ABSTRACT
The hepatitis C virus (HCV) is a major human pathogen. Genetically related viruses in animals suggest a zoonotic origin of HCV. The closest relative of HCV is found in horses (termed equine hepacivirus [EqHV]). However, low EqHV genetic diversity implies relatively recent acquisition of EqHV by horses, making a derivation of HCV from EqHV unlikely. To unravel the EqHV evolutionary history within equid sister species, we analyzed 829 donkeys and 53 mules sampled in nine European, Asian, African, and American countries by molecular and serologic tools for EqHV infection. Antibodies were found in 278 animals (31.5%), and viral RNA was found in 3 animals (0.3%), all of which were simultaneously seropositive. A low RNA prevalence in spite of high seroprevalence suggests a predominance of acute infection, a possible difference from the mostly chronic hepacivirus infection pattern seen in horses and humans. Limitation of transmission due to short courses of infection may explain the existence of entirely seronegative groups of animals. Donkey and horse EqHV strains were paraphyletic and 97.5 to 98.2% identical in their translated polyprotein sequences, making virus/host cospeciation unlikely. Evolutionary reconstructions supported host switches of EqHV between horses and donkeys without the involvement of adaptive evolution. Global admixture of donkey and horse hepaciviruses was compatible with anthropogenic alterations of EqHV ecology. In summary, our findings do not support EqHV as the origin of the significantly more diversified HCV. Identification of a host system with predominantly acute hepacivirus infection may enable new insights into the chronic infection pattern associated with HCV.
IMPORTANCE The evolutionary origins of the human hepatitis C virus (HCV) are unclear. The closest animal-associated relative of HCV occurs in horses (equine hepacivirus [EqHV]). The low EqHV genetic diversity implies a relatively recent acquisition of EqHV by horses, limiting the time span for potential horse-to-human infections in the past. Horses are genetically related to donkeys, and EqHV may have cospeciated with these host species. Here, we investigated a large panel of donkeys from various countries using serologic and molecular tools. We found EqHV to be globally widespread in donkeys and identify potential differences in EqHV infection patterns, with donkeys potentially showing enhanced EqHV clearance compared to horses. We provide strong evidence against EqHV cospeciation and for its capability to switch hosts among equines. Differential hepacivirus infection patterns in horses and donkeys may enable new insights into the chronic infection pattern associated with HCV.
KEYWORDS: equine hepacivirus, hepatitis C virus, donkey, evolution, pathogenesis
INTRODUCTION
Hepatitis C virus (HCV) is a major human pathogen infecting approximately 140 million people worldwide (1). HCV belongs to the genus Hepacivirus that comprises seven geographically distinct genotypes which likely evolved over considerable time spans (2–6). The evolutionary origins of HCV have remained obscure (6). Recent studies identified numerous hepaciviruses (HVs) in bats, rodents, monkeys, and peridomestic animals (7, 8). Considering the absence of HCV-related viruses in higher primates (9), as well as the existence of genetically diversified nonprimate HVs, mammals other than primates may have shaped primordial HCV evolution (10). The lack of cosegregation of HVs with mutually related animal hosts, as well as the detection of potential recombination events between some HV lineages, suggest low barriers against cross-host transmission (10–13). However, whether any of the animal species known to carry HVs represents a direct reservoir for HCV is unclear (14).
The equine HV (EqHV; originally described as canine HV and subsequently as nonprimate HV) (7, 8), constitutes the closest animal-associated relative of HCV among the HVs known so far (7, 13). Sporadic infections of dogs (15–17) support a broad host range of EqHV that may have enabled infection of humans with EqHV in the past. Transmission may have been aided by close contact of humans and horses since the domestication of horses about 5,500 years ago (18). However, the strikingly low genetic variation of EqHV in horses suggests a rather short evolutionary history (6), with limited opportunity for horse-human transition.
The genus Equus comprising all contemporary horses, donkeys, and zebras likely originated about 4.5 million years ago (19). Detection of EqHV homologues in equine sister species may aid elucidating the evolutionary history of this HV. The globally most widespread equine beyond domestic horses (Equus ferus caballus, ca. 59 million heads) is the domesticated donkey (E. asinus asinus, ca. 44 million heads, according to the Food and Agriculture Organization of the United Nations [FAO], FAOSTAT 2014 database). Donkeys have been tested for HV in limited numbers, such as 116 donkeys from the United Kingdom (17, 20), 30 mules and 5 donkeys from Brazil (21), and 8 mules and 6 donkeys from China (22), as well as a commercially available donkey serum from the United States (23), all with negative results. Here we investigated a considerably larger panel of donkey sera from various countries using serologic and molecular tools. We found EqHV to be globally widespread in donkeys and able to switch hosts among equines.
RESULTS
Wide-reaching exposure of donkeys to EqHV.
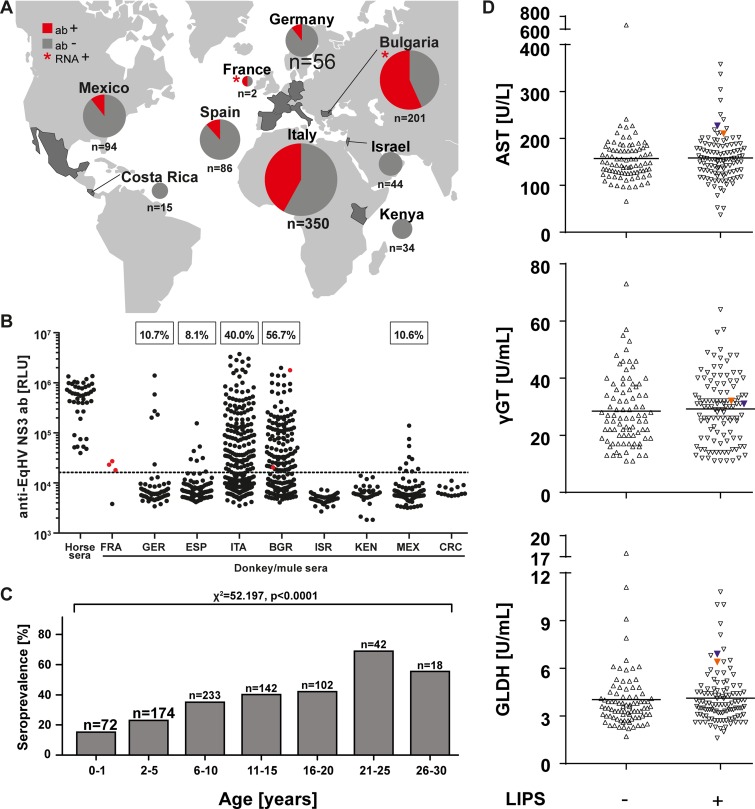
Donkey sera (n = 829) were collected in five European countries (Germany, Spain, Italy, Bulgaria, and France), as well as in Asia (Israel), Africa (Kenya), and Latin America (Costa Rica and Mexico), between 1974 and 2016 (Table 1). For three countries (France, Germany, and Italy), sampling was conducted in multiple years, and details of the annual sample characteristics in these countries are displayed in Table 2. In addition, 53 mule sera were sampled in Bulgaria in 2015. All 882 donkey and mule sera were analyzed for the presence of antibodies against the viral NS3 domain by a luciferase immunoprecipitation system (LIPS) (24, 25). Three sampling sites (Israel, Kenya, and Costa Rica) showed no serologic evidence for EqHV infection, whereas all other countries yielded positive test results (Fig. 1A). As shown in Table 1 and Fig. 1B, seroprevalence rates ranged between 8.1 and 10.7% in Germany, Spain, and Mexico. Seroprevalence rates in Italy and Bulgaria were significantly higher at 40.0 to 56.7% (corrected χ2 = 62.8 and χ2 = 109.1 [P < 0.0001] for Italy and Bulgaria compared to all other countries, respectively). Furthermore, within a specific country the seroprevalence rates varied between sampling years and hinted at the occurrence of focal EqHV epidemics, e.g., leading to 100% of EqHV-seropositive animals in Italy in 2015 (Table 2). However, the underlying factors responsible for the variations in seroprevalence are unknown. LIPS signal intensities from seropositive donkeys were comparable to those from seropositive horses, suggesting validity of the assay used for testing (Fig. 1B). Female donkeys were significantly more likely to be seropositive than male donkeys (35.0 versus 28.0%; corrected χ2 = 4.1 [P = 0.044]; risk ratio, 1.25 [lower and upper bounds, 1.01 to 1.54]; Table 1). Seroprevalence increased significantly with animal age from 20.7% in young animals (0 to 5 years of age) to 55.5% in older animals (25 to 30 years) (Fig. 1C).
TABLE 1.
Sample characteristics
| Country | Sampling period (yr) | n | Ab+ (%) | No. of animals/total no. of animals (%) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gender |
Age in yrs |
|||||||||||
| Jack | Jenny | Unknown | 0–5 | 6–10 | 11–15 | 16–20 | 21–30 | Unknown | ||||
| France | 1974/1979 | 2 | 1 (50.0) | 1/2 (50.0) | 1/2 (50.0) | |||||||
| Germany | 2007/2008/2015 | 56 | 6 (10.7) | 0/10 (0) | 6/46 (13.0) | 0/8 (0) | 4/14 (28.6) | 0/5 (0) | 0/4 (0) | 0/2 (0) | 2/23 (8.7) | |
| Spain | 2011 | 86 | 7 (8.1) | 3/38 (7.9) | 4/44 (9.1) | 0/4 (0) | 2/32 (6.3) | 3/27 (11.1) | 1/11 (9.1) | 0/10 (0) | 1/2 (50.0) | 0/4 (0) |
| Italy | 2004-2015 | 350 | 140 (40.0) | 14/52 (26.9) | 125/286 (43.7) | 1/12 (8.3) | 42/140 (30.0) | 48/108 (44.4) | 21/48 (43.8) | 20/38 (52.6) | 9/16 (56.3) | |
| Bulgaria | 2015 | 201 | 114 (56.7) | 69/113 (61.1) | 45/88 (51.1) | 3/12 (25.0) | 23/36 (63.9) | 33/58 (56.9) | 23/46 (50.0) | 29/39 (74.4) | 3/10 (30.0) | |
| Israel | 2014 | 44 | 0 (0) | 0/29 (0) | 0/15 (0) | 0/5 (0) | 0/9 (0) | 0/2 (0) | 0/2 (0) | 0/26 (0) | ||
| Kenya | 2015 | 34 | 0 (0) | 0/17 (0) | 0/6 (0) | 0/11 (0) | 0/34 (0) | |||||
| Mexico | 2016 | 94 | 10 (10.6) | 4/53 (7.5) | 6/41 (14.6) | 4/41 (9.8) | 4/33 (12.1) | 2/17 (11.8) | 0/2 (0) | 0/1 (0) | ||
| Costa Rica | 2016 | 15 | 0 (0) | 0/9 (0) | 0/6 (0) | 0/8 (0) | 0/6 (0) | 0/1 (0) | ||||
| Total | 882 | 278 (31.5) | 90/321 (28.0) | 186/532 (35.0) | 2/29 (6.9) | 51/246 (20.7) | 82/233 (35.2) | 57/142 (40.1) | 43/102 (42.2) | 39/60 (65.0) | 6/99 (6.1) | |
TABLE 2.
Annual donkey sample characteristics for France, Germany and Italy
| Country | Sampling period (yr) | n | Ab+ (%) | No. of animals/total no. of animals (%) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gender |
Age range in yrs |
|||||||||||
| Jack | Jenny | Unknown | 0–5 | 6–10 | 11–15 | 16–20 | 21–30 | Unknown | ||||
| France | 1974 | 1 | 0 (0) | 0/1 (0) | 0/1 (0) | |||||||
| 1979 | 1 | 1 (100.0) | 1/1 (100.0) | 1/1 (100.0) | ||||||||
| Germany | 2007 | 39 | 5 (12.8) | 0/9 (0) | 5/30 (16.7) | 0/7 (0) | 4/13 (30.8) | 0/5 (0) | 0/3 (0) | 0/2 (0) | 1/9 (11.1) | |
| 2008 | 3 | 0 (0) | 0/1 (0) | 0/2 (0) | 0/1 (0) | 0/1 (0) | 0/1 (0) | |||||
| 2015 | 14 | 1 (7.1) | 1/14 (7.1) | 1/14 (7.1) | ||||||||
| Italy | 2004–2009 | 38 | 5 (13.2) | 0/13 (0) | 4/13 (30.8) | 1/12 (8.3) | 1/15 (6.7) | 3/12 (25.0) | 0/7 (0) | 1/4 (25.0) | ||
| 2013 | 294 | 117 (39.8) | 11/36 (30.6) | 106/258 (41.1) | 36/120 (30.0) | 45/96 (46.9) | 10/30 (33.3) | 18/33 (54.5) | 8/15 (53.3) | |||
| 2015 | 18 | 18 (100.0) | 3/3 (100.0) | 15/15 (100.0) | 5/5 (100.0) | 11/11 (100.0) | 1/1 (100.0) | 1/1 (100.0) | ||||
FIG 1.
EqHV infection patterns. (A) Anti-EqHV antibody (ab) detection depicted in pie charts (red = positive). Asterisks, origin of the EqHV-RNA positive animals. (B) LIPS ratios of control sera form horses and donkeys; Bulgaria includes as well 53 sera from mules. EqHV-RNA positive donkey and mule sera are indicated in red. All three positive sera form France originate from one animal; no seroprevalence rate for this country is indicated due to the low sample size. Dotted line, cutoff (16,249.2 RLU). (C) Seroprevalence rates in different age groups. (D) Aspartate aminotransferase (AST), gamma-glutamyl transferase (γGGT), and glutamate dehydrogenase (GLDH) levels were determined in the sera of Bulgarian donkeys. Sera are shown according to their LIPS status, and RNA-positive samples are given in orange and blue.
Molecular detection of EqHV in donkeys.
To allow sensitive molecular detection of EqHV genetic variants in donkeys, all samples were tested using two different nested reverse transcription-PCR (RT-PCR) assays. The first assay targeted specifically the EqHV 5′-untranslated region (5′ UTR) commonly used for HV detection (26) and a second assay targeted the NS3 domain that is more conserved among diverse HVs than the 5′ UTR (11). One donkey from France (sampled in 1979, age and gender unknown), one donkey from Bulgaria (sampled in 2015, a 10-year-old male), and one mule from Bulgaria (sampled in 2015, a 16-year-old female) tested positive for EqHV RNA using the 5′-UTR-based assay (0.3% of all 882 donkey and mule sera). No additional specimens tested positive for HVs using the NS3-based assay, arguing against infection of donkeys with diverse HVs beyond EqHV.
Comparison of EqHV infection patterns between equine species.
Our data enabled comparisons of EqHV infection patterns between donkeys and horses. First, viral loads, which are a quantitative marker of virus replication, were similar between equine host species infected with EqHV. Viral loads in the RNA-positive specimens from this study ranged from 8.4 × 105 to 3.7 × 107 genome copies/ml of serum, as determined by strain-specific real-time RT-PCR assays. These viral loads were similar to viral loads observed in horses (20, 23, 25), suggesting similar infection intensities in both equine species. Furthermore, the detection of viral RNA at comparable loads in the French sera sampled in 1979 and Bulgarian sera sampled in 2015 implicated suitability of the non-recently sampled specimens for viral RNA detection.
Viral clearance is typically delayed in HV infection, including infection with EqHV in horses (7). In our study, three serial individual specimens taken at different time points over 2 weeks (May-June 1979) were available from the RNA-positive donkey sampled in France. All three specimens, as well as both individual specimens from Bulgaria, tested positive both for EqHV antibodies (indicated as red dots in Fig. 1B) and RNA, providing evidence against immediate antibody-mediated EqHV clearance in donkeys. However, the co-occurrence of viremia and antibodies as a sign of delayed clearance was apparently much lower in donkeys at 1.1% (3 of 278 antibody-positive animals) than in horses at 2 to 30% (17, 20, 24, 25, 27, 28).
Predominantly acute resolving infections were compatible with a generally lower RNA detection rate in donkeys than in horses. Combining all available data from previous studies on horses (17, 20–25, 27, 28), 148 of 2,172 horses tested positive for EqHV RNA (6.8% [range, 0.9 to 35.5%]) compared to only 3 of 1,047 donkeys or mules when combining the data from this study with previous studies (17, 20–22) (0.3%; corrected χ2 = 65.9, P < 0.00001). The low number of RNA-positive donkeys could not be explained by a putatively low exposure of donkeys to EqHV, since seroprevalence in donkeys was high at 28.3% (278 of 982 donkeys combining this and the only previous serological study [17]), although still significantly lower than in horses at 34.9% (469 of 1,343 horses from all previous studies performing serological analyses; corrected χ2 = 11.1, P < 0.0009). The EqHV seroprevalence increased with the age of donkeys, which was comparable to a study on EqHV in German horses (25) but in contrast to another study on EqHV in Japanese horses (28). Finally, female donkeys were more likely to be seropositive for EqHV than male donkeys. A similar distribution was not observed for horses in two previous studies, one showing no gender-associated differences and another one showing a higher EqHV burden in male horses (25, 28).
Next, we investigated the clinical relevance of EqHV infection in donkeys by determination of aspartate aminotransferase (AST; reference value <536 U/liter), gamma-glutamyl transferase (γGGT; <69 U/liter), and glutamate dehydrogenase (GLDH; <8.2 U/liter) levels in serum of all Bulgarian donkeys (n = 201) as markers of liver damage. As depicted in Fig. 1D, liver enzymes concentrations were mainly within the reference range (29) and were comparable between the seropositive and seronegative groups, including the RNA-positive animals (indicated in color in Fig. 1D), which is in line with the reported subclinical course of infection in horses.
Cross-species transmission of EqHV.
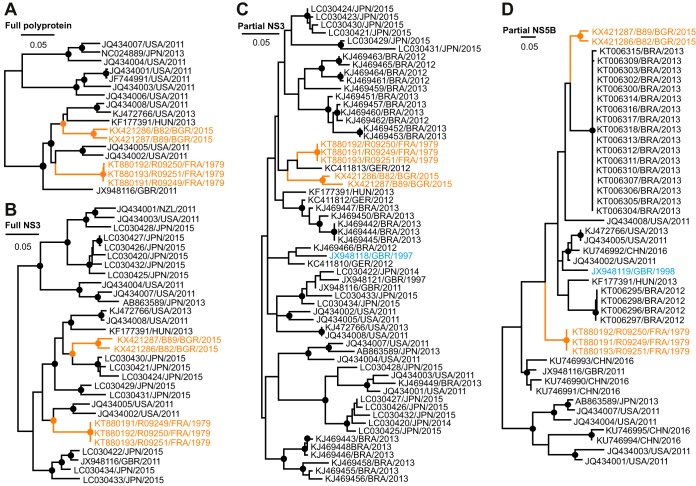
The full viral polyprotein genes were determined for all donkey EqHV strains, including those from the three serial bleedings from the French donkeys and those from the Bulgarian donkey and mule. The polyprotein genes encompassed 8,832 nucleotides from the French donkey EqHV strain, as well as 8,835 and 8,841 nucleotides from the Bulgarian donkey and mule, respectively. Polyprotein length and organization was identical in all cases to that observed before in EqHV from horses with the presence of all typical domains in the order C-E1-E2-p7-NS2-NS3-NS4A/NS4B-NS5A/NS5B. Maximum-likelihood (ML) phylogenetic reconstructions based on the complete polyprotein gene were highly robust, as suggested by high bootstrap support for clusters based on 1,000 replicates. In these ML phylogenetic reconstructions, the novel donkey HVs from France and Bulgaria formed two distinct viral lineages that were not monophyletic. In addition, these donkey HV lineages were interspersed between EqHV from horses and did not cluster in sister relationships to EqHV strains from horses (Fig. 2A). The close phylogenetic relationship between EqHV strains from horses and from donkeys or mules was compatible, with a narrow genetic distance of only 1.8 to 2.5% of the translated polyprotein genes of these strains. Of note, even upon inclusion of the novel donkey viruses, the EqHV patristic distance was only 6.2% on amino acid level in the translated polyprotein gene compared to 33.1% within HCV (calculated using 189 genotype 1 to 7 reference sequences from the Los Alamos National Laboratory [http://hcv.lanl.gov]). However, most of the previous studies on EqHV in horses characterized only short regions of the viral genome. Therefore, we repeated ML reconstructions using different data sets aiming at inclusion of the complete available EqHV genetic diversity without losing too much genetic information. As expected, statistical support for grouping of basal and intermediate nodes was low for the partial NS3 (helicase/protease) and NS5B (RNA-dependent RNA polymerase) domains commonly analyzed in EqHV studies. However, these reconstructions resulted in similar phylogenies, as shown for the complete polyprotein sequences with regard to the phylogenetic relationships between EqHV strains from donkeys and horses (Fig. 2B to D).
FIG 2.
Phylogenetic relationships of EqHV, including the novel donkey hepaciviruses. (A) Maximum-likelihood (ML) phylogeny based on the nucleotide sequences encoding for the complete EqHV polyprotein, including the newly described donkey EqHV strains (orange). Bootstrap values larger than 75% are depicted as filled circles. Taxon designations indicate GenBank accession numbers, country and year of sampling. (B to D) ML phylogenies based on the complete NS3 (1,872 nucleotides), partial NS3 (293 nucleotides), and partial NS5B (261 nucleotides), respectively. Cyan, noncontemporary strains from two horses. Partial NS3 sequences for which fewer than 200 nucleotides were characterized were not included in the analysis shown in panel C to avoid further loss of genomic information and robustness of phylogenetic reconstruction.
To investigate whether potential cross-species transmission was associated with molecular adaptation, we tested for differential selection among horse and donkey EqHV lineages using codon substitution models that allow for various nonsynonymous/synonymous substitution rate ratios (dN/dS) among branches (30). Branches leading to the two common ancestors of donkey EqHV strains showed a lower dN/dS ratio (0.02) compared to the dN/dS ratio among all other branches in the complete genome data set (0.04), indicating no detectable episodic adaptive signal underlying the transmission of EqHV strains from horses to donkeys. Identical results were obtained for the data set encompassing the full NS3, for which a larger number of horse EqHV sequences were available (Fig. 2B), with a dN/dS ratio of 0.0036 in branches leading to donkey EqHV strains compared to 0.0128 among other branches. An analysis using BUSTED confirmed the absence of any signal of gene-wide episodic diversifying selection along the branches leading to the two donkey clades. A FUBAR analysis to identify site-specific selection only indicated two positively selected sites in the complete polyprotein evolutionary history, which do not appear to be related to equine-to-donkey adaptation because the donkey viruses do not share a particular amino acid residue on those positions. In conclusion, the dN/dS ratios suggested that no host adaptation is needed for the mutual infection of horses and donkeys with EqHV.
Intra- and interhost EqHV evolution.
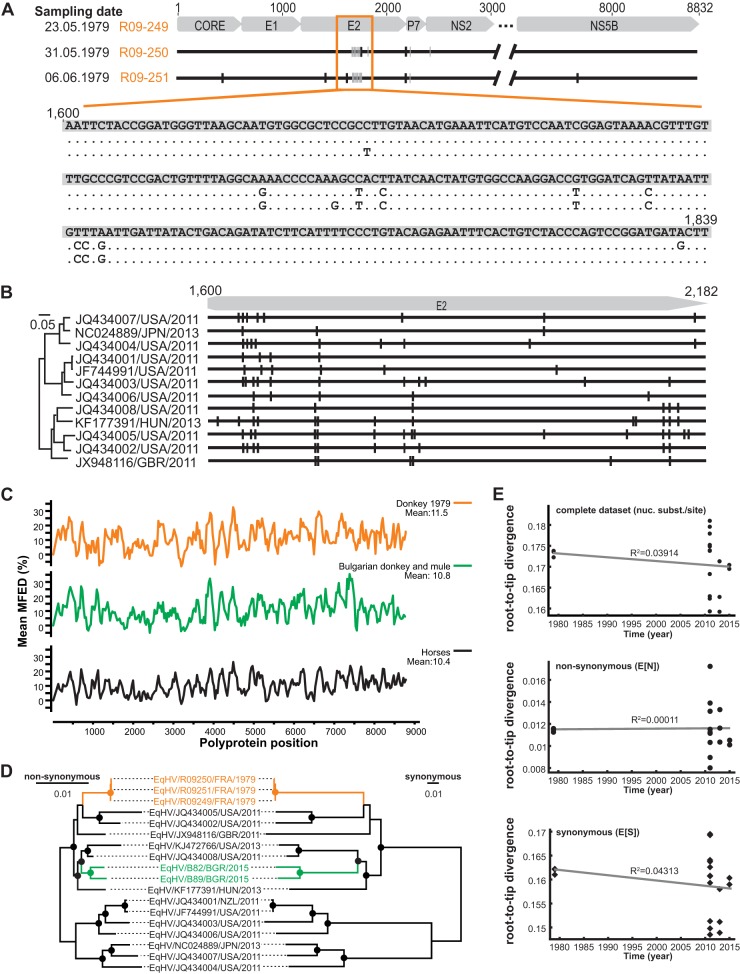
In order to determine EqHV intrahost evolutionary patterns, the complete polyprotein gene sequences of the three serial bleedings available from the French donkey were analyzed. Intrahost variability within this viral gene spanning 8,832 nucleotides was 0.17% (15 substitutions) over 2 weeks (between 23 May and 6 June 1979; Fig. 3A). Similar to HCV, most mutations and in particular the majority of nonsynonymous mutations occurred in the antigenic E2 envelope protein (31), consistent with immune pressure influencing EqHV evolution in the infected animal. However, the majority of the observed mutations did not map to the N-terminal hypervariable E2 region described for HCV (31) but accumulated in the C-terminal region of the E2 gene. To investigate whether indeed EqHV generally differs from HCV in the distribution of nonsynonymous mutations in the E2 gene, the homologous domains of 12 EqHV strains infecting horses were analyzed. As shown in Fig. 3B, EqHV strains infecting horses were similar to HCV in that 72 nonsynonymous mutations accumulated in the N-terminal region of E2, compared to only 28 nonsynonymous mutations in the C-terminal region. The different pattern observed in the EqHV-infected donkey is thus likely due to the small data set available, but potential differences of genomic variability among EqHV hosts cannot be excluded at this point. Finally, reversion of two mutations was detected across the serial bleedings (in the viral E2 and NS2 domains, Fig. 3A), which again is similar to intrahost evolution patterns observed in HCV (32). The predicted similarities in EqHV and HCV evolutionary patterns in combination with the low EqHV patristic distance suggested a limited time of EqHV evolution in equines compared to HCV in humans.
FIG 3.
EqHV evolutionary patterns. (A) Analysis of EqHV polyprotein sequences from three consecutive samples of one EqHV RNA-positive donkey sampled in France. Gray bars, synonymous substitutions; black bars, nonsynonymous substitutions. (B) On the right are the locations of nonsynonymous mutations in the E2 genes of EqHV strains infecting horses. On the left is indicated the ML phylogeny of the translated sequences as before. (C) Mean folding energy differences (MFED) for complete polyprotein sequences of EqHV strains representing both donkey EqHV lineages and all available EqHV polyprotein sequences shown in Fig. 2A. (D) Complete polyprotein ML phylogenies with branch lengths reestimated using either nonsynonymous or synonymous substitutions. Bootstrap values larger than 75% are depicted as filled circles. (E) Root-to-tip divergence plots based on ML trees shown in panel C and Fig. 2A.
However, viral evolution may be limited by noncoding constraints such as genome-scale ordered RNA structures (GORS). Albeit the level of predicted mean folding energy differences (MFEDs; a measure of GORS) across the polyprotein-coding region was slightly higher in donkey EqHV strains than the mean MFEDs within horse EqHV strains, the overall EqHV MFED patterns showed similarities between both equine species in terms of the presence and the extent of predicted stem-loops (Fig. 3C). The overall levels of MFEDs ranging up to 11.5% in our analyses were comparable to previous analyses of EqHV (15, 24) and higher than the 8.5% described before for human HCV (33), which may imply a stronger impact of GORS on EqHV than on HCV evolution (6). However, it seems unlikely that GORS alone can account for the drastic differences between EqHV and HCV genetic diversity.
Lack of temporal signal in EqHV.
The donkey HVs sequences from 1979 represent the oldest EqHV strains described so far. In order to investigate whether these sequences could serve to calibrate the molecular clock of EqHV evolution, root-to-tip distances were analyzed as a function of sampling time. To further investigate whether the temporal signal in EqHV was potentially influenced by evolutionary pressure, root-to-tip distances were compared for complete polyprotein gene trees comprising only nonsynonymous (NS) or synonymous (S) substitutions (Fig. 3D). The complete polyprotein-based tree, as well as the trees with branch lengths reestimated in either NS or S substitutions lacked a molecular clock signal, as visualized by plotting root-to-tip divergence against year of sampling (Fig. 3E). Of note, a lack of temporal signal upon inclusion of the 1979 donkey EqHV strains was consistent with the apical phylogenetic position of two EqHV strains sampled from horses in 1997 and 1998 (20) (shown in cyan in Fig. 2C and D). Unfortunately, only a partial NS3 sequence is available for the 1997 EqHV and only a partial NS5B sequence for the 1998 EqHV strain, preventing their inclusion in our temporal analyses.
DISCUSSION
In this study, we describe wide-reaching infection of donkey populations with EqHV and analyze two divergent donkey EqHV lineages from contemporary and noncontemporary samples.
If EqHV existed with donkeys for prolonged time spans, one could expect that donkeys globally would show signs of infection. However, although infection with EqHV was widespread and frequent according to our data, three populations in Kenya, Israel, and Costa Rica were entirely seronegative. Although this may be linked to the relatively smaller sample sizes (n = 15 to 44), some seropositive animals could be expected in these populations given the 8.1 to 56.7% seroprevalence in other donkey populations. The absence of EqHV infection in these three populations is consistent with the absence of serological signs of EqHV infection in 100 English donkeys (17). The most parsimonious explanation is that EqHV was neither present in the founders of these populations, nor introduced subsequently. Alternative explanations include the extinction of EqHV in these populations together with their hosts. However, the subclinical course of infection of EqHV suggested by the high seroprevalence rates in animals of all ages, the comparable biochemical profiles of seropositive and seronegatvie donkeys, and the limited clinical impact of EqHV on experimentally infected horses (25) do not support the high health costs of EqHV infection in donkeys.
Although the transmission routes of EqHV remain unclear, parenteral transmission is the most likely route based on in vivo infection experiments and comparisons to HCV (25, 34). Our data support frequent horizontal transmission in EqHV-infected populations, potentially aided by human interference, e.g., vaccination or transfusion by veterinarians (28). The higher seroprevalence we found in female donkeys may be compatible with a relevant occurrence of sexual transmission in EqHV. This would be different from HCV, for which sexual transmission is very infrequent (35), and for which detection rates and viral loads are much lower in semen than in blood (summarized in reference 36). Hypothetically, the absence of higher EqHV seroprevalence in female horses than in male horses (25, 28) may be obscured by anthropogenic intervention. Another factor aiding higher seroprevalence in female donkeys may be putatively larger groups held together, compared to more solitary male donkeys. This hypothesis would be consistent with recently described herd-specific EqHV strains from horses in Germany, suggesting focal horizontal and vertical transmission (37). Experimental infections, comparative testing of horse and donkey semen, and additional epidemiological data from both equine species will be necessary to elucidate how EqHV and HCV transmission modes may differ. Furthermore, the reason for the high variability of RNA-positive EqHV infections in horses (17, 20, 24, 25, 27, 28) is not clear yet. The only factors, which have been noticed so far are horse racing and attendance at equestrian sports (21, 22, 25, 28).
The genetic relatedness of donkeys and horses likely facilitated the cross-species transmission events suggested by our data (38). Hypothetically, the similarities in the time of domestication of horses and donkeys 5,000 to 6,000 years ago (18, 39) would have facilitated host shifts between the two equine species. However, the geographically most relevant area for the domestication of horses was likely the Eurasian steppe (18), compared to northeastern Africa for donkeys (40), narrowing the time span of frequent co-occurrence of these two species to more recent times. It would thus be interesting to analyze ancient donkey species for evidence of ancestral EqHV strains, including the wild African ass (E. africanus), which is an evolutionary old species that likely contributed to the development of the widespread domestic donkey (41). However, only few individuals exist nowadays within this species classified as “critically endangered” by the International Union for the Conservation of Nature.
Our phylogenetic analyses provide clear evidence against a potential coevolutionary relationship between EqHV and different equine hosts, which diverged millions of years ago (18, 19). The recent evolutionary history of EqHV thus narrows the time window for putative equine-to-human transmission in the past as an explanation for the origins of HCV (10). Of note, absence of past EqHV infections of humans is consistent with absence of signs of present EqHV infection in different human cohorts (17, 42, 43). A short evolutionary association between equine hosts and EqHV is also consistent with the highly diverse HV lineages found in the genetically related hosts belonging to the order Artiodactyla (cattle). The perissodactylan and artiodactylan lineages clearly did not cospeciate with their hosts (12), and whether both of them are the result of independent cross-species HV transmission events or whether unique host associations can be found for either the perissodactylan or the artiodactylan lineage remains to be determined. Immediate experimental approaches include testing of related host species, e.g., zebras for the Perissodactyla and livestock species like sheep or goats for the Artiodactyla.
Lack of deep-branching monophyletic clusters of EqHV strains from different regions compared to the existence of geographically distinct HCV genotypes (2) are compatible with global virus admixture through human interference, i.e., transport of infected animals or animal products over wide geographic distances. The observation of viral admixture in equids is paralleled by the occurrence of closely related HVs in cattle in Ghana and Germany (12, 44). Probably, the distribution of cattle has undergone anthropogenic change in an extent similar to that of equids. An unrestricted exchange of EqHV strains among horses and donkeys suggested by our phylogenetic data is consistent with the inability to calibrate a molecular clock using EqHV strains sampled in 1979. Of note, our results do not exclude that a clock-like signal may have existed in EqHV ancestors that evolved prior to the viruses analyzed in this study. Similarly, a 40-year interval may be generally insufficient to analyze the EqHV molecular clock. Interestingly, although investigations of the HCV molecular clock have met considerable difficulties (31), a recent study was able to reconcile phylogeny and sampling dates of archived HCV strains from 1953 (45). An interval spanning several decades is thus not generally unsuitable for HV molecular clock analyses. Although we cannot exclude the existence of potentially more diverse EqHV lineages in donkeys, our large sample reached almost half of that of the combined previous studies into horses and extended all of the latter in geographic extent, suggesting the robustness of our evolutionary reconstructions. Limitations of our study that can be circumvented in future prospective studies include inhomogeneous sampling across sites, lack of knowledge on medical treatment and health status of donkeys, as well as their contact to horses.
Finally, EqHV infection patterns in horses and donkeys may differ in the potentially higher ability of donkeys to clear EqHV infection. The first hints at possible explanations originate from strikingly different EqHV RNA and antibody detection rates between different horse breeds. More frequent EqHV infection may be linked to the frequency of veterinary examinations, since valuable race horses and thoroughbreds seem to be particularly often infected by EqHV (21, 25, 28). Alternatively, differences in immune responses influencing viral clearance may occur between different horse breeds, although a generally higher susceptibility to viral infections in thoroughbreds is not supported by data on equine influenza (46). However, our data permit hypotheses on differential immune control of EqHV by different equine species, since donkeys may differ in their immune capacity from horses more than horse breeds from each other (47). Again, alternative explanations that remain to be explored include less intense veterinary handling of donkeys than in more valuable horse species. Beyond investigations of EqHV ecology, our data suggest a unique opportunity to comparatively investigate hepaciviral pathogenesis in a natural host. Here, infection courses can be directly compared by experimentally infecting horses and donkeys with identical EqHV strains, without the need to conduct highly restricted experimental infections of chimpanzees with HCV lacking the simultaneous infection of the human counterpart (48).
In conclusion, our study highlights the impact of evolutionarily guided investigations into viral ecology and offers new possibilities to elucidate factors involved in the development of chronic HV infections.
MATERIALS AND METHODS
Sample collection.
Donkey sera were collected based on availability in France, Germany, Spain, Italy, Bulgaria, Israel, Kenya, Mexico, and Costa Rica from 1974 to 2016. Animal sera were stored at −20 or −80°C prior to analysis. In addition, 53 mule samples were collected in Bulgaria in 2015. Samples were either collected as part of routine examinations (Germany, Italy, Costa Rica, and France) or under permits issued by the responsible authorities. The permit numbers were as follows: Mexico, SICUAE FMVZ-UNAM F. García-Lacy 12042013; Kenya, IACUC 2015.8; Spain, BOJA55-20/2012; Israel, KSVM-VTH/5_2013; Italy, protocol 45/2013/CEISA/COM; and Bulgaria, FVM 15/15. Host designations were assessed for all EqHV RNA-positive specimens from France from 1979 by characterization of the mitochondrial COI gene as described before (49).
Luciferase immunoprecipitation system.
All samples were analyzed for the presence of anti-NS3 antibodies by the previously described LIPS (24). Briefly, sera were diluted 1:10 in buffer A and incubated for 1 h on a rotary shaker. Renilla-NS3 fusion proteins were expressed in Cos1 cells, and 107 relative light units (RLU) were added per well to the diluted sera in a 96-well plate. After incubation for 1 h on a rotary shaker, antibody-antigen complexes were immunoprecipitated by A/G beads and the RLU were determined. Each sample was measured in duplicate wells. The cutoff was calculated by the mean values of wells containing only buffer A, the Renilla-NS3 fusion protein and A/G beads plus three standard deviations as described previously (24). A positive control containing anti-EqHV antibody-positive horse serum was included in each run.
Detection of EqHV RNA.
For the detection of hepaciviral RNA a hemi-nested RT-PCR assay targeting the 5′ UTR was developed based on all available EqHV 5′-UTR sequences. The primer sequences were as follows: HCV-F150, GSWSCYYCYAGGICCMCCCC; HCV-R371, CTCRTGIISYAIGGTCTACRAGRCC; and HCV-R342, GGIGCICTCGCAAGCRYGCCYATCA (I = inosine, S = C/G, W = A/T, Y = C/T, M = A/C, and R = A/G). The limits of detection were determined as the number in probit analyses conducted with SPSS V23 (IBM, Ehningen, Germany) using eight replicates per RNA concentration as described previously (26). The 95% lower limit of detection of the EqHV 5′-UTR assay was 5.7 × 102 RNA copies per reaction (range, 3.8 × 102 to 1.2 × 103), which was well below the commonly observed viral loads in EqHV-infected horses (25). The HV NS3-based assay was described previously (11). Cross-tables were calculated using EpiInfo V7 (http://www.cdc.gov/epiinfo/index.html) and an online tool (http://quantpsy.org/chisq/chisq.htm). Sequencing of the complete EqHV polyprotein genes was performed by amplifying genome-spanning islets with degenerate broadly reactive oligonucleotides as described previously (11). Viral loads were determined by strain-specific quantitative real-time RT-PCR (oligonucleotide sequences available upon request) with photometrically quantified in vitro cRNA transcripts used for calculation of the standard curve as described previously (11).
In silico analyses.
Statistical analyses were done using SPSS V23 (IBM). Sequences were aligned with MAFFT (Geneious 6.1.8). Maximum-likelihood phylogenetic analyses were calculated in MEGA6 (50) and RAxML (51) using a general time reversible model with a discrete gamma distribution and a proportion of invariable sites and 1,000 bootstrap replicates. To estimate branch lengths in synonymous and nonsynonymous substitutions per site, a codon substitution model was applied in HypHy (52) that allows for branch-specific synonymous and nonsynonymous substitution rates (53). PAML (54) was used to fit a codon substitution model that allowed for a different nonsynonymous/synonymous substitution rate ratio (ω) on the branches leading to the two donkey HV common ancestors compared to the ω on the remaining branches (30). In addition, we used BUSTED (55) to search for gene-wide evidence of episodic positive selection along the branches leading to the donkey virus clades, and FUBAR (56) to identify site-specific selection patterns, both implemented in HypHy. Root-to-tip divergence was plotted against sampling time using TempEst (57). Mean folding energy differences were calculated using SSE V1.2 as described previously (12).
Accession number(s).
All polyprotein gene sequences generated in this study were submitted to GenBank under accession numbers KT880191 to KT880193 and KX421286 to KX421287.
ACKNOWLEDGMENTS
We thank Monika Eschbach-Bludau, Sebastian Brünink, and Tobias Bleicker (University of Bonn Medical Centre, Bonn, Germany), Michael Engelmann (Twincore, Hannover, Germany), and Rocio Gonzales Barrientos and Gabriela Hernandez Mora (SENASA Costa Rica) for assistance. We are grateful to Peter D. Burbelo (NIH, Bethesda, MD) for providing the Renilla-luciferase-NS3 fusion plasmid.
S.W. was supported by the Hannover Biomedical Research School and the Centre for Infection Biology (ZIB). E.S. was supported by an intramural young investigator award from the Helmholtz Centre for Infection Research. A.M.S. was supported by a personal scholarship from the German Academic Exchange Service (DAAD). This study was funded by German Research Foundation (DFG) grants STE 1954/1-1 to E.S. and 810/1-1 to J.F.D. and an intramural grant from the University of Bonn (BONFOR) to J.F.D. TWINCORE is a joint venture between the Hannover Medical School (MHH) and the Helmholtz Centre for Infection Research (HZI). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Global Burden of Disease Study. 2015. Global, regional, and national incidence, prevalence, and years lived with disability for 301 acute and chronic diseases and injuries in 188 countries, 1990-2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet 386:743–800. doi: 10.1016/S0140-6736(15)60692-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith DB, Bukh J, Kuiken C, Muerhoff AS, Rice CM, Stapleton JT, Simmonds P. 2014. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: updated criteria and genotype assignment web resource. Hepatology 59:318–327. doi: 10.1002/hep.26744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Messina JP, Humphreys I, Flaxman A, Brown A, Cooke GS, Pybus OG, Barnes E. 2015. Global distribution and prevalence of hepatitis C virus genotypes. Hepatology 61:77–87. doi: 10.1002/hep.27259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Magiorkinis G, Magiorkinis E, Paraskevis D, Ho SY, Shapiro B, Pybus OG, Allain JP, Hatzakis A. 2009. The global spread of hepatitis C virus 1a and 1b: a phylodynamic and phylogeographic analysis. PLoS Med 6:e1000198. doi: 10.1371/journal.pmed.1000198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Markov PV, Pepin J, Frost E, Deslandes S, Labbe AC, Pybus OG. 2009. Phylogeography and molecular epidemiology of hepatitis C virus genotype 2 in Africa. J Gen Virol 90:2086–2096. doi: 10.1099/vir.0.011569-0. [DOI] [PubMed] [Google Scholar]
- 6.Simmonds P. 2013. The origin of hepatitis C virus. Curr Top Microbiol Immunol 369:1–15. [DOI] [PubMed] [Google Scholar]
- 7.Scheel TK, Simmonds P, Kapoor A. 2015. Surveying the global virome: identification and characterization of HCV-related animal hepaciviruses. Antiviral Res 115:83–93. doi: 10.1016/j.antiviral.2014.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pfaender S, Brown RJ, Pietschmann T, Steinmann E. 2014. Natural reservoirs for homologs of hepatitis C virus. Emerg Microbes Infect 3:e21. doi: 10.1038/emi.2014.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Makuwa M, Souquiere S, Telfer P, Leroy E, Bourry O, Rouquet P, Clifford S, Wickings EJ, Roques P, Simon F. 2003. Occurrence of hepatitis viruses in wild-born non-human primates: a 3 year (1998-2001) epidemiological survey in Gabon. J Med Primatol 32:307–314. doi: 10.1046/j.1600-0684.2003.00042.x. [DOI] [PubMed] [Google Scholar]
- 10.Pybus OG, Theze J. 2016. Hepacivirus cross-species transmission and the origins of the hepatitis C virus. Curr Opin Virol 16:1–7. doi: 10.1016/j.coviro.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 11.Drexler JF, Corman VM, Muller MA, Lukashev AN, Gmyl A, Coutard B, Adam A, Ritz D, Leijten LM, van Riel D, Kallies R, Klose SM, Gloza-Rausch F, Binger T, Annan A, Adu-Sarkodie Y, Oppong S, Bourgarel M, Rupp D, Hoffmann B, Schlegel M, Kummerer BM, Kruger DH, Schmidt-Chanasit J, Setien AA, Cottontail VM, Hemachudha T, Wacharapluesadee S, Osterrieder K, Bartenschlager R, Matthee S, Beer M, Kuiken T, Reusken C, Leroy EM, Ulrich RG, Drosten C. 2013. Evidence for novel hepaciviruses in rodents. PLoS Pathog 9:e1003438. doi: 10.1371/journal.ppat.1003438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corman VM, Grundhoff A, Baechlein C, Fischer N, Gmyl A, Wollny R, Dei D, Ritz D, Binger T, Adankwah E, Marfo KS, Annison L, Annan A, Adu-Sarkodie Y, Oppong S, Becher P, Drosten C, Drexler JF. 2015. Highly divergent hepaciviruses from African cattle. J Virol 89:5876–5882. doi: 10.1128/JVI.00393-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Theze J, Lowes S, Parker J, Pybus OG. 2015. Evolutionary and phylogenetic analysis of the hepaciviruses and pegiviruses. Genome Biol Evol 7:2996–3008. doi: 10.1093/gbe/evv202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pybus OG, Gray RR. 2013. Virology: the virus whose family expanded. Nature 498:310–311. doi: 10.1038/498310a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kapoor A, Simmonds P, Gerold G, Qaisar N, Jain K, Henriquez JA, Firth C, Hirschberg DL, Rice CM, Shields S, Lipkin WI. 2011. Characterization of a canine homolog of hepatitis C virus. Proc Natl Acad Sci U S A 108:11608–11613. doi: 10.1073/pnas.1101794108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.El-Attar LM, Mitchell JA, Brooks Brownlie H, Priestnall SL, Brownlie J. 2015. Detection of non-primate hepaciviruses in UK dogs. Virology 484:93–102. doi: 10.1016/j.virol.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lyons S, Kapoor A, Schneider BS, Wolfe ND, Culshaw G, Corcoran B, Durham AE, Burden F, McGorum BC, Simmonds P. 2014. Viraemic frequencies and seroprevalence of non-primate hepacivirus and equine pegiviruses in horses and other mammalian species. J Gen Virol 95:1701–1711. doi: 10.1099/vir.0.065094-0. [DOI] [PubMed] [Google Scholar]
- 18.Outram AK, Stear NA, Bendrey R, Olsen S, Kasparov A, Zaibert V, Thorpe N, Evershed RP. 2009. The earliest horse harnessing and milking. Science 323:1332–1335. doi: 10.1126/science.1168594. [DOI] [PubMed] [Google Scholar]
- 19.Orlando L, Ginolhac A, Zhang G, Froese D, Albrechtsen A, Stiller M, Schubert M, Cappellini E, Petersen B, Moltke I, Johnson PL, Fumagalli M, Vilstrup JT, Raghavan M, Korneliussen T, Malaspinas AS, Vogt J, Szklarczyk D, Kelstrup CD, Vinther J, Dolocan A, Stenderup J, Velazquez AM, Cahill J, Rasmussen M, Wang X, Min J, Zazula GD, Seguin-Orlando A, Mortensen C, Magnussen K, Thompson JF, Weinstock J, Gregersen K, Roed KH, Eisenmann V, Rubin CJ, Miller DC, Antczak DF, Bertelsen MF, Brunak S, Al-Rasheid KA, Ryder O, Andersson L, Mundy J, Krogh A, Gilbert MT, Kjaer K, Sicheritz-Ponten T, Jensen LJ, Olsen JV, Hofreiter M, Nielsen R, Shapiro B, Wang J, Willerslev E. 2013. Recalibrating Equus evolution using the genome sequence of an early Middle Pleistocene horse. Nature 499:74–78. doi: 10.1038/nature12323. [DOI] [PubMed] [Google Scholar]
- 20.Lyons S, Kapoor A, Sharp C, Schneider BS, Wolfe ND, Culshaw G, Corcoran B, McGorum BC, Simmonds P. 2012. Nonprimate hepaciviruses in domestic horses, United Kingdom. Emerg Infect Dis 18:1976–1982. doi: 10.3201/eid1812.120498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gemaque BS, Junior Souza de Souza A, do Carmo Pereira Soares M, Malheiros AP, Silva AL, Alves MM, Gomes-Gouvea MS, Pinho JR, Ferreira de Figueiredo H, Ribeiro DB, Souza da Silva J, Moraes LA, Ribeiro AS, Pereira WL. 2014. Hepacivirus infection in domestic horses, Brazil, 2011-2013. Emerg Infect Dis 20:2180–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu G, Sun L, Xu T, He D, Wang Z, Ou S, Jia K, Yuan L, Li S. 2016. First description of hepacivirus and pegivirus infection in domestic horses in China: a study in Guangdong Province, Heilongjiang Province, and Hong Kong District. PLoS One 11:e0155662. doi: 10.1371/journal.pone.0155662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scheel TK, Kapoor A, Nishiuchi E, Brock KV, Yu Y, Andrus L, Gu M, Renshaw RW, Dubovi EJ, McDonough SP, Van de Walle GR, Lipkin WI, Divers TJ, Tennant BC, Rice CM. 2015. Characterization of nonprimate hepacivirus and construction of a functional molecular clone. Proc Natl Acad Sci U S A 112:2192–2197. doi: 10.1073/pnas.1500265112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burbelo PD, Dubovi EJ, Simmonds P, Medina JL, Henriquez JA, Mishra N, Wagner J, Tokarz R, Cullen JM, Iadarola MJ, Rice CM, Lipkin WI, Kapoor A. 2012. Serology-enabled discovery of genetically diverse hepaciviruses in a new host. J Virol 86:6171–6178. doi: 10.1128/JVI.00250-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pfaender S, Cavalleri JM, Walter S, Doerrbecker J, Campana B, Brown RJ, Burbelo PD, Postel A, Hahn K, Anggakusuma Riebesehl N, Baumgartner W, Becher P, Heim MH, Pietschmann T, Feige K, Steinmann E. 2015. Clinical course of infection and viral tissue tropism of hepatitis C virus-like nonprimate hepaciviruses in horses. Hepatology 61:447–459. doi: 10.1002/hep.27440. [DOI] [PubMed] [Google Scholar]
- 26.Drexler JF, Kupfer B, Petersen N, Grotto RM, Rodrigues SM, Grywna K, Panning M, Annan A, Silva GF, Douglas J, Koay ES, Smuts H, Netto EM, Simmonds P, Pardini MI, Roth WK, Drosten C. 2009. A novel diagnostic target in the hepatitis C virus genome. PLoS Med 6:e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanaka T, Kasai H, Yamashita A, Okuyama-Dobashi K, Yasumoto J, Maekawa S, Enomoto N, Okamoto T, Matsuura Y, Morimatsu M, Manabe N, Ochiai K, Yamashita K, Moriishi K. 2014. Hallmarks of hepatitis C virus in equine hepacivirus. J Virol 88:13352–13366. doi: 10.1128/JVI.02280-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsuu A, Hobo S, Ando K, Sanekata T, Sato F, Endo Y, Amaya T, Osaki T, Horie M, Masatani T, Ozawa M, Tsukiyama-Kohara K. 2015. Genetic and serological surveillance for non-primate hepacivirus in horses in Japan. Vet Microbiol 179:219–227. doi: 10.1016/j.vetmic.2015.05.028. [DOI] [PubMed] [Google Scholar]
- 29.Burden FA, Hazell-Smith E, Mulugeta G, Patrick V, Trawford R, Brooks-Brownlie HW. 2016. Reference intervals for biochemical and haematological parameters in mature domestic donkeys (Equus asinus) in the UK. Equine Vet Educ 28:134–139. doi: 10.1111/eve.12512. [DOI] [Google Scholar]
- 30.Yang Z. 1998. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol Biol Evol 15:568–573. doi: 10.1093/oxfordjournals.molbev.a025957. [DOI] [PubMed] [Google Scholar]
- 31.Gray RR, Parker J, Lemey P, Salemi M, Katzourakis A, Pybus OG. 2011. The mode and tempo of hepatitis C virus evolution within and among hosts. BMC Evol Biol 11:131. doi: 10.1186/1471-2148-11-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pfafferott K, Gaudieri S, Ulsenheimer A, James I, Heeg M, Nolan D, John M, Rauch A, Mallal S, Lucas A, Klenerman P, Diepolder HM, Lucas M. 2011. Constrained pattern of viral evolution in acute and early HCV infection limits viral plasticity. PLoS One 6:e16797. doi: 10.1371/journal.pone.0016797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simmonds P, Tuplin A, Evans DJ. 2004. Detection of genome-scale ordered RNA structure (GORS) in genomes of positive-stranded RNA viruses: implications for virus evolution and host persistence. RNA 10:1337–1351. doi: 10.1261/rna.7640104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramsay JD, Evanoff R, Wilkinson TE Jr, Divers TJ, Knowles DP, Mealey RH. 2015. Experimental transmission of equine hepacivirus in horses as a model for hepatitis C virus. Hepatology 61:1533–1546. doi: 10.1002/hep.27689. [DOI] [PubMed] [Google Scholar]
- 35.Thomas DL. 2013. Global control of hepatitis C: where challenge meets opportunity. Nat Med 19:850–858. doi: 10.1038/nm.3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turner SS, Gianella S, Yip MJ-S, van Seggelen WO, Gillies RD, Foster AL, Barbati ZR, Smith DM, Fierer DS. 2016. Shedding of hepatitis C virus in semen of human immunodeficiency virus-infected men. Open Forum Infect Dis 3:ofw057. doi: 10.1093/ofid/ofw057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gather T, Walter S, Todt D, Pfaender S, Brown RJ, Postel A, Becher P, Moritz A, Hansmann F, Baumgaertner W, Feige K, Steinmann E, Cavalleri JV. 2016. Vertical transmission of hepatitis C virus-like nonprimate hepacivirus in horses. J Gen Virol 97:e1–12. [DOI] [PubMed] [Google Scholar]
- 38.Longdon B, Brockhurst MA, Russell CA, Welch JJ, Jiggins FM. 2014. The evolution and genetics of virus host shifts. PLoS Pathog 10:e1004395. doi: 10.1371/journal.ppat.1004395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rossel S, Marshall F, Peters J, Pilgram T, Adams MD, O'Connor D. 2008. Domestication of the donkey: timing, processes, and indicators. Proc Natl Acad Sci U S A 105:3715–3720. doi: 10.1073/pnas.0709692105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beja-Pereira A, England PR, Ferrand N, Jordan S, Bakhiet AO, Abdalla MA, Mashkour M, Jordana J, Taberlet P, Luikart G. 2004. African origins of the domestic donkey. Science 304:1781. doi: 10.1126/science.1096008. [DOI] [PubMed] [Google Scholar]
- 41.Kimura B, Marshall FB, Chen S, Rosenbom S, Moehlman PD, Tuross N, Sabin RC, Peters J, Barich B, Yohannes H, Kebede F, Teclai R, Beja-Pereira A, Mulligan CJ. 2011. Ancient DNA from Nubian and Somali wild ass provides insights into donkey ancestry and domestication. Proc R Soc B Biol Sci 278:50–57. doi: 10.1098/rspb.2010.0708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pfaender S, Walter S, Todt D, Behrendt P, Doerrbecker J, Wolk B, Engelmann M, Gravemann U, Seltsam A, Steinmann J, Burbelo PD, Klawonn F, Feige K, Pietschmann T, Cavalleri JM, Steinmann E. 2015. Assessment of cross-species transmission of hepatitis C virus-related non-primate hepacivirus in a population of humans at high risk of exposure. J Gen Virol 96:2636–2642. doi: 10.1099/vir.0.000208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Levi JE, Cabral SP, Nishiya A, Ferreira S, Romano CM, Polite MB, Pereira RA, Mota MA, Kutner JM. 2014. Absence of nonprimate hepacivirus-related genomes in blood donors seroreactive for hepatitis C virus displaying indeterminate blot patterns. J Viral Hepat 21:e164–166. doi: 10.1111/jvh.12252. [DOI] [PubMed] [Google Scholar]
- 44.Baechlein C, Fischer N, Grundhoff A, Alawi M, Indenbirken D, Postel A, Baron AL, Offinger J, Becker K, Beineke A, Rehage J, Becher P. 2015. Identification of a novel hepacivirus in domestic cattle from Germany. J Virol 89:7007–7015. doi: 10.1128/JVI.00534-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gray RR, Tanaka Y, Takebe Y, Magiorkinis G, Buskell Z, Seeff L, Alter HJ, Pybus OG. 2013. Evolutionary analysis of hepatitis C virus gene sequences from 1953. Philos Trans R Soc Lond B Biol Sci 368:20130168. doi: 10.1098/rstb.2013.0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nyaga PN, Wiggins AD, Priester WA. 1980. Special Issue on animal and human influenzas epidemiology of equine influenza, risk by age, breed and sex. Comp Immunol Microbiol Infect Dis 3:67–73. doi: 10.1016/0147-9571(80)90040-5. [DOI] [PubMed] [Google Scholar]
- 47.Huang J, Zhao Y, Bai D, Shiraigol W, Li B, Yang L, Wu J, Bao W, Ren X, Jin B, Zhao Q, Li A, Bao S, Bao W, Xing Z, An A, Gao Y, Wei R, Bao Y, Bao T, Han H, Bai H, Bao Y, Zhang Y, Daidiikhuu D, Zhao W, Liu S, Ding J, Ye W, Ding F, Sun Z, Shi Y, Zhang Y, Meng H, Dugarjaviin M. 2015. Donkey genome and insight into the imprinting of fast karyotype evolution. Sci Rep 5:14106. doi: 10.1038/srep14106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bukh J. 2012. Animal models for the study of hepatitis C virus infection and related liver disease. Gastroenterology 142:1279–1287. doi: 10.1053/j.gastro.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 49.Alcaide M, Rico C, Ruiz S, Soriguer R, Munoz J, Figuerola J. 2009. Disentangling vector-borne transmission networks: a universal DNA barcoding method to identify vertebrate hosts from arthropod bloodmeals. PLoS One 4:e7092. doi: 10.1371/journal.pone.0007092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pond SL, Frost SD, Muse SV. 2005. HyPhy: hypothesis testing using phylogenies. Bioinformatics 21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- 53.Lemey P, Kosakovsky Pond SL, Drummond AJ, Pybus OG, Shapiro B, Barroso H, Taveira N, Rambaut A. 2007. Synonymous substitution rates predict HIV disease progression as a result of underlying replication dynamics. PLoS Comput Biol 3:e29. doi: 10.1371/journal.pcbi.0030029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang Z. 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci 13:555–556. [DOI] [PubMed] [Google Scholar]
- 55.Murrell B, Weaver S, Smith MD, Wertheim JO, Murrell S, Aylward A, Eren K, Pollner T, Martin DP, Smith DM, Scheffler K, Kosakovsky-Pond SL. 2015. Gene-wide identification of episodic selection. Mol Biol Evol 32:1365–1371. doi: 10.1093/molbev/msv035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murrell B, Moola S, Mabona A, Weighill T, Sheward D, Kosakovsky Pond SL, Scheffler K. 2013. FUBAR: a fast, unconstrained Bayesian approximation for inferring selection. Mol Biol Evol 30:1196–1205. doi: 10.1093/molbev/mst030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rambaut A, Lam TT, Max Carvalho L, Pybus OG. 2016. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen). Virus Evol 2:e1–e7. [DOI] [PMC free article] [PubMed] [Google Scholar]