragment were compared using our original software, TraceViewer (http://www.ige.tohoku.ac.jp/joho/traceviewer/), to precisely determine the size of the nicked ssDNA fragment that was labeled with FAM.
Bioinformatic analysis.
The DNA sequences were compared by the MAFFT program (39), and the secondary structure prediction for the oriT region was performed using the software package MFOLD (40). A homology search was carried out using the BLAST programs that are available at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/BLAST/).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a grant-in-aid (no. 15H04471) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, by a grant from the Institute for Fermentation, Osaka (IFO), Japan, and in part by a Grant for Environmental Research Projects from The Sumitomo Foundation, Japan.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02359-16.
REFERENCES
- 1.Koraimann G, Wagner MA. 2014. Social behavior and decision making in bacterial conjugation. Front Cell Infect Microbiol 4:54. doi: 10.3389/fcimb.2014.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cabezon E, Ripoll-Rozada J, Pena A, de la Cruz F, Arechaga I. 2015. Towards an integrated model of bacterial conjugation. FEMS Microbiol Rev 39:81–95. doi: 10.1111/1574-6976.12085. [DOI] [PubMed] [Google Scholar]
- 3.Smillie C, Garcillan-Barcia MP, Francia MV, Rocha EP, de la Cruz F. 2010. Mobility of plasmids. Microbiol Mol Biol Rev 74:434–452. doi: 10.1128/MMBR.00020-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garcillan-Barcia MP, Francia MV, de la Cruz F. 2009. The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol Rev 33:657–687. doi: 10.1111/j.1574-6976.2009.00168.x. [DOI] [PubMed] [Google Scholar]
- 5.de la Cruz F, Frost LS, Meyer RJ, Zechner EL. 2010. Conjugative DNA metabolism in Gram-negative bacteria. FEMS Microbiol Rev 34:18–40. doi: 10.1111/j.1574-6976.2009.00195.x. [DOI] [PubMed] [Google Scholar]
- 6.Draper O, Cesar CE, Machon C, de la Cruz F, Llosa M. 2005. Site-specific recombinase and integrase activities of a conjugative relaxase in recipient cells. Proc Natl Acad Sci U S A 102:16385–16390. doi: 10.1073/pnas.0506081102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paterson ES, Iyer VN. 1997. Localization of the nic site of IncN conjugative plasmid pCU1 through formation of a hybrid oriT. J Bacteriol 179:5768–5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meyer R. 1989. Site-specific recombination at oriT of plasmid-R1162 in the absence of conjugative transfer. J Bacteriol 171:799–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cesar CE, Machon C, de la Cruz F, Llosa M. 2006. A new domain of conjugative relaxase TrwC responsible for efficient oriT-specific recombination on minimal target sequences. Mol Microbiol 62:984–996. doi: 10.1111/j.1365-2958.2006.05437.x. [DOI] [PubMed] [Google Scholar]
- 10.Tsuda M, Ohtsubo Y, Yano H. 2014. Mobile catabolic genetic elements in pseudomonads, p 83–103. In Nojiri H, Tsuda M, Fukuda M, Kamagata Y (ed), Biodegradative bacteria: how bacteria degrade, survive, adapt, and evolve. Springer Japan, Tokyo, Japan. [Google Scholar]
- 11.Seo J, Kang SI, Kim M, Han J, Hur HG. 2011. Flavonoids biotransformation by bacterial non-heme dioxygenases, biphenyl and naphthalene dioxygenase. Appl Microbiol Biotechnol 91:219–228. doi: 10.1007/s00253-011-3334-z. [DOI] [PubMed] [Google Scholar]
- 12.Miyazaki R, Ohtsubo Y, Nagata Y, Tsuda M. 2008. Characterization of the traD operon of naphthalene-catabolic plasmid NAH7: a host-range modifier in conjugative transfer. J Bacteriol 190:6281–6289. doi: 10.1128/JB.00709-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inoue K, Miyazaki R, Ohtsubo Y, Nagata Y, Tsuda M. 2013. Inhibitory effect of Pseudomonas putida nitrogen-related phosphotransferase system on conjugative transfer of IncP-9 plasmid from Escherichia coli. FEMS Microbiol Lett 345:102–109. doi: 10.1111/1574-6968.12188. [DOI] [PubMed] [Google Scholar]
- 14.Tsuda M, Iino T. 1990. Naphthalene degrading genes on plasmid NAH7 are on a defective transposon. Mol Gen Genet 223:33–39. doi: 10.1007/BF00315794. [DOI] [PubMed] [Google Scholar]
- 15.Sota M, Yano H, Ono A, Miyazaki R, Ishii H, Genka H, Top EM, Tsuda M. 2006. Genomic and functional analysis of the IncP-9 naphthalene-catabolic plasmid NAH7 and its transposon Tn4655 suggests catabolic gene spread by a tyrosine recombinase. J Bacteriol 188:4057–4067. doi: 10.1128/JB.00185-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greated A, Lambertsen L, Williams PA, Thomas CM. 2002. Complete sequence of the IncP-9 TOL plasmid pWW0 from Pseudomonas putida. Environ Microbiol 4:856–871. doi: 10.1046/j.1462-2920.2002.00305.x. [DOI] [PubMed] [Google Scholar]
- 17.Lambertsen LM, Molin S, Kroer N, Thomas CM. 2004. Transcriptional regulation of pWW0 transfer genes in Pseudomonas putida KT2440. Plasmid 52:169–181. doi: 10.1016/j.plasmid.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 18.Tsuda M, Iino T. 1988. Identification and characterization of Tn4653, a transposon covering the toluene transposon Tn4651 on TOL plasmid pWW0. Mol Gen Genet 213:72–77. doi: 10.1007/BF00333400. [DOI] [PubMed] [Google Scholar]
- 19.Parker C, Becker E, Zhang X, Jandle S, Meyer R. 2005. Elements in the co-evolution of relaxases and their origins of transfer. Plasmid 53:113–118. doi: 10.1016/j.plasmid.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 20.Lucas M, Gonzalez-Perez B, Cabezas M, Moncalian G, Rivas G, de la Cruz F. 2010. Relaxase DNA binding and cleavage are two distinguishable steps in conjugative DNA processing that involve different sequence elements of the nic site. J Biol Chem 285:8918–8926. doi: 10.1074/jbc.M109.057539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moncalian G, Grandoso G, Llosa M, de la Cruz F. 1997. oriT-processing and regulatory roles of TrwA protein in plasmid R388 conjugation. J Mol Biol 270:188–200. doi: 10.1006/jmbi.1997.1082. [DOI] [PubMed] [Google Scholar]
- 22.Carballeira JD, Gonzalez-Perez B, Moncalian G, de la Cruz F. 2014. A high security double lock and key mechanism in HUH relaxases controls oriT-processing for plasmid conjugation. Nucleic Acids Res 42:10632–10643. doi: 10.1093/nar/gku741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Llosa M, Grandoso G, de la Cruz F. 1995. Nicking activity of TrwC directed against the origin of transfer of the IncW plasmid R388. J Mol Biol 246:54–62. doi: 10.1006/jmbi.1994.0065. [DOI] [PubMed] [Google Scholar]
- 24.Gao Q, Luo Y, Deonier RC. 1994. Initiation and termination of DNA transfer at F plasmid oriT. Mol Microbiol 11:449–458. doi: 10.1111/j.1365-2958.1994.tb00326.x. [DOI] [PubMed] [Google Scholar]
- 25.Llosa M, Bolland S, Grandoso G, de la Cruz F. 1994. Conjugation-independent, site-specific recombination at the oriT of the IncW plasmid R388 mediated by TrwC. J Bacteriol 176:3210–3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Llosa M, Bolland S, de la Cruz F. 1991. Structural and functional analysis of the origin of conjugal transfer of the broad-host-range IncW plasmid R388 and comparison with the related IncN plasmid R46. Mol Gen Genet 226:473–483. doi: 10.1007/BF00260661. [DOI] [PubMed] [Google Scholar]
- 27.Fekete RA, Frost LS. 2000. Mobilization of chimeric oriT plasmids by F and R100-1: role of relaxosome formation in defining plasmid specificity. J Bacteriol 182:4022–4027. doi: 10.1128/JB.182.14.4022-4027.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Francia MV, Clewell DB. 2002. Transfer origins in the conjugative Enterococcus faecalis plasmids pAD1 and pAM373: identification of the pAD1 nic site, a specific relaxase and a possible TraG-like protein. Mol Microbiol 45:375–395. doi: 10.1046/j.1365-2958.2002.03007.x. [DOI] [PubMed] [Google Scholar]
- 29.Pansegrau W, Ziegelin G, Lanka E. 1988. The origin of conjugative IncP plasmid transfer: interaction with plasmid-encoded products and the nucleotide sequence at the relaxation site. Biochim Biophys Acta 951:365–374. doi: 10.1016/0167-4781(88)90108-X. [DOI] [PubMed] [Google Scholar]
- 30.Martini MC, Albicoro FJ, Nour E, Schluter A, van Elsas JD, Springael D, Smalla K, Pistorio M, Lagares A, Del Papa MF. 2015. Characterization of a collection of plasmid-containing bacteria isolated from an on-farm biopurification system used for pesticide removal. Plasmid 80:16–23. doi: 10.1016/j.plasmid.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 31.Encinas D, Garcillan-Barcia MP, Santos-Merino M, Delaye L, Moya A, de la Cruz F. 2014. Plasmid conjugation from Proteobacteria as evidence for the origin of xenologous genes in Cyanobacteria. J Bacteriol 196:1551–1559. doi: 10.1128/JB.01464-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shintani M, Matsui K, Inoue J, Hosoyama A, Ohji S, Yamazoe A, Nojiri H, Kimbara K, Ohkuma M. 2014. Single-cell analyses revealed transfer ranges of IncP-1, IncP-7, and IncP-9 plasmids in a soil bacterial community. Appl Environ Microbiol 80:138–145. doi: 10.1128/AEM.02571-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 34.Choi KH, Kumar A, Schweizer HP. 2006. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J Microbiol Methods 64:391–397. doi: 10.1016/j.mimet.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 35.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86. doi: 10.1016/S0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- 36.Choi KH, Gaynor JB, White KG, Lopez C, Bosio CM, Karkhoff-Schweizer RR, Schweizer HP. 2005. A Tn7-based broad-range bacterial cloning and expression system. Nat Methods 2:443–448. doi: 10.1038/nmeth765. [DOI] [PubMed] [Google Scholar]
- 37.Dennis JJ, Zylstra GJ. 1998. Plasposons: modular self-cloning minitransposon derivatives for rapid genetic analysis of gram-negative bacterial genomes. Appl Environ Microbiol 64:2710–2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eckert RL. 2001. DNA sequencing by the chemical method. Curr Protoc Mol Biol 17:7.5.1–7.5.11. [DOI] [PubMed] [Google Scholar]
- 39.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moncalian G, Valle M, Valpuesta JM, de la Cruz F. 1999. IHF protein inhibits cleavage but not assembly of plasmid R388 relaxosomes. Mol Microbiol 31:1643–1652. doi: 10.1046/j.1365-2958.1999.01288.x. [DOI] [PubMed] [Google Scholar]
- 42.Yanisch-Perron C, Vieira J, Messing J. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 43.Studier FW, Moffatt BA. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol 189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 44.Dunn NF, Gunsalus IC. 1973. Transmissible plasmid coding early enzymes of naphthalene oxidation in Pseudomonas putida. J Bacteriol 114:974–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ono A, Miyazaki R, Sota M, Ohtsubo Y, Nagata Y, Tsuda M. 2007. Isolation and characterization of naphthalene-catabolic genes and plasmids from oil-contaminated soil by using two cultivation-independent approaches. Appl Microbiol Biotechnol 74:501–510. doi: 10.1007/s00253-006-0671-4. [DOI] [PubMed] [Google Scholar]
- 46.Yano H, Miyakoshi M, Ohshima K, Tabata M, Nagata Y, Hattori M, Tsuda M. 2010. Complete nucleotide sequence of TOL plasmid pDK1 provides evidence for evolutionary history of IncP-7 catabolic plasmids. J Bacteriol 192:4337–4347. doi: 10.1128/JB.00359-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stanier RY, Palleroni NJ, Doudoroff M. 1966. The aerobic pseudomonads: a taxonomic study. J Gen Microbiol 43:159–271. doi: 10.1099/00221287-43-2-159. [DOI] [PubMed] [Google Scholar]
- 48.Newman LM, Wackett LP. 1995. Purification and characterization of toluene 2-monooxygenase from Burkholderia cepacia G4. Biochemistry 34:14066–14076. doi: 10.1021/bi00043a012. [DOI] [PubMed] [Google Scholar]
- 49.Wang H, Sun H, Wei D. 2013. Discovery and characterization of a highly efficient enantioselective mandelonitrile hydrolase from Burkholderia cenocepacia J2315 by phylogeny-based enzymatic substrate specificity prediction. BMC Biotechnol 13:14. doi: 10.1186/1472-6750-13-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mukaihara T, Tamura N, Murata Y, Iwabuchi M. 2004. Genetic screening of Hrp type III-related pathogenicity genes controlled by the HrpB transcriptional activator in Ralstonia solanacearum. Mol Microbiol 54:863–875. doi: 10.1111/j.1365-2958.2004.04328.x. [DOI] [PubMed] [Google Scholar]
- 51.Imai R, Nagata Y, Senoo K, Wada H, Fukuda M, Takagi M, Yano K. 1989. Dehydrochlorination of gamma-hexachlorocyclohexane (gamma-BHC) by gamma-BHC-assimilating Pseudomonas paucimobilis. Agric Biol Chem 53:2015–2017. doi: 10.1080/00021369.1989.10869597. [DOI] [Google Scholar]
- 52.Ito M, Prokop Z, Klvana M, Otsubo Y, Tsuda M, Damborsky J, Nagata Y. 2007. Degradation of beta-hexachlorocyclohexane by haloalkane dehalogenase LinB from gamma-hexachlorocyclohexane-utilizing bacterium Sphingobium sp. MI1205. Arch Microbiol 188:313–325. doi: 10.1007/s00203-007-0251-8. [DOI] [PubMed] [Google Scholar]
- 53.Tabata M, Ohhata S, Kawasumi T, Nikawadori Y, Kishida K, Sato T, Ohtsubo Y, Tsuda M, Nagata Y. 2016. Complete genome sequence of a gamma-hexachlorocyclohexane degrader, Sphingobium sp. strain TKS, isolated from a gamma-hexachlorocyclohexane-degrading microbial community. Genome Announc 4:e00247-16. doi: 10.1128/genomeA.00247-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meade HM, Long SR, Ruvkun GB, Brown SE, Ausubel FM. 1982. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J Bacteriol 149:114–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yen KM, Gunsalus IC. 1982. Plasmid gene organization: naphthalene/salicylate oxidation. Proc Natl Acad Sci U S A 79:874–878. doi: 10.1073/pnas.79.3.874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Taylor LA, Rose RE. 1988. A correction in the nucleotide sequence of the Tn903 kanamycin resistance determinant in pUC4K. Nucleic Acids Res 16:358. doi: 10.1093/nar/16.1.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Heeb S, Itoh Y, Nishijyo T, Schnider U, Keel C, Wade J, Walsh U, O'Gara F, Haas D. 2000. Small, stable shuttle vectors based on the minimal pVS1 replicon for use in gram-negative, plant-associated bacteria. Mol Plant Microbe Interact 13:232–237. doi: 10.1094/MPMI.2000.13.2.232. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.