ABSTRACT
We recently reported that the segment-specific noncoding regions (NCRs) of the hemagglutinin (HA) and neuraminidase (NA) segments are subtype specific, varying significantly in sequence and length at both the 3′ and 5′ ends. Interestingly, we found that nucleotides CC at positions 13 and 14 at the 3′ end and GUG at positions 14 to 16 at the 5′ end (termed 14′ and 16′ to distinguish them from 3′ positions) are absolutely conserved among all HA subtype-specific NCRs. These HA segment-specific NCR nucleotides are located in the extended duplex region of the viral RNA promoter. In order to understand the significance of these highly conserved HA segment-specific NCR nucleotides in the virus life cycle, we performed extensive mutagenesis on the HA segment-specific NCR nucleotides and studied their functional significance in regulating influenza A virus replication in the context of the HA segment with both RNP reconstitution and virus infection systems. We found that the base pairing of the 3′-end 13 position with the 5′-end 14′ position (3′13-5′14′) position is critical for RNA promoter activity while the identity of the base pair is critical in determining HA segment packaging. Moreover, the identity of the residue at the 3′-end 14 position is functionally more important in regulating virus genome packaging than in regulating viral RNA synthesis. Taken together, these results demonstrated that the HA segment-specific NCR nucleotides in the extended duplex region of the promoter not only form part of the promoter but also play a key role in controlling virus selective genome packaging.
IMPORTANCE The segment-specific complementary nucleotides (13 to 15 in the 3′ end and 14′ to 16′ in the 5′ end) in the extended duplex region of the influenza virus RNA promoter vary significantly among different segments and have rarely been studied. Here, we performed mutagenesis analysis of the highly conserved HA segment-specific nucleotides in the extended duplex region and examined their effects on virus replication in the context of the influenza A/WSN/33 (WSN) virus infection. We found that these HA segment-specific nucleotides not only act as a part of the RNA promoter but also play a critical role in HA segment packaging. Therefore, we showed experimentally, for the first time, the requirement of the nucleotides in the extended duplex region for the RNA promoter and also identified specific noncoding residues in regulating HA segment packaging. This work has implications for the development of attenuated vaccine strains and for elucidation the mechanisms of the virus genome packaging.
KEYWORDS: HA segment-specific noncoding nucleotides, influenza A virus, viral RNA promoter, genome packaging
INTRODUCTION
Influenza A virus has long been a major pathogen causing significant concerns to human health. Further understanding the molecular basis for the influenza A virus replication strategies is still of great importance to prevent and control influenza epidemics or pandemics. The genome of influenza A virus consists of eight single-stranded RNA segments (virion RNA, vRNA) in the negative sense, designated PB2, PB1, PA, hemagglutinin (HA), nucleoprotein (NP), neuraminidase (NA), M, and NS segments (1). Unlike the single-stranded RNA viruses, the eight vRNA segments of influenza A virus contain different noncoding regions (NCRs) varying from 17 to 60 nucleotides at the two extremities of their open reading frames (ORFs) (1, 2). The noncoding region of each segment consists of a highly conserved promoter region followed by the segment-specific NCR at the 3′ and 5′ ends (1, 3, 4). An evolutionary analysis of the influenza virus NCR has demonstrated that the segment-specific noncoding region has extremely low divergence and that its evolutionary rate is lower than that of the coding region (5). It is well known that the NCRs of influenza virus play important roles during synthesis of the three species of viral RNAs (vRNA, cRNA, and mRNA) and during selective genome packaging (6, 7). However, the detailed mechanisms need further clarification.
The promoter region of the influenza virus NCR is formed by the terminal 12 and 13 nucleotides (nt) at 3′ and 5′ NCRs that are highly conserved among different RNA segments and virus strains (8). According to the two-dimensional (2D) corkscrew model proposed for the vRNA promoter and recent advances in three-dimensional (3D) viral ribonucleoprotein (vRNP) structure analyses, the noncoding nucleotides at the 3′ and 5′ ends fold back and base pair to form a duplex region followed by terminal stem-loop structures that are bound by the heterotrimeric RNA-dependent RNA polymerase (PB2, PB1, and PA) (9–14). The duplex region, formed by the nucleotides at positions 10 to 12 in the 3′ terminus (3′-UCC-5′) and positions 11′ to 13′ in the 5′ terminus (5′-AGG-3′) (nucleotide positions at the 5′ end of the promoter are designated with a prime to distinguish them from positions at the 3′ end) (3, 15), has been shown to be necessary for cap snatching (16), transcription initiation (10), polyadenylation (17), and virus gene packaging (18). According to the sequence of the segment-specific NCR, the duplex region is most likely to be further extended by 1 to 4 base pairs by segment-specific nucleotides, and the identities of the base pairs vary among different segments (8, 19). The biological significance of the extended duplex region for each segment during virus replication is still unclear.
We recently performed systematic bioinformatics analyses and found that nucleotides in the HA segment-specific NCRs are also subtype specific and vary significantly in sequence and length among 16 different HA subtypes. Interestingly, we found that among these HA subtype-specific NCRs, nucleotides CC at 3′-end positions 13 and 14 and GUG at 5′-end positions 14′ to 16′, located in the extended duplex region of the viral RNA promoter, are absolutely conserved (4, 15, 20). In order to understand the biological significance of these HA segment-specific nucleotides, we conducted a mutagenic analysis on these HA segment-specific nucleotides and examined their effects on viral RNA synthesis and virion RNA packaging. We found that these nucleotides play not only an important role in ensuring the activities of viral RNA promoter but also a critical role in determining the packaging efficiency of the HA segment into progeny viruses during virus multiplication in cell culture.
RESULTS
Nucleotides at positions 13 to 14 at the 3′ end and positions 14′ to 16′ at the 5′ end are highly conserved among all subtype-specific NCRs of HA segments.
We have recently bioinformatically analyzed the NCRs of all eight segments of influenza A viruses and found that the segment-specific NCRs of HA and NA segments are also subtype specific (4). We also noticed that the nucleotides at positions 13 to 14 at the 3′ end and 14′ to 16′ at the 5′ end are highly conserved among all subtype-specific NCRs of the HA segment (referred to as HA segment-specific NCR nucleotides) but not in the NA segment (4, 15). In the corkscrew model of influenza A virus promoter, a duplex region is formed by Watson-Crick base pairing between 3′-end positions 10 to 12 and 5′-end positions 11′ to 13′ (2, 3, 21) (Fig. 1A). The evolutionary analyses of NCRs of influenza viruses have shown that the duplex region could be extended by one to four Watson-Crick base pairs by segment-specific NCRs: 4 bp for the PA segment; 3 bp for the PB2, NP, NA, M and NS segments; 2 bp for the PB1 segment; and only 1 bp for the HA segment (Table 1). According to the sequence, the duplex region of all HA segments is extended by only a C-G base pairing of the 3′-end 13 position with the 5′-end 14′ position (3′13-5′14′) (Fig. 1A). The nucleotide C at the 3′-end position 14 is not paired with the U at the 5′-end position 15′. The G at the 5′-end position 16′ is facing either U or A at the 3′-end position 15 (4, 15, 20).
FIG 1.
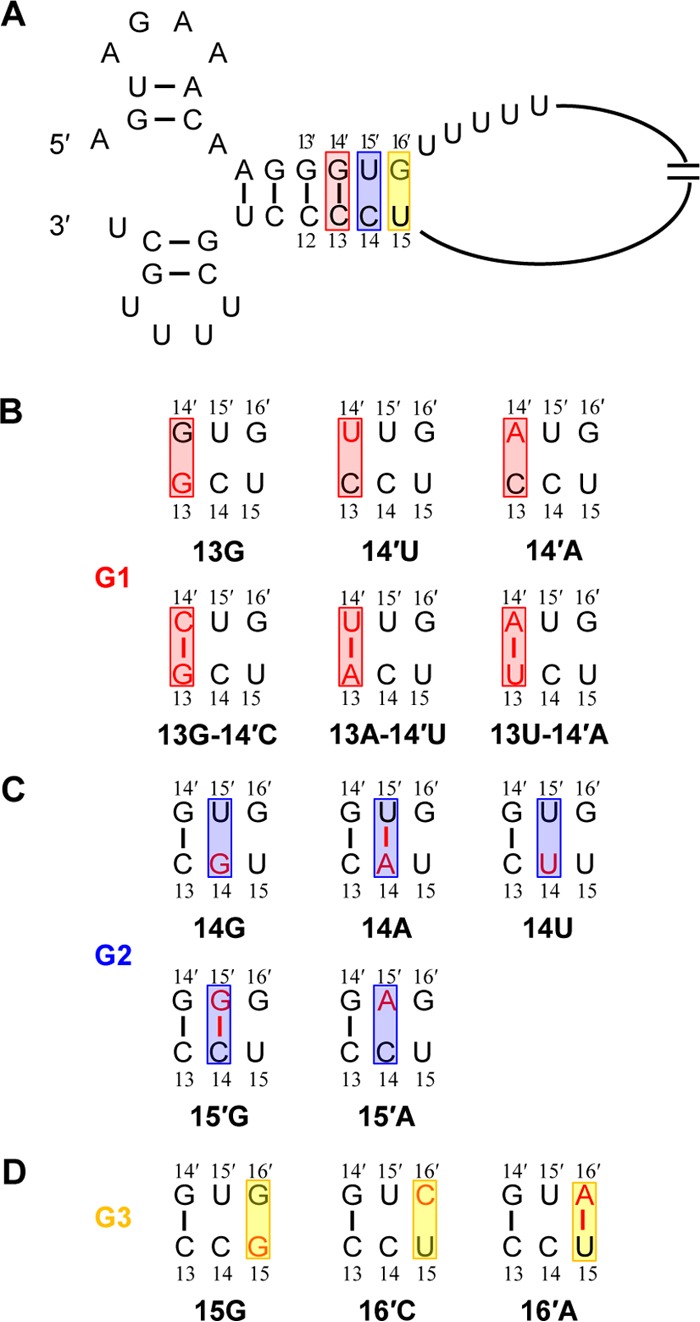
The HA segment-specific nucleotides in the extended duplex region of the viral RNA promoter and the mutagenesis strategies used in this study. (A) Representation of HA segment-specific nucleotides next to the viral RNA promoter in the corkscrew configuration. The common stretch of uridine residues is also shown. The extended duplex region potentially formed by the HA segment-specific nucleotides is highlighted with red, blue, and yellow rectangles at the 13-14′, 14-15′, and 15-16′, positions, respectively. Base pairs are indicated with a connecting line. (B to D) Mutations introduced at the HA segment-specific nucleotide sites. Mutations made at the 3′-end position 13 and/or 5′-end position 14′ belong to the G1 mutants. Mutations made at the 3′-end position 14 and/or 5′-end position 15′ belong to the G2 mutants. Mutations made at 3′-end position 15 and/or 5′-end position 16′ belong to the G3 mutants.
TABLE 1.
Conservative nucleotides and base pairs at 3′-end positions 13 to 15 and 5′-end positions 14′ to 16′
| Segment and/or subtype(s) | 3′-end positions 13–15 | 5′-end positions 14′–16′ | Base pairing at the indicated positionb |
||
|---|---|---|---|---|---|
| 13-14′ | 14-15′ | 15-16′ | |||
| PB2 | AGU | UCG | √ | √ | × |
| PB1 | GUU | CAU | √ | √ | × |
| PA | AUG | UAC | √ | √ | √ |
| NP | CAU | GUA | √ | √ | √ |
| M | AUC | UAC | √ | √ | × |
| NS | CAC | GUG | √ | √ | √ |
| HA | |||||
| 1–16 | CCXa | GUG | √ | × | √ or × |
| NA | |||||
| 1, 2, 4, 5, 8 | UCA | AGU | √ | √ | √ |
| 6, 7 | CAC | GUG | √ | √ | √ |
| 3 | ACG | UGC | √ | √ | √ |
| 9 | CAG | GUC | √ | √ | √ |
X represents either A or U at position 15 in the 3′ end that is variable among different HA subtype segments.
The presence (√) or absence (×) of base pairing is indicated.
Mutagenesis of the HA segment-specific NCR nucleotides and examinations of their effects on virus RNA synthesis and protein production.
Since the HA segment-specific NCR nucleotides are located in the extended duplex region of the promoter, we mutated these nucleotides and analyzed their effects on viral RNA synthesis and protein production initially. We divided mutants into three groups (named G1, G2, and G3) according to their locations in forming potential Watson-Crick base pairs (Fig. 1A). All mutations made at the 3′-end position 13 and/or 5′-end position 14′ belong to the G1 mutants. Three point mutations, 13G, 14′U, and 14′A, were constructed to destroy the base pairs. To reform alternative base pairs, additional double mutants were generated by introducing a complementary base at the corresponding positions (13G-14′C, 13A-14′U, and 13U-14′A) (Fig. 1B). The G2 mutants contain the mutations made at either the 3′-end position 14 or 5′-end position 15′. We introduced 14G, 14A, 14U, 15′G, and 15′A, in which a Watson-Crick base pair could be formed at position 14-15′ for the 14A and 15′G mutants (Fig. 1C). In the G3 mutants, we constructed two point mutations (16′C and 16′A) at 5′-end position 16′ and a U-to-G mutation (15G) at 3′-end position 15 (Fig. 1D).
We then examined the capabilities of these mutants on viral RNA synthesis and protein production in an RNP reconstitution system. The pPol I-HA-WT (where Pol I is polymerase I and WT is wild type)-based RNA-expressing plasmids containing the corresponding mutations shown in Fig. 1 were individually cotransfected with pcDNA-PB2, pcDNA-PB1, pcDNA-PA, and pcDNA-NP plasmid in 293T cells. At 24 h posttransfection, the total RNAs of the cells were extracted, and levels of three species of viral RNAs (mRNA, cRNA, and vRNA) were detected by primer extension analysis. In the meantime, we also performed Western blot analysis to examine the levels of HA production with the above crude lysates of the transfected cells.
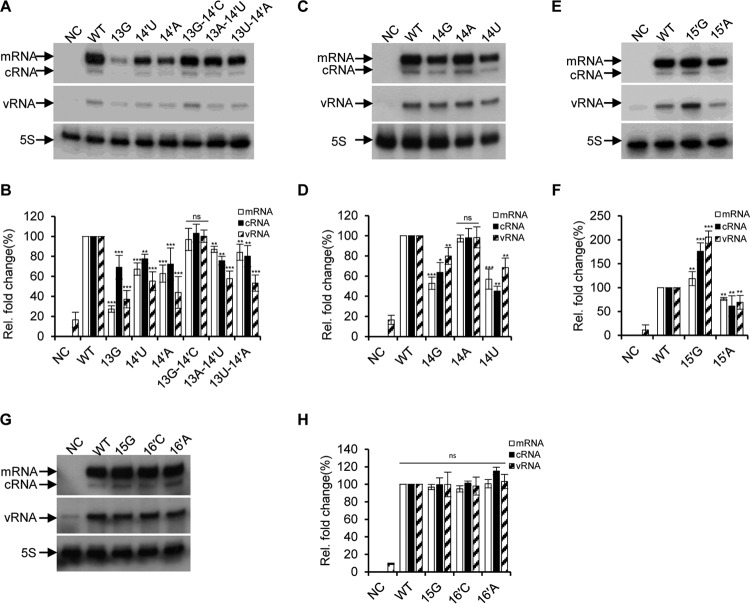
Among the group 1 (G1) mutants, the 13G, 14′U, and 14′A mutants that disrupted the 13C-14′G base pairing resulted in significantly reduced RNA synthesis of their corresponding RNPs. In contrast, the double mutants that reformed the base pair at positions 13 and 14′ (13G-14′C, 13A-14′U, and 13U-14′A) rescued RNA synthesis more or less. It can be seen that the 13G-14′C mutant completely recovered the RNA levels to the wild-type level, while the 13A-14′U and 13U-14′A mutants increased the levels of RNA synthesis in comparison with that of the single 14′U and 14′A mutants (Fig. 2A and B). Primer extension of the G2 mutants showed that 14G, 14U, and 15′A mutants, which could not form a base pair at the 14-15′ position resulted in reduced viral RNA (mRNA, cRNA, and vRNA) synthesis. In contrast, the 14A and 15′G mutants that allowed formation of a base pair at the 14-15′ position showed significantly increased viral RNA levels: the 14A showed comparable levels to that of the wild type, and 15′G mediated even higher RNA levels (especially for cRNA and vRNA levels) than the wild-type levels (Fig. 2C to F). In comparison with the G1 and G2 mutants, all G3 mutants made at the 3′-end 15 position and 5′-end 16′ position did not affect viral RNA synthesis (Fig. 2G and H).
FIG 2.
Effects of nucleotide substitutions at the 3′-end positions 13 to 15 and 5′-end positions 14′ to 16′ on RNA synthesis in an RNP reconstitution system. (A, C, E, and G) Primer extension analysis to assess the RNA levels derived from wild-type and mutant templates as showed in Fig. 1 in a PR8-derived RNP reconstitution system. The wild-type and mutant RNPs were reconstituted in 293T cells, and the levels of mRNA, cRNA, and vRNA were detected by a primer extension assay at 24 h posttransfection. The vRNA levels in negative-control (NC) lanes represent the viral RNA template transcribed from the input pPolI plasmid. (B, D, F and H) Quantification of the levels of mRNA, cRNA, and vRNA in panels A, C, E, and G by phosphorimager analysis. Values were normalized against cellular 5S RNA. The data are presented as means ± standard deviations of three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, nonsignificant.
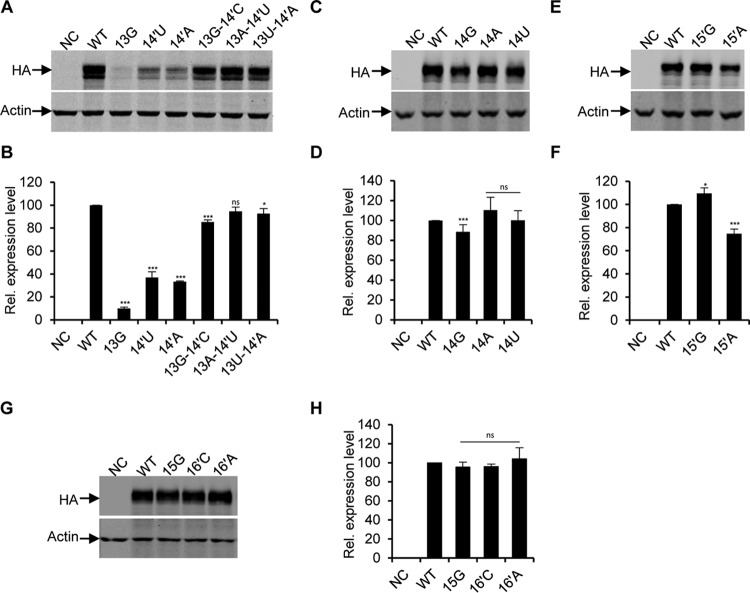
In order to further examine the levels of protein synthesis, we performed Western blotting on protein samples from the above RNP transfected cells. We found that the levels of HA protein expressed by the above RNPs are generally consistent with the mRNA levels (Fig. 3), except that a slight reduction (<10%) in HA protein synthesis was observed for the 13G-14′C mutants. These results imply that mutations made in the extended duplex region generally are not involved in regulating gene expression at the translation level. Taking these observations together, we concluded that the base pairing between the 13-14′ position is primarily important for the promoter activity, while a base pairing at 14-15′ is favored, and a base pairing at 15-16′ is not required for viral RNA synthesis.
FIG 3.
Effects of nucleotide substitutions at 3′-end positions 13 to 15 and 5′-end positions 14′ to 16′ on protein productions in the RNP reconstitution system. (A, C, E, and G) Western blotting to examine the HA proteins expressed by the wild-type and mutant RNPs (the same as above). The wild-type and mutant RNPs were reconstituted in 293T cells individually, cells were harvested at 24 h posttransfection, and lysates were analyzed by Western blotting; β-actin was detected as a loading control. (B, D, F, and H) Quantification of the HA protein levels obtained from the experiments shown in panels A, C, E, and G by densitometry analysis, respectively. The values were expressed as percentages relative to the level of the HA protein expressed from the wild-type RNP system. Data in panels B, D, F, and H represent the means ± standard deviations of three independent experiments. *, P < 0.05; ***, P < 0.001; ns, nonsignificant.
The effects of the HA-specific NCR nucleotides on virus replication.
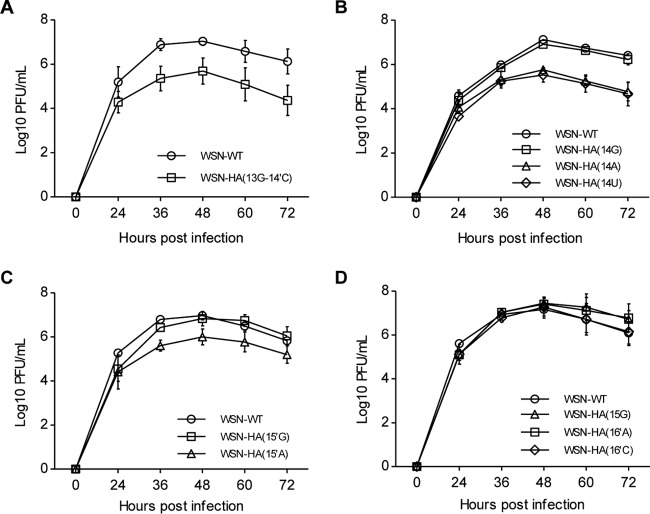
We then further examined the virus replication efficiencies with the HA segment-specific NCR nucleotide mutants in the context of virus infection. We attempted to rescue recombinant influenza A viruses containing all G1 to G3 mutations within an influenza A/WSN/33 (WSN) virus background (22). After performing five independent rescue experiments, we found that, among all G1, G2, and G3 mutants, none of the G1 mutants except for the 13G-14′C could be rescued, and all G2 and G3 mutants could be rescued. The inability to rescue most of the G1 mutants was further confirmed within the influenza A/PuertoRico/8/34 (PR8) virus rescue system (data not shown). These results indicate that nucleotides at the 13-14′ position are more critical than the nucleotides at other positions within the extended duplex region for virus survival. We then examined virus growth efficiencies of all rescued mutant viruses in MDCK cells. It can be seen in Fig. 4 that the growth in MDCK cells of the only rescued G1 mutant, WSN-HA(13G-14′C) virus, was reduced by approximately 1 log unit compared to that of wild-type WSN (maximum titer of 4.9 × 105 versus 1.1 × 107) (Fig. 4A). Among the G2 mutants, we also found that virus containing a 14A, 14U, or 15′A mutation [WSN-HA(14A), WSN-HA(14U), or WSN-HA(15′A)] was attenuated approximately 1 log compared to the level of the wild-type virus, while viruses containing the 14G or 15′G mutation replicated as efficiently as wild-type virus (Fig. 4B and C). In contrast, all mutations made at the 3′-end position 15 and 5′-end position 16′ had no detectable effect on virus growth (Fig. 4D).
FIG 4.
Effects of nucleotide substitutions at 3′-end positions 13 to 15 and 5′-end positions 14′ to 16′ on virus replication efficiency. (A to D) Growth curves of the wild-type WSN virus and recombinant viruses with the indicated mutation(s). A total of 5 × 105 MDCK cells were infected with the indicated viruses at an MOI of 0.001. The numbers of infectious particles in supernatants were determined at the indicated time points by plaque assay in MDCK cells. The data represent means ± standard deviations of three independent experiments.
Considering the effects of the G1, G2, and G3 mutants on viral RNA synthesis, the most surprises came from the 13A-14′U, 13U-14′A, 13G-14′C, and 14A mutants. They showed viral RNA synthesis capabilities comparable to those of the wild type, whereas the 13A-14′U and 13U-14′A mutations were fatal while the 13G-14′C and 14A mutants reduced virus replication efficiencies significantly (Fig. 4A and B). Therefore, we concluded that in addition to the effects on viral RNA synthesis, the mutations at the 13-14′ and 14 positions could also be involved in regulating other aspects of the virus life cycle.
The effects of the HA segment-specific NCR nucleotides on virion RNA packaging.
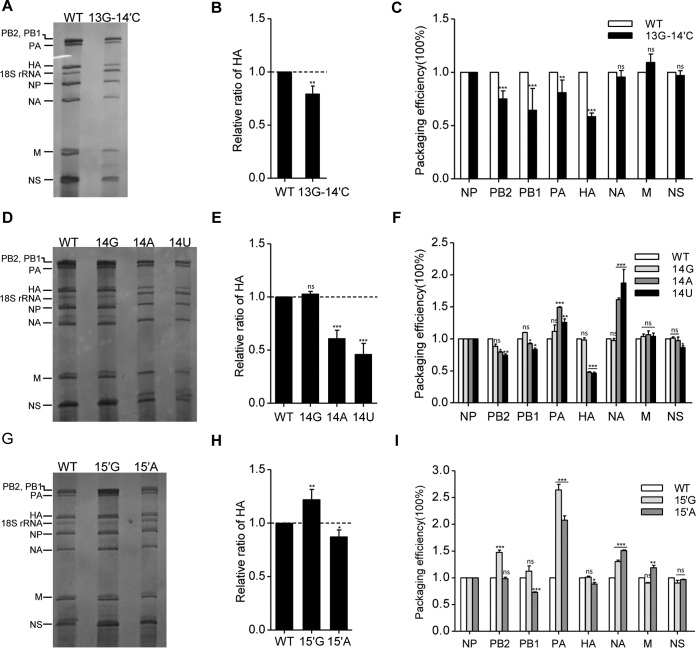
Our previous studies have shown that HA subtype-specific NCRs play a critical role in regulating HA segment packaging efficiency into virions (4, 23, 24). In this study, we observed that the effects of some mutations on RNA synthesis in the RNP reconstitution system were not consistent with their effects on virus replication efficiencies, such as the 13G-14′C, 14A, and 14U mutants. Therefore, in order to further clarify the reasons for the attenuation of the mutant viruses, the effects of the mutations on virus genome packaging efficiency were analyzed by virion RNA staining and reverse transcription-quantitative PCR (RT-qPCR) from purified viruses (4, 23). The silver staining of the virion RNAs obtained from WSN-WT and WSN-HA(13G-14′C) virus showed that the HA vRNA content of the WSN-HA(13G-14′C) virus was significantly reduced from that of the WSN-WT virus (Fig. 5A and B). To further investigate whether the mutation affects the packaging efficiency of other segments, we analyzed the virion RNA by RT-qPCR. Consistent with the RNA silver staining result, the HA packaging efficiency was significantly reduced with WSN-HA(13G-14′C) virus in comparison with that of the WSN-WT virus. Meanwhile, we observed a slight reduction in the packaging of the PB2, PB1, and PA segments (Fig. 5C).
FIG 5.
Effects of nucleotide substitutions at 3′-end positions 13 and 14 and 5′-end positions 14′ and 15′ on virus genome packaging. (A, D, and G) Analysis of the packaging efficiency of each individual vRNA with silver staining gel. Wild-type and mutant viruses [WSN-HA(13G-14′C), WSN-HA(14G), WSN-HA(14A), WSN-HA(14U), WSN-HA(15′G), and WSN-HA(15′A)] were amplified on MDCK cells and purified by ultracentrifugation through a 30% sucrose cushion. Then, the virion vRNAs were extracted with TRIzol reagent, analyzed on a 2.8% denaturing polyacrylamide gel, and visualized by silver staining. (B, E, and H) Statistical analysis of the HA vRNA packaging efficiencies from the experiments shown in panels A, D, and G by densitometry analysis. The band intensities were quantified using ImageJ software. The HA/NP vRNA ratio in the WT virus was set as 1. The relative HA/NP vRNA ratios in mutant viruses were calculated (means and standard deviations of three independent experiments). (C, F, and I) Statistical analysis of packaging efficiency of each individual vRNA by RT-qPCR. The data represent means ± standard deviations of three independent experiments. The relative ratio of each vRNA segment to NP vRNA was calculated. The standard deviations were derived from three independent virus preparations, with vRNA levels quantified in duplicate. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, nonsignificant.
We then further examined the effects of G2 mutants (mutation constructed at the 3′-end position 14 and 5′-end position 15′) on virus genome packaging. The mutant viruses [WSN-HA(14G), WSN-HA(14A), and WSN-HA(14U)] containing point mutations at 3′-end position 14 showed differential effects on HA segment packaging in both virion RNA staining and RT-qPCR analysis. It can be seen that WSN-HA(14A) and WSN-HA(14U) viruses showed significant defects in HA vRNA packaging while the WSN-HA(14G) virus did not (Fig. 5D to F). In conjunction with the reduction of HA segment packaging, we also observed slight reductions in PB1 and PB2 segment packaging and a slight increase in PA segment packaging with the WSN-HA(14A) and WSN-HA(14U) viruses. In contrast, the WSN-HA(14G) virus behaved similarly to the WSN-WT virus. These results suggest that the nucleotide identity at the 3′-end 14 position plays a critical role in regulating HA segment packaging. In the case of the mutant viruses [WSN-HA(15′G) and WSN-HA(15′A)], we observed only a slight reduction on HA segment packaging with the WSN-HA(15′A) mutant while we observed a slight increase in HA segment packaging with the WSN-HA(15′G) mutant. Surprisingly, we found that both viruses with a mutation at the 5′-end 15′ position increased the level of the PA segment dramatically (Fig. 5G to I). Taken together, these results suggested that conservative nucleotides at the 3′-end 13 and 14 positions and the 5′-end 14′ and 15′ positions of the HA segment play critical roles in mediating HA segment incorporation. Mutations in HA segments may also affect packaging of three polymerase gene segments.
DISCUSSION
The noncoding region of influenza A viruses contains two parts: the highly conserved promoter region followed by the segment-specific region. The two parts have been shown to play respective biological functions during virus replication (25–29). The promoter region is responsible for binding viral RNA polymerase and initiating viral RNA synthesis, while the segment-specific region, together with the terminal coding regions, has been shown to act as the packaging signal for selective virus genome packaging (4, 7, 23). However, the exact biological function(s) of a few segment-specific nucleotides which extend the duplex region of the promoter for 1 to 4 bp remains vague. In the case of the HA segments, the duplex region is extended by only 1 bp with the HA segment-specific NCR nucleotides. Our extensive mutational analyses revealed that the Watson-Crick base pairing at the 3′13-5′14′ position is required for significant viral RNA syntheses and HA protein production. On top of that, we found that the identities of the base pairing at 3′13-5′14′ position is critical in determining HA segment packaging efficiency (i.e., WT 13C-14′G is ideal; 13G-14′C is acceptable with about a 20% reduction; 13A-14′U and 13U-14′A are unacceptable). Moreover, we showed that the identity of the residue at 3′ position 14 is involved in regulating both viral RNA synthesis and virus genome packaging whereas the identities at 3′ position 14 are more functionally important in regulating virus genome packaging than in regulating viral RNA synthesis. These results, for the first time, not only emphasize the important roles of the nucleotides adjoining the promoter region and the segment-specific region but also reveal that the nucleotides at the beginning of the segment-specific NCR play key roles in controlling virus genome packaging.
The recently solved polymerase-promoter crystal structure with recombinant bat influenza A polymerase and two synthetic RNA oligonucleotides confirmed that the duplex region is indeed present and plays critical roles for promoter activity. The duplex region of the promoter emerges at the interface of all three subunits and projects away from the polymerase complex (13, 30). Based on this structural information, our extensive mutagenesis of the HA segment-specific NCR nucleotides at positions 3′ 13 to 15 and 5′ 14′ to 16′ were unlikely to change the structure of the promoter. However, we could not rule out the possibilities that the noncanonical base pairs may take effects in our mutagenesis study. On the other hand, considering that different identities and numbers of potential Watson-Crick base pairings in the extended duplex region vary among different segments, the detailed 3D structure may also vary among the eight segments. Within the context of the HA segment, here we experimentally showed that the fourth base pair of the duplex region at the 3′13-5′14′ position is critical for the viral RNA synthesis. In contrast, the fifth base pairing at 3′14-5′15′ is favored, but not critical, and the sixth base pairing at 3′15-5′16′ is not required for viral RNA synthesis. Meanwhile, the identities of the base pair are also able to modulate promoter activity to certain extents. Whether these results are also true for other segments regarding the promoter activities needs further investigation.
The packaging signals reported for the HA vRNA of influenza A virus (WSN, H1N1) in the coding region have been identified as 9 or 45 nucleotides at the 3′ end and 80 nucleotides at the 5′ end (23, 24). However, specific nucleotides in the NCR involved in HA vRNA packaging were hard to define. In this study, we identified, for the first time, the HA segment-specific nucleotides that connect the promoter and the HA subtype-specific NCR that are important for HA vRNA packaging. In particular, the G-C base pairing at the 13-14′ position is required for the packaging of the HA segment. In addition, we observed that a reduction in HA packaging often accompanies variations in PB2, PB1, and PA. We previously also reported that the substitution of HA subtype-specific NCRs led to reduced HA segment packaging but significantly increased PB1 and PA segment incorporations (4). These observations imply a link between HA and the PB2, PB1, and PA during their selective packaging. However, the underlying mechanism for these observations is still unknown. A study of the NP segment packaging proposed that the NCR serves as an incorporation signal and that the terminal coding sequence serves as the bundling signal (31). Whether the HA segment-specific NCR nucleotides play a role in virus genome bundling needs further clarification.
It has been reported that the levels of the vRNA in infected cells are not directly correlated with the packaging efficiency of the vRNA (23, 32). In line with this idea, when we examined the virus replication efficiencies with the rescued G1, G2, and G3 mutant viruses, we found that the capacities for viral RNA synthesis were not consistent with their effects on virus replication efficiencies. In the case of the 13A-14′U and 13U-14′A mutants, the three RNA species could be synthesized to significant levels whereas they could not be rescued. In addition, we observed that the 13G-14′C and 14A mutant showed wild-type-like viral RNA synthesis capacities but significantly reduced virus replication efficiencies. In contrast, the 14G mutant showed a reduced viral RNA synthesis capacity but wild-type-like virus replication efficiency. On the other hand, the vRNAs were produced by the 15′G mutation at significantly higher levels than the wild-type level, while the virus replication of this mutant virus is similar to that of the wild-type virus. According to the packaging efficiencies of these HA segment mutants, we concluded that the virus replication efficiency is more closely correlated with the segment packaging efficiencies than with their viral RNA synthesis capabilities. Of course, this conclusion is based on a prerequisite that the minimum levels of RNA synthesis capacities must be retained for genome packaging. This conclusion also explains that, during long-term evolution, the HA segment may prefer to use suboptimal mismatched nucleotides at the 3′14-5′15′ positions in order to ensure efficient segment packaging rather than maximum viral RNA synthesis. On the other hand, we also observed that the ratios of the three virus RNA species was changed with certain mutations made in the extended duplex region, for example, 13G, 14G, and 15′G mutations. The underlying mechanism of this needs further clarification. In addition, it should also be pointed out that, among the eight segments, only the NA segment at the extended duplex region is polymorphic (Table 1). The reason for this polymorphism is worth further investigation. Another interesting observation within this region is that an A14U substitution in the 3′ end of the M segment has recently been found to be involved in modulating alternative splicing of M segment mRNAs in NS1 deleted virus (33).
In summary, we studied the role(s) of the HA segment-specific NCR nucleotides located in the extended duplex region of the viral RNA promoter for influenza virus replication. We found that these nucleotides play dual roles in maintaining viral RNA promoter activities and in HA segment packaging. Thus, our studies not only clarified the promoter requirement in the extended duplex region in the context of the HA segment but also identified specific noncoding residues involved in regulating virus genome packaging. These findings may facilitate the development of attenuated vaccine strains and the elucidation of virus genome packaging mechanisms.
MATERIALS AND METHODS
Cells, plasmids, and antibodies.
Human embryonic kidney 293T cells and Madin-Darby canine kidney (MDCK) cells were cultured as described previously (4). The RNP reconstitution system plasmids (pcDNA-PB2-PR8, pcDNA-PB1-PR8, pcDNA-PA-PR8, pcDNA-NP-PR8, and pPolI-HA-PR8) of influenza A/PuertoRico/8/34 (PR8) virus were provided by Ervin Fodor (University of Oxford, United Kingdom). The eight plasmids for generation of recombinant A/WSN/33 (H1N1) viruses (pHW-PB2-WSN, pHW-PB1-WSN, pHW-PA-WSN, pHW-HA-WSN, pHW-NP-WSN, pHW-NA-WSN, pHW-M-WSN, and pHW-NS-WSN) have been described previously (22). Mutations described in G1, G2, and G3 were introduced into the vRNA-encoding plasmid pPolI-HA-PR8 and reverse genetic plasmid pHW-HA-WSN by site-directed mutagenesis using a QuikChange site-directed mutagenesis kit (Stratagene). Rabbit anti-HA monoclonal antibody (86001-RM01) was purchased from Sino Biological, Inc. (China). Mouse anti-β-actin monoclonal antibody was purchased from Cell Signaling Technology.
Virus rescue and viral growth kinetics.
Recombinant viruses (the wild-type WSN and mutant viruses) were rescued by the previously described eight-plasmid rescue system (22). The rescued viruses were plaque purified and fully sequenced. The viral growth kinetics of the wild-type and mutant viruses were performed in MDCK cells at a multiplicity of infection (MOI) of 0.001, and PFU titers were determined by a standard plaque assay in MDCK cells.
Primer extension and Western blotting.
To reconstitute RNPs, 1 × 106 293T cells in 35-mm dishes were transfected in suspension with 500 ng each of protein expression plasmids encoding PB2, PB1, PA, and NP of PR8 virus (pcDNA-PB2-PR8, pcDNA-PB1-PR8, pcDNA-PA-PR8, and pcDNA-NP-PR8), along with 500 ng of pPolI-SapI-Rib-based RNA-expressing plasmids, as needed, using 6 μl of Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. At 24 h posttransfection, total RNAs or proteins from the transfected cells were extracted and analyzed by primer extension or Western blot analysis.
The primer extension analysis was carried out as described previously (34). The primers used for detecting HA (PR8) vRNA and mRNA/cRNA were 5′-AGTTCACTGGTGCTTTTGGTC-3′ and 5′-TGTCACATTCTTCTCGAGCAC-3′. The primer used for detecting 5S RNA was 5′-TCCCAGGCGGTCTCCCATCC-3′. Western blotting was carried out by standard procedures with IRDye secondary antibodies (Li-Cor Biosciences, Lincoln, NE). Protein expression levels were visualized with an Odyssey Infrared Imaging System (Li-Cor Biosciences, USA). The relative protein expression level was analyzed using the integrated software of the Odyssey system.
RNA packaging assays.
The packaging efficiencies of virus genome RNAs were examined as described previously (4). Briefly, MDCK cells were infected with virus at an MOI of 0.001. Forty-eight hours later, the supernatants were collected and purified by ultracentrifugation through a 30% (wt/wt) sucrose cushion, and the RNA of the virus pellet was extracted using TRIzol reagent (Invitrogen). The content of each segment of RNA was further examined by silver staining and RT-qPCR as described previously (4, 23).
Statistical analysis.
GraphPad Prism, version 5, software was used for statistical analysis. One-way analysis of variance (ANOVA) was used to determine the significance of two-group comparisons.
ACKNOWLEDGMENTS
This work was supported by grants from the Chinese Academy of Medical Sciences (2016-12M-1-014), National Natural Science Foundation of China (31070152 and 81501351), and Chinese Science and Technology Key Projects of China (2013ZX10004601-002), to T.D.
REFERENCES
- 1.Palese P, Shaw M. 2013. Orthomyxoviridae, p 1151–1185. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed Lippincott Williams and Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Robertson JS. 1979. 5′ And 3′ terminal nucleotide sequences of the RNA genome segments of influenza virus. Nucleic Acids Res 6:3745–3757. doi: 10.1093/nar/6.12.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fodor E. 2013. The RNA polymerase of influenza A virus: mechanisms of viral transcription and replication. Acta Virol 57:113–122. doi: 10.4149/av_2013_02_113. [DOI] [PubMed] [Google Scholar]
- 4.Zhao L, Peng Y, Zhou K, Cao M, Wang J, Wang X, Jiang T, Deng T. 2014. New insights into the nonconserved noncoding region of the subtype-determinant hemagglutinin and neuraminidase segments of influenza a viruses. J Virol 88:11493–11503. doi: 10.1128/JVI.01337-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Furuse Y, Oshitani H. 2011. Evolution of the influenza A virus untranslated regions. Infect Genet Evol 11:1150–1154. doi: 10.1016/j.meegid.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 6.Luytjes W, Krystal M, Enami M, Parvin JD, Palese P. 1989. Amplification, expression, and packaging of foreign gene by influenza virus. Cell 59:1107–1113. doi: 10.1016/0092-8674(89)90766-6. [DOI] [PubMed] [Google Scholar]
- 7.Hutchinson EC, von Kirchbach JC, Gog JR, Digard P. 2010. Genome packaging in influenza A virus. J Gen Virol 91:313–328. doi: 10.1099/vir.0.017608-0. [DOI] [PubMed] [Google Scholar]
- 8.Desselberger U, Racaniello VR, Zazra JJ, Palese P. 1980. The 3′ and 5′-terminal sequences of influenza A, B and C virus RNA segments are highly conserved and show partial inverted complementarity. Gene 8:315–328. doi: 10.1016/0378-1119(80)90007-4. [DOI] [PubMed] [Google Scholar]
- 9.Fodor E, Seong BL, Brownlee GG. 1993. Photochemical cross-linking of influenza A polymerase to its virion RNA promoter defines a polymerase binding site at residues 9 to 12 of the promoter. J Gen Virol 74:1327–1333. doi: 10.1099/0022-1317-74-7-1327. [DOI] [PubMed] [Google Scholar]
- 10.Fodor E, Pritlove DC, Brownlee GG. 1994. The influenza virus panhandle is involved in the initiation of transcription. J Virol 68:4092–4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tiley LS, Hagen M, Matthews JT, Krystal M. 1994. Sequence-specific binding of the influenza virus RNA polymerase to sequences located at the 5′ ends of the viral RNAs. J Virol 68:5108–5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tomescu AI, Robb NC, Hengrung N, Fodor E, Kapanidis AN. 2014. Single-molecule FRET reveals a corkscrew RNA structure for the polymerase-bound influenza virus promoter. Proc Natl Acad Sci U S A 111:E3335–E3342. doi: 10.1073/pnas.1406056111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pflug A, Guilligay D, Reich S, Cusack S. 2014. Structure of influenza A polymerase bound to the viral RNA promoter. Nature 516:355–360. doi: 10.1038/nature14008. [DOI] [PubMed] [Google Scholar]
- 14.Reich S, Guilligay D, Pflug A, Malet H, Berger I, Crepin T, Hart D, Lunardi T, Nanao M, Ruigrok RW, Cusack S. 2014. Structural insight into cap-snatching and RNA synthesis by influenza polymerase. Nature 516:361–366. doi: 10.1038/nature14009. [DOI] [PubMed] [Google Scholar]
- 15.Suzuki Y, Kobayashi Y. 2013. Evolution of complementary nucleotides in 5′ and 3′ untranslated regions of influenza A virus genomic segments. Infect Genet Evol 13:175–179. doi: 10.1016/j.meegid.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 16.Hagen M, Chung TD, Butcher JA, Krystal M. 1994. Recombinant influenza virus polymerase: requirement of both 5′ and 3′ viral ends for endonuclease activity. J Virol 68:1509–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poon LL, Pritlove DC, Fodor E, Brownlee GG. 1999. Direct evidence that the poly(A) tail of influenza A virus mRNA is synthesized by reiterative copying of a U track in the virion RNA template. J Virol 73:3473–3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsu MT, Parvin JD, Gupta S, Krystal M, Palese P. 1987. Genomic RNAs of influenza viruses are held in a circular conformation in virions and in infected cells by a terminal panhandle. Proc Natl Acad Sci U S A 84:8140–8144. doi: 10.1073/pnas.84.22.8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stoeckle MY, Shaw MW, Choppin PW. 1987. Segment-specific and common nucleotide sequences in the noncoding regions of influenza B virus genome RNAs. Proc Natl Acad Sci U S A 84:2703–2707. doi: 10.1073/pnas.84.9.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J, Peng Y, Zhao L, Cao M, Hung T, Deng T. 2015. Influenza A virus utilizes a suboptimal Kozak sequence to fine-tune virus replication and host response. J Gen Virol 96:756–766. doi: 10.1099/vir.0.000030. [DOI] [PubMed] [Google Scholar]
- 21.Fodor E, Brownlee GG. 2002. Influenza virus replication, p 1–29. In Potter CW. (ed), Influenza. Elsevier Science, New York, NY. [Google Scholar]
- 22.Hoffmann E, Neumann G, Kawaoka Y, Hobom G, Webster RG. 2000. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc Natl Acad Sci U S A 97:6108–6113. doi: 10.1073/pnas.100133697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marsh GA, Hatami R, Palese P. 2007. Specific residues of the influenza A virus hemagglutinin viral RNA are important for efficient packaging into budding virions. J Virol 81:9727–9736. doi: 10.1128/JVI.01144-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watanabe T, Watanabe S, Noda T, Fujii Y, Kawaoka Y. 2003. Exploitation of nucleic acid packaging signals to generate a novel influenza virus-based vector stably expressing two foreign genes. J Virol 77:10575–10583. doi: 10.1128/JVI.77.19.10575-10583.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Enami M, Luytjes W, Krystal M, Palese P. 1990. Introduction of site-specific mutations into the genome of influenza virus. Proc Natl Acad Sci U S A 87:3802–3805. doi: 10.1073/pnas.87.10.3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neumann G, Hobom G. 1995. Mutational analysis of influenza virus promoter elements in vivo. J Gen Virol 76:1709–1717. doi: 10.1099/0022-1317-76-7-1709. [DOI] [PubMed] [Google Scholar]
- 27.Parvin JD, Palese P, Honda A, Ishihama A, Krystal M. 1989. Promoter analysis of influenza virus RNA polymerase. J Virol 63:5142–5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seong BL, Brownlee GG. 1992. A new method for reconstituting influenza polymerase and RNA in vitro: a study of the promoter elements for cRNA and vRNA synthesis in vitro and viral rescue in vivo. Virology 186:247–260. doi: 10.1016/0042-6822(92)90079-5. [DOI] [PubMed] [Google Scholar]
- 29.Yamanaka K, Ogasawara N, Yoshikawa H, Ishihama A, Nagata K. 1991. In vivo analysis of the promoter structure of the influenza virus RNA genome using a transfection system with an engineered RNA. Proc Natl Acad Sci U S A 88:5369–5373. doi: 10.1073/pnas.88.12.5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Te Velthuis AJ, Fodor E. 2016. Influenza virus RNA polymerase: insights into the mechanisms of viral RNA synthesis. Nat Rev Microbiol 14:479–493. doi: 10.1038/nrmicro.2016.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goto H, Muramoto Y, Noda T, Kawaoka Y. 2013. The genome-packaging signal of the influenza A virus genome comprises a genome incorporation signal and a genome-bundling signal. J Virol 87:11316–11322. doi: 10.1128/JVI.01301-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muramoto Y, Takada A, Fujii K, Noda T, Iwatsuki-Horimoto K, Watanabe S, Horimoto T, Kida H, Kawaoka Y. 2006. Hierarchy among viral RNA (vRNA) segments in their role in vRNA incorporation into influenza A virions. J Virol 80:2318–2325. doi: 10.1128/JVI.80.5.2318-2325.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng M, Wang P, Song W, Lau SY, Liu S, Huang X, Mok BW, Liu YC, Chen Y, Yuen KY, Chen H. 2015. An A14U substitution in the 3′ noncoding region of the M segment of viral RNA supports replication of influenza virus with an NS1 deletion by modulating alternative splicing of M segment mRNAs. J Virol 89:10273–10285. doi: 10.1128/JVI.00919-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fodor E, Crow M, Mingay LJ, Deng T, Sharps J, Fechter P, Brownlee GG. 2002. A single amino acid mutation in the PA subunit of the influenza virus RNA polymerase inhibits endonucleolytic cleavage of capped RNAs. J Virol 76:8989–9001. doi: 10.1128/JVI.76.18.8989-9001.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]