ABSTRACT
RNA silencing is an innate antiviral immunity response of plants and animals. To counteract this host immune response, viruses have evolved an effective strategy to protect themselves by the expression of viral suppressors of RNA silencing (VSRs). Most potyviruses encode two VSRs, helper component-proteinase (HC-Pro) and viral genome-linked protein (VPg). The molecular biology of the former has been well characterized, whereas how VPg exerts its function in the suppression of RNA silencing is yet to be understood. In this study, we show that infection by Turnip mosaic virus (TuMV) causes reduced levels of suppressor of gene silencing 3 (SGS3), a key component of the RNA silencing pathway that functions in double-stranded RNA synthesis for virus-derived small interfering RNA (vsiRNA) production. We also demonstrate that among 11 TuMV-encoded viral proteins, VPg is the only one that interacts with SGS3. We furthermore present evidence that the expression of VPg alone, independent of viral infection, is sufficient to induce the degradation of SGS3 and its intimate partner RNA-dependent RNA polymerase 6 (RDR6). Moreover, we discover that the VPg-mediated degradation of SGS3 occurs via both the 20S ubiquitin-proteasome and autophagy pathways. Taken together, our data suggest a role for VPg-mediated degradation of SGS3 in suppression of silencing by VPg.
IMPORTANCE Potyviruses represent the largest group of known plant viruses and cause significant losses of many agriculturally important crops in the world. In order to establish infection, potyviruses must overcome the host antiviral silencing response. A viral protein called VPg has been shown to play a role in this process, but how it works is unclear. In this paper, we found that the VPg protein of Turnip mosaic virus (TuMV), which is a potyvirus, interacts with a host protein named SGS3, a key protein in the RNA silencing pathway. Moreover, this interaction leads to the degradation of SGS3 and its interacting and functional partner RDR6, which is another essential component of the RNA silencing pathway. We also identified the cellular pathways that are recruited for the VPg-mediated degradation of SGS3. Therefore, this work reveals a possible mechanism by which VPg sabotages host antiviral RNA silencing to promote virus infection.
KEYWORDS: RDR6, RNA silencing, SGS3, VPg, VSR, autophagy, potyvirus, ubiquitination
INTRODUCTION
To establish systemic infection, an invading virus must remodel and recruit host elements and overcome resistance responses through complex molecular virus-host interactions (1–3). One such essential antiviral response is RNA silencing (4–6), a cellular mechanism conserved from lower eukaryotes to mammals to regulate endogenous gene expression at the transcriptional and posttranscriptional levels (7, 8). To activate antiviral RNA silencing, the viral double-stranded RNAs (dsRNAs), which may originate during viral replication, as a result of internal pairing of viral RNA molecules, or through de novo synthesis by the host RNA-dependent RNA polymerase 6 (RDR6)/suppressor of gene silencing 3 (SGS3) complex using viral genomic fragments as the template, are processed by Dicer-like (DCL) proteins into small 20- to 24-nucleotide (nt) RNA duplexes termed virus-derived short interfering RNAs (vsiRNAs). These vsiRNAs are incorporated into Argonaute (AGO) proteins, the key component of the RNA-induced silencing complex (RISC) that directly possesses RNase activity and catalyzes the cleavage of homologous RNAs. SGS3/RDR6 bodies are also required for the amplification of RNA silencing through the biogenesis of secondary siRNAs and for the production of endogenous trans-acting siRNA (ta-siRNA) (9–12). Plants with dysfunctional RNA silencing pathways by the knockout of RNA silencing components show enhanced susceptibility to viral infections (11, 13–17). A majority of plant viruses have evolved one or more proteins, termed viral suppressors of RNA silencing (VSRs), to suppress host RNA silencing. VSRs have been shown to suppress RNA silencing at virtually all steps of RNA silencing (6). Many VSRs suppress RNA silencing via sequestration of virus-derived siRNAs and inactivation of AGO proteins (mainly AGO1) (18–20). In contrast, some other VSRs, such as V2 of Tomato yellow leaf curl virus (TYLCV), P2 of Rice stripe virus (RSV), TGB1 of Plantago asiatica mosaic virus (PlAMV), and the class 1 RNase III endoribonuclease (RNase 3) of Sweet potato chlorotic stunt virus (SPCSV), target other RNA silencing components, e.g., SGS3, to attenuate plant antivirus immunity (21–24).
The genus Potyvirus within the family Potyviridae consists of a group of plant-infecting viruses with a positive-sense single-stranded RNA genome. Potyviruses account for >30% of known plant viruses and include many agriculturally important viruses such as Turnip mosaic virus (TuMV), Potato virus A (PVA), Potato virus Y (PVY), Soybean mosaic virus (SMV), Tobacco etch virus (TEV), and Plum pox virus (PPV), which cause significant losses in many economic crops. The genome of typical potyviruses encodes a long open reading frame (ORF) and another relatively short ORF that results from RNA polymerase slippage in the P3 coding sequence (25–29). Upon translation, these two polyproteins are proteolytically processed by three viral protease domains into 11 mature viral proteins. More recently, another short ORF, termed PISPO (pretty interesting sweet potato potyvirus ORF), also resulting from transcriptional slippage, was found to be embedded in the P1 coding regions of two sweet potato potyviruses (26, 30, 31). Among the 11 common potyvirus proteins, HC-Pro and VPg have been shown to be VSRs (32–34). The molecular mechanisms underlying the functional role of HC-Pro as a VSR have been relatively well understood (35–46). It still remains unclear how VPg interferes with RNA silencing.
Potyvirus VPg is a relatively small protein (∼21 kDa) that is covalently linked to the viral RNA through a phosphodiester bond between a conserved serine or tyrosine residue and the 5′-terminal uridine nucleotide of viral RNA (47). Biochemical and bioinformatics analyses showed that VPg is an intrinsically disordered protein (48–51). This structural flexibility confers the possibility of forming interaction complexes with different proteins to function diversely. Indeed, accumulated evidence suggests that VPg interacts with itself, most other potyvirus proteins (reviewed in references 52 and 53), and many host factors, e.g., alpha heat shock protein (α-HSP), Arabidopsis RNA helicase-like 8 (AtRH8), eukaryotic initiation factor 4E (eIF4E) and its isoform eIF(iso)4E, eIF4G, eIF(iso)4G, fibrillarin, OBERON1 (OBE1), OBE2, poly(A) binding protein 2 (PABP2), PABP4, RRP6, and SGS3 (34, 54–60).
In the present study, we report that TuMV infection induces SGS3 degradation in plants. We furthermore demonstrate that SGS3 interacts with TuMV VPg and VPgs of other potyviruses and that VPg alone is sufficient to induce the degradation of SGS3 and its intimate interaction partner RDR6. Moreover, we present evidence that VPg-mediated degradation of SGS3 and RDR6 occurs via both the 20S proteasome and autophagy pathways. Taken together, these data suggest that potyvirus VPg suppresses host RNA silencing, possibly via the interaction of SGS3, to mediate the degradation of SGS3 and its interaction proteins such as RDR6.
RESULTS
TuMV infection causes degradation of the SGS3 protein.
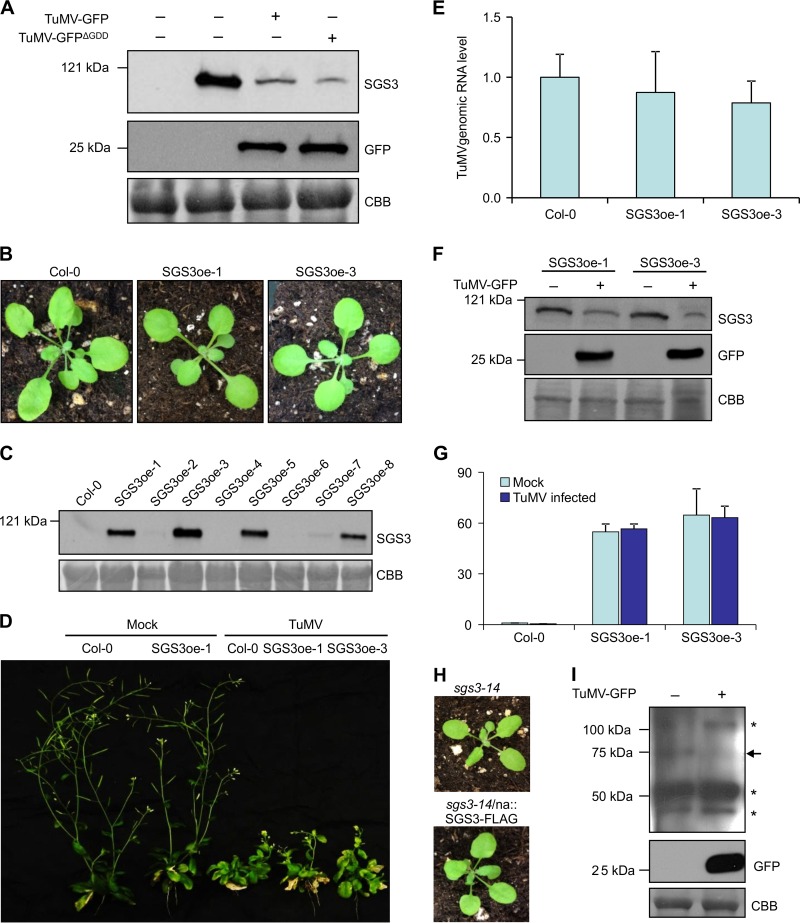
Previous studies suggested the involvement of SGS3 in potyvirus infections (11, 60). To further elucidate its functional mechanism, we used TuMV as a model virus in this study. At first, we conducted a transient-expression assay by agroinfiltration of Nicotiana benthamiana leaves with agrobacteria harboring plant expression vectors containing coding sequences of N-terminally FLAG- and 4×Myc-tagged Arabidopsis SGS3 (FLAG-4×Myc-SGS3) under the control of the Cauliflower mosaic virus (CaMV) 35S promoter alone or together with a wild-type green fluorescent protein (GFP)-tagged TuMV infectious clone (TuMV-GFP) or a replication-defective clone (TuMV-GFPΔGDD) in which the viral RNA-dependent RNA polymerase (RdRp) was made dysfunctional by mutating the three key residues (Gly-Asp-Asp) into Ala. Agroinfiltration of TuMV-GFP into compatible host plants such as N. benthamiana and Arabidopsis allows the establishment of local and systemic infection, whereas the introduction of TuMV-GFPΔGDD leads to the transient expression of all viral proteins (including mutated RdRp) only in agroinfiltrated leaf tissues due to the 35S promoter. The accumulation of the SGS3 protein was evaluated by Western blotting with anti-Myc antibodies at 60 h postinoculation (hpi). A protein with the predicted molecular mass of FLAG-4×Myc-SGS3 was detected in protein extracts from N. benthamiana leaves infiltrated with 35S::FLAG-4×Myc-SGS3, whereas no corresponding protein was detected in protein extracts from mock-treated N. benthamiana leaves (Fig. 1A), suggesting that this protein is the recombinant FLAG-4×Myc-SGS3 protein. However, the intensity of FLAG-4×Myc-SGS3 in the protein extracts from N. benthamiana leaves coinfiltrated with FLAG-4×Myc-SGS3 and TuMV-GFP was significantly reduced in comparison to that from leaves infiltrated with FLAG-4×Myc-SGS3 alone. Interestingly, the intensity of FLAG-4×Myc-SGS3 was also significantly reduced when FLAG-4×Myc-SGS3 was coexpressed with TuMV-GFPΔGDD (Fig. 1A), suggesting that the reduction of the intensity of SGS3 is independent of viral replication.
FIG 1.
TuMV infection induces SGS3 degradation in N. benthamiana and Arabidopsis. (A) TuMV-GFP and TuMV-GFPΔGDD were transiently coexpressed with FLAG-4×Myc-SGS3 in N. benthamiana leaves by Agrobacterium infiltration. The FLAG-4×Myc-SGS3 protein and GFP were detected at 60 hpi. (Top) Western blotting of total protein extracts with Myc antibodies; (middle) Western blotting of total protein extracts with GFP antibodies; (bottom) Coomassie bright blue (CBB) staining showing equal loading of total protein extracts. (B) Photograph of 3-week-old wild-type (Col-0) and SGS3-overexpressing transgenic (SGS3oe-1 and SGS3oe-3) plants. (C) Western blot detection of FLAG-4×Myc-SGS3 in eight independent transgenic Arabidopsis lines expressing FLAG-4×Myc-SGS3 with Myc antibodies. FLAG-4×Myc-SGS3 was strongly detected in lines SGS3oe-1, -3, -5, and -8. Weak expression was evident in lines SGS3oe-2 and -7. No FLAG-4×Myc-SGS3 was detectable in SGS3oe-4 and -6. (D) Phenotype of TuMV-infected transgenic Arabidopsis plants overexpressing SGS3 (SGS3oe-1 and SGS3oe-3) at 20 dpi. (E) The relative TuMV genomic RNA levels shown in panel D were determined by qRT-PCR. Mean genomic RNA levels of TuMV on Col-0 plants were normalized to 1.0. Bars represent means ± standard deviations of data from five biological replicates. (F) Western blot detection of SGS3 and GFP in SGS3oe-1 and SGS3oe-3 with Myc (top) and GFP (middle) antibodies. The bottom panel shows equal loading of total proteins by CBB staining. (G) The expression levels of SGS3 shown in panel D were determined by RT-PCR. The expression levels of SGS3 in mock-infected plants were normalized to 1.0. Bars represent means ± standard deviations of data from five biological replicates. (H) Phenotype of 3-week-old Arabidopsis sgs3-14 plants transformed with na::SGS3-FLAG. (I) Western blot detection of SGS3 and GFP in na::SGS3-FLAG plants with SGS3 (top) and GFP (middle) antibodies. The position of FLAG-SGS3 is indicated by an arrow. Stars indicate the cross-reacting bands of SGS3 antibodies. CBB staining shows equal loading of the samples (bottom).
To further test this assumption, we generated transgenic plants overexpressing FLAG-4×Myc-SGS3 under the control of the CaMV 35S promoter in an Arabidopsis Col-0 background (Fig. 1B). A total of eight (SGS3oe-1 to -8) independent homological transgenic lines were obtained. Quantitative reverse transcription-PCR (qRT-PCR) results confirmed that SGS3 was overexpressed in all these lines in comparison to the expression level in wild-type plants. Western blotting with Myc antibodies detected FLAG-4×Myc-SGS3 in six (SCGSoe-1, -2, -3, -5, -7, and -8) of the eight lines (Fig. 1C). All the transgenic lines showed a normal leaf phenotype similar to that of wild-type Arabidopsis Col-0 (Fig. 1B), suggesting that the overexpression of SGS3 does not affect normal plant development under the given growth conditions. Four-week-old seedlings of the homozygous SGS3oe-1 and SGS3oe-3 lines, which have relatively higher SGS3 expression levels among the eight transgenic lines, were mechanically inoculated with TuMV-GFP. Both wild-type and SGS3-overexpressing plants developed typical TuMV symptoms, including a substantial reduction in stature, yellowish leaves, curling stem, and defective flowers, at 20 days postinfection (dpi), and no obvious differences between TuMV-infected wild-type and TuMV-infected SGS3-overexpressing plants were observed (Fig. 1D). qRT-PCR revealed that there were no statistically significant differences in viral RNA levels between wild-type and SGS3-overexpressing plants in upper systematically infected leaves, although both SGS3-overexpressing lines accumulated slightly lower levels of viral RNA than those in wild-type plants (Fig. 1E). Western blotting with anti-Myc antibodies showed that the SGS3 protein level was also greatly reduced in systemically infected leaves of SGS3-overexpressing lines inoculated with TuMV-GFP compared to that in leaves of mock-inoculated SGS3-overexpressing plants (Fig. 1F). qRT-PCR was performed to determine if SGS3 transcription is affected in response to TuMV infection, leading to reduced levels of the SGS3 protein. No significant differences between the transcript levels of SGS3 in mock- and TuMV-inoculated plants were found (Fig. 1G).
Since polyclonal antibodies to SGS3 failed to detect the endogenous SGS3 protein in Arabidopsis Col-0 plants (data not shown), an Arabidopsis SGS3 knockout mutant, sgs3-14, was complemented with C-terminally FLAG-tagged SGS3 under the control its native promoter (na::SGS3-FLAG), making it possible to detect SGS3 by using antibodies to FLAG. The sgs3-14 mutant, showing typically elongated, downwardly curled leaves, contains a transfer DNA (T-DNA) insertion at its first exon, which disrupts SGS3 transcription (9). The sgs3-14 plants transformed with na::SGS3-FLAG showed a restored normal wild-type leaf phenotype (Fig. 1H), suggesting that C-terminally FLAG-tagged SGS3 is functional. The na::SGS3-FLAG-transformed plants were sap inoculated with TuMV-GFP. The SGS3 protein was then purified from virus-infected leaf tissues with anti-FLAG M2 affinity agarose at 14 dpi and detected with goat polyclonal antibodies against SGS3. Western blotting detected a protein with the predicted size in the mock-inoculated na::SGS3-FLAG-transformed plants, which was hardly detectable in TuMV-GFP-infected plants (Fig. 1I), consistent with results from TuMV-infected plants overexpressing or transiently expressing SGS3 (Fig. 1A and F) or plants transiently coexpressing SGS3 and replication-defective TuMV (Fig. 1A). Taken together, these results suggest that TuMV infection results in the degradation of SGS3, and it is TuMV protein(s) rather than its replication that causes a reduction of SGS3 levels.
VPg is the only potyvirus protein that interacts with SGS3.
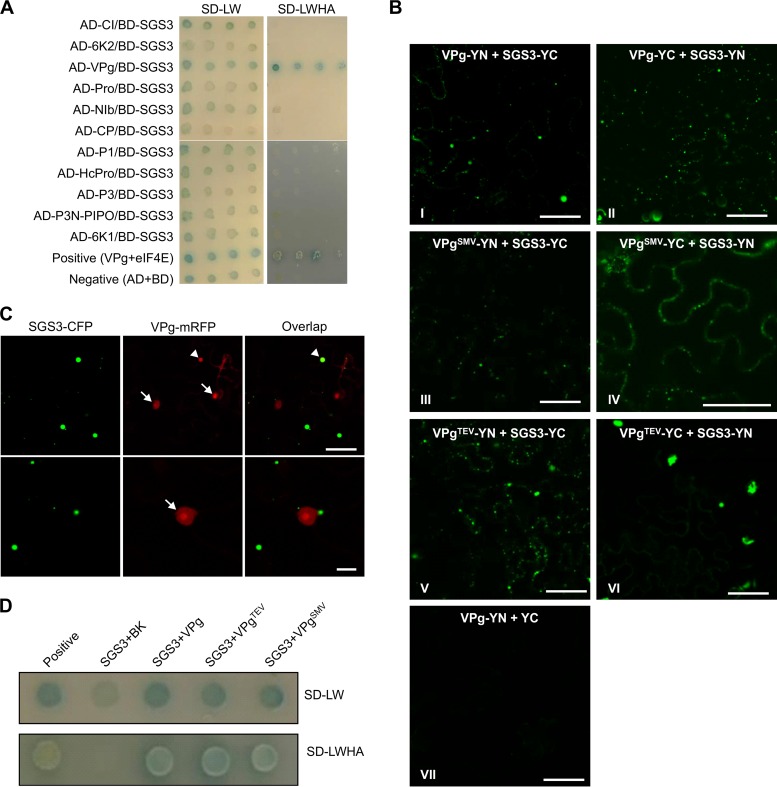
To determine which viral protein(s) is responsible for the decreased SGS3 levels, we cloned all 11 proteins encoded by TuMV and examined their ability to interact with SGS3 by yeast two-hybrid (Y2H) and bimolecular fluorescence complementation (BiFC) assays. The Y2H assay showed that VPg was the only viral protein that could rescue the growth of yeast on media lacking the four amino acids and restore β-galactosidase (GUS) activity (Fig. 2A). Neither VPg nor SGS3 showed any autoactivation or autobinding activity (data not shown). Furthermore, a VPg C-terminal fusion with either the N- or C-terminal half of yellow fluorescent protein (YFP) (VPg-YN and VPg-YC, respectively) restored YFP fluorescence when it was coexpressed with SGS3-YC and SGS3-YN, respectively (Fig. 2BI and II). In both cases, the YFP fluorescence resulting from the interaction between VPg and SGS3 showed granule-like foci in the cytosol, which resembles the subcellular localization of SGS3 (Fig. 3A) (10, 60, 61). However, no positive signals between other potyvirus proteins and SGS3 were found in the BiFC assay (data not shown). C-terminally cyan fluorescent protein (CFP)-tagged SGS3 (SGS3-CFP) was also transiently coexpressed with C-terminally monomer red fluorescent protein (mRFP)-tagged TuMV VPg (VPg-mRFP) in N. benthamiana epidermal cells. Colocalization of SGS3-CFP and VPg-mRFP was found in the large fluorescent bodies (Fig. 2C, top). No visible fluorescence from SGS3-CFP was observed in the nucleus, where the majority of VPg was localized (Fig. 2C, bottom), suggesting that VPg interacts with SGS3 in the cytoplasm.
FIG 2.
VPg is the only viral protein that interacts with SGS3. (A) Yeast two-hybrid assay for protein-protein interactions between SGS3 and TuMV-encoded proteins on selective media supplied with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). The positive and negative controls are yeast cotransformants with pGAD-VPg plus pGBK-eIF4E and pGAD-T plus pGBK-Lam, respectively. AD, GAL4 activation domain; BD, GAL4 DNA binding domain. SD, synthetic defined (SD) yeast minimal medium; SD-LW, SD medium lacking Leu and Trp; SD-LWHA, SD medium lacking Leu, Trp, His, and adenine hemisulfate. (B) BiFC assay for protein-protein interactions between SGS3 and VPg encoded by TuMV, SMV, and TEV. SGS3, TuMV VPg, SMV VPg, and TEV VPg were fused to either the C-terminal or N-terminal half of YFP and transiently expressed in N. benthamiana leaves. Fluorescence was monitored by confocal microscopy at 48 hpi. Bar, 50 μm. (C) Confocal micrographs of N. benthamiana coexpressing SGS3-CFP and VPg-mRFP at 48 hpi. Bar, 50 μm. (Bottom) Closeup images showing SGS3 bodies at the perinuclear area. White arrows, nuclear localized VPg; white arrowhead, cytoplasmic SGS3-VPg interaction body. Bar, 10 μm. (D) Yeast cells cotransformed with AD-SGS3 and BD, or VPg protein encoded by TuMV, TEV (VPgTEV), or SMV (VPgTEV) were tested as described above for panel A. The positive control is the yeast cotransformant with pGAD-VPg and pGBK-eIF4E.
FIG 3.
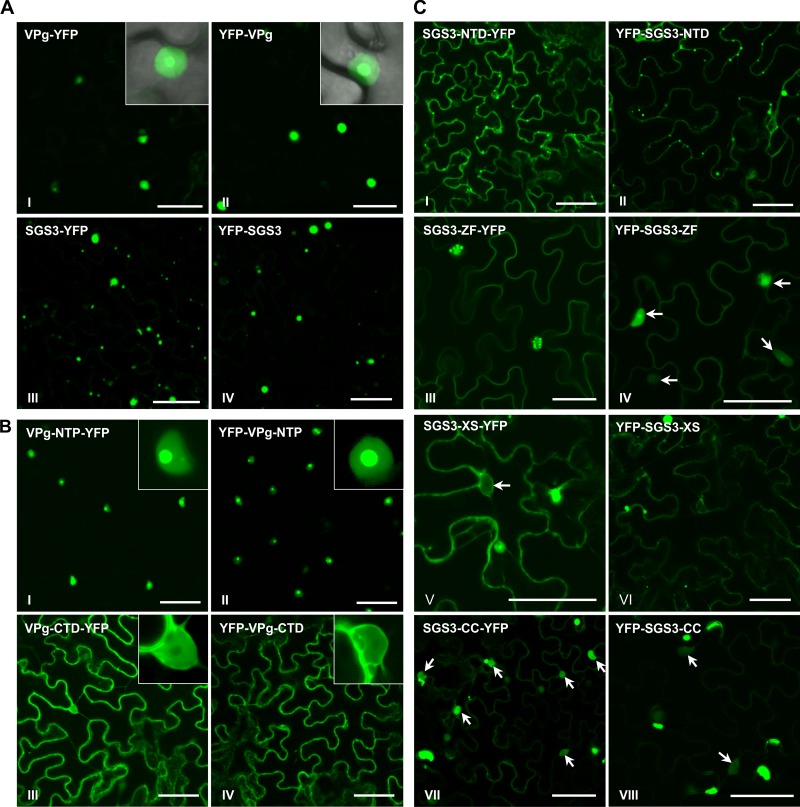
Subcellular localization of VPg, SGS3, and their domains in N. benthamiana determined by confocal microscopy. (A) Confocal micrographs of N. benthamiana leaf cells expressing N- or C-terminally YFP-fused VPg or SGS3 in N. benthamiana leaves at 48 hpi. Insets show the nuclear localization of VPg. Bars, 50 μm. (B) Confocal micrographs of N. benthamiana leaf cells expressing N- or C-terminally YFP-fused VPg-NTP or VPg-CTD at 48 hpi. Bars, 50 μm. (C) Confocal micrographs of N. benthamiana leaf cells expressing N- or C-terminally YFP-fused SGS3-NTD, SGS3-ZF, SGS3-XS, and SGS3-CC at 48 hpi. Bars, 50 μm.
To test if SGS3 also interacts with VPgs of other potyviruses, two additional VPgs, VPgs of SMV (VPgSMV) and TEV (VPgTEV), were cloned. Y2H and BiFC results showed that both VPgSMV and VPgTEV could interact with SGS3 (Fig. 2BIII through VI and D). These data suggest that the interaction of VPg and SGS3 might be evolutionarily conserved for potyviruses.
Mapping of the interaction domains of VPg and SGS3.
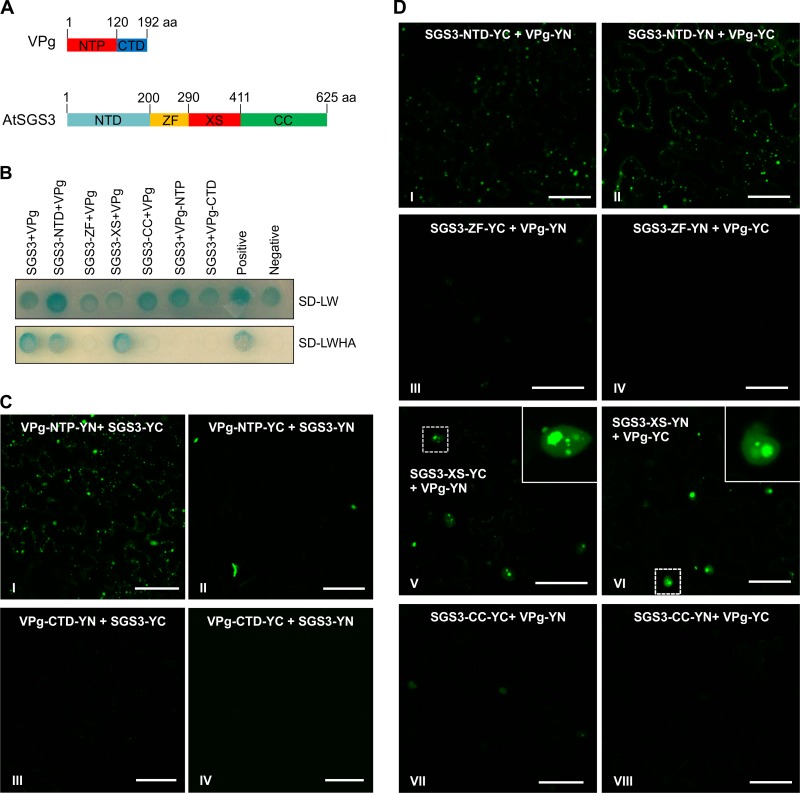
Based on the characteristics of several potyvirus VPg proteins (48–50), TuMV VPg was split into two portions, VPg-NTP (amino acids [aa] 1 to 120) and VPg-CTD (aa 121 to 192), containing the N-terminal NTP-binding domain and the C-terminal unstructured portion, respectively (Fig. 4A). Similarly to the full-length VPg protein (Fig. 3AI and II), VPg-NTP was localized predominantly to the nucleus, especially the nucleolus (Fig. 3BI and II). In contrast, the fluorescence of VPg-CTD was found mainly in the cytoplasm (Fig. 3BIII and IV). These results suggest that the nuclear localization of TuMV VPg is determined mainly by the N-terminal domain. In the Y2H assay, no interaction between SGS3 and the N terminus of VPg (VPg-NTP) or the C-terminal region of VPg (VPg-CTD) was found (Fig. 4B). BiFC analysis showed an inconsistent interaction between VPg-NTP and SGS3 (Fig. 4CI and II) and no interaction between VPg-CTD and SGS3 (Fig. 4CIII and IV). These data suggest that the entire TuMV VPg protein may be necessary for its robust interaction with SGS3.
FIG 4.
Mapping of the interaction domains in VPg and SGS3. (A) Schematic representations of SGS3 and VPg proteins. NTP, VPg N-terminal NTP-binding domain; CTD, VPg C-terminal domain; NTD, SGS3 N-terminal domain; ZF, putative SGS3 zinc finger domain; XS, SGS3 XS domain; CC, SGS3 coiled-coil domain. The numbers represent amino acid positions of domain boundaries. (B) Protein-protein interaction assays of BD-SGS3 and AD-VPg-NTP or AD-VPg-CTD and BD-VPg and AD-SGS3, AD-eIF4e, and AD-SGS3-NTD with AD-SGS3-ZF, AD-SGS3-XS, or AD-SGS3-CC in yeast cells. (C) BiFC assay of possible interactions between SGS3 and each of the two VPg domains in N. benthamiana leaf cells at 48 hpi. Bars, 50 μm. (D) BiFC assay of possible interactions between VPg and each of the four SGS3 domains in N. benthamiana leaf cells at 48 hpi. Bars, 50 μm.
To map the VPg-interacting domain(s) in SGS3, a series of truncated SGS3 mutations were constructed: SGS3-NTD (aa 1 to 200), SGS3-ZF (aa 201 to 289), SGS3-XS (aa 290 to 411), and SGS3-CC (aa 412 to 625), which contain the N-terminal domain (NTD), the zinc finger (ZF) domain, the rice gene X and SGS3 (XS) domain, and the C-terminal coiled-coil (CC) domain, respectively (Fig. 4A). SGS3-NTD was found mainly in the cytoplasm as granule-like foci (Fig. 3CI and II), whereas SGS3-ZF was localized mostly to the nucleus (Fig. 3CIII and IV). SGS3-XS was present in the cytoplasm and nucleus as diffuse fluorescence (Fig. 3CV and VI). SGS3-CC was localized to the nucleus and cytoplasm (Fig. 3CVII and VIII). Y2H analysis revealed that VPg interacted with SGS3-NTD or SGS3-XS but not with SGS3-ZF or SGS3-CC (Fig. 4B). Consistent with these results, SGS3-NTD or SGS3-XS, rather than SGS3-ZF or SGS3-CC, reconstructed the YFP signal when coexpressed with TuMV VPg in the BiFC assay (Fig. 4D). Therefore, both Y2H and BiFC assays mapped the VPg-interacting domains to two distant regions, i.e., SGS3-NTD and SGS3-XS. Interestingly, although the SGS3-XS domain is present mainly in the cytoplasm (Fig. 3B), the BiFC signal from its interaction with VPg was found mostly in the nucleus (Fig. 4DV and VI), suggesting that there is a specific motif in SGS3 or a mechanism to prevent its trafficking to the nucleus.
Transient expression of VPg induces the degradation of SGS3 and its intimate partner RDR6.
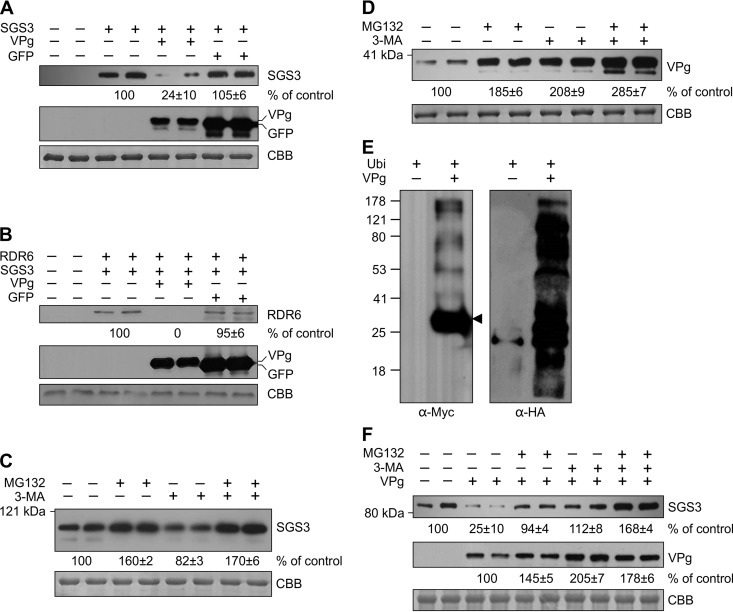
As VPg is the only viral protein that interacts with SGS3, we speculated that VPg mediates the reduction of SGS3 under TuMV infection. We thus tested this assumption by transiently expressing FLAG-4×Myc-SGS3 alone or together with VPg-3× hemagglutinin (3×HA) or GFP-3×HA in N. benthamiana leaves. Western blotting with anti-Myc antibodies at 48 hpi showed that the level of the SGS3 protein was greatly reduced when it was coexpressed with VPg-3×HA (Fig. 5A, top). In contrast, no reduction of SGS3 levels was found when it was coexpressed with GFP-3×HA (Fig. 5A, top).
FIG 5.
VPg-mediated degradation of SGS3. (A) FLAG-4×Myc-SGS3 was transiently coexpressed with VPg-3×HA or GFP-3×HA in N. benthamiana leaf cells. (Top and middle) Total protein was extracted at 48 hpi and detected with Myc antibodies (top) or HA antibodies (middle). The relative amount of the SGS3 protein is indicated. (Bottom) Equal protein loading by CBB staining. (B) FLAG-4×Myc-RDR6 was transiently coexpressed with VPg-3×HA and SGS3-CFP in N. benthamiana leaves. Infiltrated leaf areas were harvested for protein extraction at 60 hpi. (Top) RDR6 was purified with anti-FLAG M2 affinity agarose and detected with Myc antibodies. Numbers indicate the relative amounts of the RDR6 protein. VPg and GFP were directly detected with HA antibodies. (Bottom) Equal protein loading as determined by CBB staining. (C and D) FLAG-4×Myc-SGS3 (C) or FLAG-4×Myc-VPg (D) was transiently expressed in N. benthamiana leaves by Agrobacterium infiltration. At 48 hpi, the infiltrated leaf areas were treated with MG132, 3-MA, or MG132 and 3-MA for an additional 12 h. Total protein was extracted, separated by SDS-PAGE, and detected with Myc antibodies. The relative amounts of SGS3 or VPg are indicated. The bottom panel shows equal protein loading by CBB staining. (E) FLAG-4×Myc-VPg was transiently coexpressed with HA-Ub in N. benthamiana leaves by Agrobacterium infiltration. VPg was purified at 60 hpi with anti-FLAG M2 affinity agarose, separated by SDS-PAGE, and then detected with anti-Myc antibodies or anti-HA antibodies. (F) FLAG-4×Myc-VPg was transiently coexpressed with FLAG-4×Myc-SGS3 in N. benthamiana leaves. At 48 hpi, the infiltrated leaf areas were treated with MG132, 3-MA, or MG132 and 3-MA for an additional 12 h. Total protein was extracted, separated by SDS-PAGE, and detected with Myc antibodies. The relative amount of SGS3 or VPg is indicated. The bottom panel shows equal protein loading as determined by CBB staining. Each experiment was repeated at least three times.
It is well known that SGS3 forms heterogeneous complexes with RDR6 (10). To test if VPg further affects RDR6 stability, FLAG-4×Myc-RDR6 was expressed alone or together with VPg-3×HA in N. benthamiana leaves via Agrobacterium infiltration. C-terminally CFP-tagged SGS3 (SGS3-CFP) was included in both combinations to ensure VPg-SGS3-RDR6 tripartite interactions. FLAG-4×Myc-RDR6 and VPg-3×HA were detected with polyclonal antibodies against c-Myc and HA, respectively, at 60 hpi. Clearly, the level of RDR6 was hardly detectable when it was coexpressed with VPg and SGS3 but was not significantly affected when it was coexpressed with VPg and GFP (Fig. 5B). These data suggest that VPg can also induce RDR6 degradation and that VPg-mediated RDR6 degradation requires SGS3.
VPg-mediated degradation of SGS3 occurs via the 20S proteasome and autophagy pathways.
To explore the pathways that are recruited for VPg-mediated SGS3 degradation, we examined the 20S proteasome and autophagy pathways. Carbobenzoxy-l-leucyl-l-leucyl-l-leucinal (MG132) and 3-methyladenine (3-MA) are the drugs that specifically inhibit the 20S proteasome and autophagy pathways, respectively (62, 63). The protein level of FLAG-4×Myc-SGS3 was increased by 60% in N. benthamiana leaf tissues treated with MG132 but reduced by 18% in leaf tissues treated with 3-MA (Fig. 5C), suggesting that the turnover of SGS3 is dependent mainly on the ubiquitin (Ub)-proteasome pathway, and treatment with 3-MA can slightly stimulate the ubiquitin-proteasome pathway. Either MG132 or 3-MA treatment significantly increased the level of FLAG-4×Myc-VPg (Fig. 5D), suggesting that VPg may be targeted by both the 20S proteasome and autophagy pathways for degradation. To further confirm whether VPg is targeted by the 20S proteasome pathway, we expressed FLAG-4×Myc-VPg together with N-terminally HA-tagged Ub in N. benthamiana leaves. At 48 hpi, the VPg protein was purified with agarose beads covalently linked with a monoclonal antibody against the FLAG tag and then detected by a polyclonal antibody against Myc and HA. Besides a major band corresponding to the FLAG-4×Myc-VPg monomer, several additional bands with higher molecular weights were detected by anti-Myc antibodies (Fig. 5E). Consistently, HA signals were detected in the protein extract from leaf tissues coexpressing FLAG-4×Myc-VPg and HA-Ub, whereas only a weak signal could be detected in the protein extract from the mock-treated leaves (Fig. 5E). Therefore, the 20S proteasome pathway indeed targets VPg. We further tested the effects of the two drugs on VPg or SGS3 accumulation in N. benthamiana leaves coexpressing SGS3 and VPg. As shown in Fig. 5F, treatment with either MG132 or 3-MA compromised the degradation of not only VPg but also SGS3. Taken together, these results suggest that VPg-mediated degradation of SGS3 likely occurs through both the ubiquitin-proteasome and autophagy pathways.
VPg fails to completely degrade SGS3.
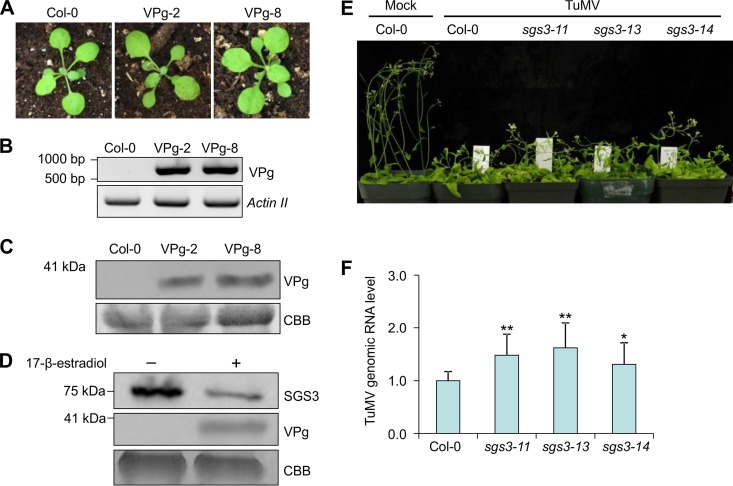
Since the TuMV VPg protein can cause SGS3 degradation, we wondered if transgenic Arabidopsis plants expressing TuMV VPg could mimic the phenotype of SGS3 knockout mutants. Transgenic plants expressing FLAG-4×Myc-VPg under the control of the 35S promoter were generated (Fig. 6A). Two independent transgenic lines (VPg-2 and VPg-8) showing the highest expression levels of VPg as evaluated by RT-PCR and Western blotting were selected for subsequent study (Fig. 6B and C). VPg-2 and VPg-8 did not exhibit discernible development phenotypes different from that of wild-type Col-0 plants (Fig. 6A). We also evaluated the levels of primary transcripts of ta-siRNA targets in transgenic plants, including Auxin response transcription factor 3 (ARF3) (locus tag At2g33860), ARF4 (At5g60450), Heat-induced TAS1 target 1 (HTT1) (At4g29770), and HTT2 (At5g18040). Theoretically, the overaccumulation of primary transcripts of ta-siRNA targets should be observed when the ta-siRNA biogenesis pathway is completely impaired (9). No significant differences in primary transcript levels of ARF3, ARF4, HTT1, or HTT2 between wild-type and transgenic Arabidopsis plants overexpressing VPg were detected (data not shown), implying that the ectopic expression of VPg did not completely degrade endogenous SGS3. To further confirm this assumption, we generated transgenic sgs3-14/na::SGS3-FLAG/XVE::FLAG-VPg plants by transforming homological sgs3-14/na::SGS3-FLAG plants with N-terminally FLAG-tagged VPg under the control of an estrogen-inducible promoter (XVE::FLAG-VPg). The expression of FLAG-VPg in sgs3-14/na::SGS3-FLAG/XVE::FLAG-VPg plants was detected after treatment with 2 μM 17-β-estradiol for 16 h (Fig. 6D, middle). The SGS3 protein was enriched with anti-FLAG M2 affinity agarose and detected by using rabbit anti-FLAG antibodies. SGS3 was detectable in sgs3-14/na::SGS3-FLAG/XVE::FLAG-VPg plants either treated with 17-β-estradiol or not (Fig. 6D, top). The amount of SGS3 was largely reduced upon the addition of 17-β-estradiol compared to the sample from the nontreatment control (Fig. 6D, top). These results further confirmed that VPg is unable to completely degrade endogenous SGS3.
FIG 6.
Knockout of SGS3 promotes TuMV replication. (A) Phenotype of 3-week-old VPg transgenic Arabidopsis plants overexpressing VPg. (B) Detection of VPg transcripts in transgenic plants by RT-PCR. The full coding sequence of VPg was amplified by RT-PCR. A fragment of the Arabidopsis Actin II gene was amplified as an RNA loading control. (C) Western blotting of protein extracts from transgenic plants overexpressing VPg with Myc antibodies. The bottom panel shows equal protein loading as determined by CBB staining. (D, top) Comparison of SGS3 in sgs3-14/na::SGS3-FLAG/XVE::FLAG-VPg plants after treatment with 17-β-estradiol for 16 h with rabbit antibodies against FLAG. (Middle) Expression of VPg monitored by Western blotting with anti-FLAG antibody. (Bottom) Equal protein loading as determined by CBB staining. (E) Phenotypes of TuMV-infected wild-type and SGS3 knockout Arabidopsis plants at 20 dpi. (F) Relative viral genomic RNA levels in TuMV-infected wild-type and SGS3 knockout Arabidopsis plants at 20 dpi. Actin II was used as an internal control. The mean genomic RNA level of TuMV in Col-0 plants was normalized to 1.0. Bars represent means ± standard deviations of data from 10 biological replicates, each containing three technical repeats. ** indicates a P value of <0.01, and * indicates a P value of <0.1 (Student t test).
The influence of SGS3 on TuMV infectivity was evaluated previously by Northern blotting using an SGS3 knockout mutant (sgs3-1) (11). To quantify viral genomic RNA accumulation and compare the levels of TuMV accumulation in wild-type and sgs3 mutant plants, we inoculated 4-week-old wild-type and SGS3 knockout (sgs3-11, sgs3-13, and sgs3-14) plants with TuMV-GFP by sap inoculation. PCR and sequencing confirmed that sgs3-11 plants contain a G-to-A mutation at nt 222, which disrupts the splicing of SGS3 transcripts, whereas sgs3-13 plants have a T-DNA insertion in the third exon. At 20 dpi, both wild-type and SGS3 knockout plants developed typical TuMV symptoms, as described above (Fig. 1D), and no obvious differences between TuMV-infected wild-type and SGS3 knockout plants were observed (Fig. 6E). qRT-PCR analysis showed that all three SGS3 knockout lines accumulated higher levels of viral RNA than did wild-type plants (Fig. 6F). Statistical analysis showed that the genomic RNA level of TuMV in SGS3 knockout plants was significantly higher than that in wild-type plants. These results suggest that knockout of SGS3 promotes TuMV infection.
DISCUSSION
The RDR6/SGS3 complex plays essential roles in antiviral immunity in virus-infected plants (9–11, 64, 65). Several viral proteins, e.g., PlAMV TGB1, TYLCV V2, SPCSV RNase 3, and RSV P2, have been found to target SGS3 to enhance host susceptibility (21–24). These VSRs have evolved distinct mechanisms to inhibit SGS3 functions. For example, TYLCV V2 inhibits SGS3 RNA binding activity by directly competing for SGS3 substrates (66), whereas PlAMV TGB1 directly sequesters SGS3/RDR6 bodies by coaggregation with them (23). In this work, we report that TuMV VPg interacts with SGS3 and mediates its degradation. This conclusion is supported by several findings in this study: (i) infection of TuMV or expression of replication-defective TuMV leads to the degradation of SGS3 (Fig. 1A), (ii) the VPg protein is the only viral protein that interacts with SGS3 (Fig. 2A), and (iii) VPg alone is sufficient to mediate the degradation of SGS3 and RDR6 (in the presence of SGS3) (Fig. 5A and B). These data provide new insights into the molecular mechanism by which VPg functions as a VSR.
In a recent study, Rajamäki et al. discovered that PVA VPg interacts with SGS3 of potato and Arabidopsis, and this interaction appears beneficial for PVA infection (60). Here, we further show that SGS3 of Arabidopsis interacts not only with TuMV VPg but also VPgs of two additional potyviruses tested, e.g., TEV and SMV. These four potyviruses are diverse in their nucleotide and amino acid sequences and host range. This addresses the possibility that the VPg-SGS3 interaction is a general feature of potyviruses. Thus, VPg-mediated degradation of SGS3 and its RNA silencing functional partner RDR6 might also be a conserved event during potyvirus infection.
In this study, treatment with MG132 or 3-MA inhibited the degradation of VPg (Fig. 5D) or VPg-mediated SGS3 degradation (Fig. 5F), suggesting that the degradation of VPg or the VPg-SGS3 complex occurs via both the ubiquitin-proteasome and autophagy pathways. In TuMV-infected cells, VPg is present either as a soluble form (free VPg or 6-kDa protein 2 [6K2]-free intermediate precursors) or as a membrane protein (VPg precursors containing 6K2) (67, 68). Thus, the ubiquitin-proteasome and autophagy pathways may target VPg-SGS3 complexes at different subcellular locations. Consistent with data from this study, both the ubiquitination and autophagy pathways have been implicated in viral infections in previous studies (2, 69).
Although VPg is localized predominantly to the nucleus, our data showed that the VPg-SGS3 interaction takes place in the cytoplasm (Fig. 2B), where SGS3/RDR6 bodies are normally located. Deletion analyses suggest that VPg interacts with both the NTD and the XS domain of SGS3. The XS domain of SGS is required for RNA binding activity (66, 70). Thus, our data address the possibility that VPg may also function by inhibiting the binding of SGS3 to target RNA.
Several plant viruses, e.g., TuMV, RSV, and Citrus tristeza virus (CTV), encode more than one VSR, which enables them to target more than one RNA silencing component to compromise host RNA silencing (21, 71, 72). In addition to VPg (34), the majority of potyviruses encode a strong VSR, HC-Pro (32, 33). HC-Pro apparently functions through the sequestration of virus-derived siRNAs (37, 38, 43–46). VPg seems to play a minor role in suppressing host RNA silencing during TuMV infection (34). A recent study showed that silencing of SGS3 reduces the amount of PVA RNA only slightly (60). In this study, we found that VPg is unable to completely eliminate SGS3 in the transient-expression assay, in transgenic plants, and in TuMV-infected plants (Fig. 1); that transgenic plants expressing VPg could not induce phenotypes that mimic those of SGS3 knockout mutants (Fig. 6A); and that knockout of SGS3 slightly promotes TuMV genomic RNA accumulation. During TuMV infection, RDR1, rather than RDR6, plays a major role in secondary viral siRNA biogenesis in Arabidopsis (73). Perhaps this explains why VPg is a less competent VSR in potyvirus infection and why silencing of SGS3, the partner of RDR6, only slightly affects TuMV and PVA infections. However, knockdown of RDR6 in N. benthamiana causes hyperaccumulation of PPV and PVY genomic RNAs (74, 75). This discrepancy suggests that the RDR6/SGS3 complex affects potyvirus infections depending on a particular virus. It is well known that different potyviruses may selectively recruit different isoforms of a host factor, e.g., eIF4E, to support their infection (2, 76). It is possible that the respective complex of RDR1 and RDR6 may selectively target different potyviruses.
Recently, HC-Pro has been shown to induce cytoplasmic RNA granules that contain the host proteins acidic ribosomal protein P0, AGO1, oligouridylate binding protein 1 (UBP1), varicose (VCS), and eIF(iso)4E (77). These cytoplasmic RNA granules are associated with HC-Pro VSR activity since point mutations that compromise the VSR activity of HC-Pro concomitantly abolish its ability to generate RNA granules (77). These HC-Pro-induced cytoplasmic RNA granules are downregulated by VPg (77). Thus, there is clearly an interplay between the two VSRs. It would be interesting to test if and how these two VSRs coordinate their functional roles in suppressing RNA silencing in potyvirus infections.
MATERIALS AND METHODS
Plant materials.
All Arabidopsis thaliana and Nicotiana benthamiana plants were grown in pots at 23°C in a growth chamber under a 16-h/8-h photoperiod with 60% humidity. The sgs3-11 (ABRC stock number CS24289), sgs3-13 (SALK_039005), and sgs3-14 (SALK_001394) mutants were obtained from the Arabidopsis Biological Resource Center (ABRC) at The Ohio State University. Screening for homozygous-mutation lines was performed essentially as described previously (78, 79). Arabidopsis (ecotype Col-0) plants were transformed by the floral-dip method (80). Transformants were screened by direct spraying of solutions containing 20 mg/liter glufosinate-ammonium or 1/2× Murashige & Skoog (MS) medium supplied with 15 μg/ml hygromycin B and then further confirmed by PCR.
Plasmid constructs.
All plasmids used in this work were constructed by using Gateway technology (Invitrogen, Burlington, Ontario, Canada). TuMV genes were amplified by using the TuMV-GFP infectious clone (GenBank accession no. NC_002509) as the template. SGS3 (At5g23570) and its truncated mutants were amplified by using cDNA extracted from wild-type Arabidopsis Col-0 plants. All PCRs were performed with Phusion DNA polymerase (NEB, Pickering, Ontario, Canada). The amplified fragments were inserted into the pDONR221 or pENTR/D-TOPO entry vector (Invitrogen) and confirmed by DNA sequencing.
The entry clones with the DNA fragment of interest were then inserted into the corresponding gateway-compatible vectors by recombination with Gateway LR Clonase enzyme mix (Invitrogen). In brief, pEarleyGate-101 and -102 (81), pGWB554, and pGWB514 (82) were used to produce C-terminally YFP-, CFP-, mRFP-, or 3×HA-tagged fusion constructs. The pEarleyGate-104 or pBA-FLAG-4×Myc-DC vector (83) was used to yield N-terminally YFP- or FLAG-4×Myc-tagged constructs. Plasmids p35S-YN gateway and p35S-YC gateway (84), which also contain N-terminal HA and FLAG tags, respectively, were used to construct C-terminal YN and YC fusion constructs. pGBKT7-472 DEST and pGADT7-DEST (84) were used to construct N-terminal GAL4 activation domain (AD) or GAL4 DNA binding domain (BD) fusion constructs. pMDC7 (85) was used for the generation of the 17-β-estradiol-inducible expression vector.
To complement sgs3 mutants under the control of the SGS3 native promoter, a 1,956-bp fragment upstream of the SGS3 coding sequence was amplified, fused to the SGS3 coding sequence, and then inserted into the pENTR/D-TOPO entry vector to construct pENTR-na::SGS3. The resulting vector was recombined into gateway-compatible pEarleyGate-302 (81) to generate plasmid na::SGS3-FLAG.
Confocal microscopy.
Fully expanded leaves from 3-week-old N. benthamiana plants were infiltrated with A. tumefaciens strain GV3101 containing proper constructs at an optical density at 600 nm (OD600) of 0.2. At 2 days postagroinfiltration (dpa), the infiltrated leaf area was visualized with a Leica TCS SP2 confocal laser scanning microscope (Leica, Germany) as described previously (86). The sequential mode was used when more than one fluorescence protein was expressed.
Yeast two-hybrid assay.
A Y2H assay was performed as described previously (87). Yeast expression plasmids were introduced into yeast strain AH109 (Clontech) by lithium acetate-mediated transformation. Transformed yeast cells were plated onto selection medium lacking Trp and Leu, and putative transformants were transferred to a high-stringency selection medium lacking Trp, Leu, His, and Ade.
Protein work.
N. benthamiana or Arabidopsis leaf tissues were sampled with a hole puncher (0.8 cm in diameter). Three leaf disks from different leaves were ground into fine powder in liquid nitrogen, mixed with 40 μl protein extraction buffer (50 mM Tris-HCl [pH 7.5], 0.15 M NaCl, 1 mM EDTA, 0.1% Tween 20, 10% glycerol, 0.1% phenylmethylsulfonyl fluoride [PMSF]) (88), and vermixed for 1 min. The crude lysate was clarified by centrifugation at 12,000 × g for 1 min at 4°C. The protein concentration in the supernatant was determined by using a Bio-Rad protein assay. An equal amount of total protein was aliquoted, brought to 40 μl with protein extraction buffer, and then mixed with 10 μl 5× SDS-PAGE loading buffer. After being boiled for 5 min at 99°C, the total protein lysate was quick-chilled on ice for 2 min, centrifuged at 12,000 × g for 2 min at 4°C, and stored at −80°C until use or used directly for SDS-PAGE.
For affinity purification, about 1 g of N. benthamiana or Arabidopsis leaf tissues was ground into fine powder in liquid nitrogen and then dissolved in 5 ml protein extraction buffer. The crude lysate was clarified by passage through a double layer of Miracloth (EMD Millipore Ltd., Etobicoke, Ontario, Canada) and centrifugation at 5,000 × g for 5 min at 4°C. Twenty microliters of anti-FLAG M2 affinity agarose (Sigma-Aldrich, Oakville, Ontario, Canada) or c-Myc agarose affinity gel (Sigma-Aldrich) was added to the supernatant to enrich the Myc- or FLAG-tagged target protein by rotating incubation for 2 h at 4°C. After being washed three times with protein extraction buffer, the agarose beads were mixed with 5× SDS-PAGE loading buffer and boiled for 5 min at 99°C to release target proteins for SGS-PAGE.
Total protein was separated on 10% or 8 to 16% Mini-Protean TGX precast gels (Bio-Rad) and transferred onto a polyvinylidene fluoride membrane by using a Trans-Blot SD semidry transfer cell (Bio-Rad Laboratories Ltd., Mississauga, Ontario, Canada). The membrane was blocked by using TBST (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, and 0.05% Tween 20) with 5% nonfat dry milk at room temperature for 2 h and incubated with rabbit anti-HA (Sigma-Aldrich) at a 1:5,000 dilution, goat anti-SGS3 (aR-15; Santa Cruz Biotechnology Inc., Dallas, TX, USA) at 1:500, the rabbit anti-GFP N-terminal domain (Sigma-Aldrich) at a 1:5,000 dilution, mouse anti-FLAG tag (Sigma-Aldrich) at a 1:10,000 dilution, rabbit anti-FLAG tag (Sigma-Aldrich) at a 1:5,000 dilution, or rabbit anti-Myc (Abcam Inc., Toronto, Ontario, Canada) at a 1:5,000 dilution for 2 h at room temperature or overnight at 4°C. After being washed six times with TBST, the membranes were incubated with the proper secondary peroxidase antibodies (Sigma-Aldrich) for 2 h at room temperature. After washing with TBST, the membranes were visualized by using an Immobilon Western chemiluminescent horseradish peroxidase (HRP) substrate (Millipore) according to the manufacturer's instructions. In each experiment, a parallel gel was stained with Coomassie brilliant blue (CBB) to monitor equal loading of the samples. Relative quantification of proteins was done by densitometric analysis using GelAnalyzer 2010 software (http://www.gelanalyzer.com/) according to the instructions provided. All Western blot analyses were repeated at least three times.
RNA extraction and real-time quantitative RT-PCR.
Total RNA was isolated from Arabidopsis leaf tissues by using the RNeasy Plant minikit (Qiagen Inc., Toronto, Ontario, Canada) according to the supplier's instructions. First-strand cDNA synthesis was carried out with an oligo(dT)12–18 primer using Superscript III reverse transcriptase (Invitrogen). RT-PCR and real-time qRT-PCR were performed essentially as described previously (89). For qRT-PCR to determine viral RNA accumulation and SGS3 transcript levels, a 20-μl volume containing 4 μl of 10-fold-diluted cDNA, 5 μM each primer, and 1× SYBR green PCR mix (Bio-Rad) was used. The relative transcript abundance was estimated by using Bio-Rad CFX Manager software (Bio-Rad). SGS3 transcripts were quantified by using the primer set SGS33-RTF (5′ -CCAGCAGCAGCAGAACATTA-3′) and SGS3-RTR (5′-TCCTCGTGATGCCTCTTCTT-3′). The genomic RNA level of TuMV was determined by amplification of a 257-bp coat protein (CP) gene fragment using the primer set TuMV-CPrtF (5′-CAGGTTTGACAGACGAGCAA-3′) and TuMV-CPrtR (5′-CCAGAGGTTCCAGCGTTTAC-3′). Arabidopsis Actin II gene (At3g18780) transcripts, amplified with primers Actin2-rtF (5′-CCGGTATTGTGCTGGATTCT-3′) and Actin2-rtR (5′-TTCTCGATGGAAGAGCTGGT-3′), were used as an internal control. Except where otherwise stated, each treatment contained five biological repeats, each with three technical repeats. All experiments were repeated at least three times.
ACKNOWLEDGMENTS
We are grateful to Jamie McNeil for technical support and Alex Molnar for photography and artwork.
This work was funded in part by an A-base grant from Agriculture and Agri-Food Canada (AAFC) and a discovery grant from the Natural Sciences and Engineering Research Council of Canada (NSERC) to A.W.
REFERENCES
- 1.García JA, Pallás V. 2015. Viral factors involved in plant pathogenesis. Curr Opin Virol 11:21–30. doi: 10.1016/j.coviro.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 2.Wang A. 2015. Dissecting the molecular network of virus-plant interactions: the complex roles of host factors. Annu Rev Phytopathol 53:45–66. doi: 10.1146/annurev-phyto-080614-120001. [DOI] [PubMed] [Google Scholar]
- 3.Ivanov KI, Eskelin K, Lõhmus A, Mäkinen K. 2014. Molecular and cellular mechanisms underlying potyvirus infection. J Gen Virol 95:1415–1429. doi: 10.1099/vir.0.064220-0. [DOI] [PubMed] [Google Scholar]
- 4.Mlotshwa S, Pruss GJ, Vance V. 2008. Small RNAs in viral infection and host defense. Trends Plant Sci 13:375–382. doi: 10.1016/j.tplants.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 5.Ding SW. 2010. RNA-based antiviral immunity. Nat Rev Immunol 10:632–644. doi: 10.1038/nri2824. [DOI] [PubMed] [Google Scholar]
- 6.Csorba T, Burgyán J. 2016. Antiviral silencing and suppression of gene silencing in plants, p 1–33. In Wang A, Zhou X (ed), Current research topics in plant virology. Springer International Publishing, Cham, Switzerland. doi: 10.1007/978-3-319-32919-2_1. [DOI] [Google Scholar]
- 7.Baulcombe D. 2004. RNA silencing in plants. Nature 431:356–363. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- 8.Weiberg A, Jin H. 2015. Small RNAs—the secret agents in the plant-pathogen interactions. Curr Opin Plant Biol 26:87–94. doi: 10.1016/j.pbi.2015.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS. 2004. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev 18:2368–2379. doi: 10.1101/gad.1231804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumakura N, Takeda A, Fujioka Y, Motose H, Takano R, Watanabe Y. 2009. SGS3 and RDR6 interact and colocalize in cytoplasmic SGS3/RDR6-bodies. FEBS Lett 583:1261–1266. doi: 10.1016/j.febslet.2009.03.055. [DOI] [PubMed] [Google Scholar]
- 11.Mourrain P, Béclin C, Elmayan T, Feuerbach F, Godon C, Morel JB, Jouette D, Lacombe AM, Nikic S, Picault N, Rémoué K, Sanial M, Vo TA, Vaucheret H. 2000. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101:533–542. doi: 10.1016/S0092-8674(00)80863-6. [DOI] [PubMed] [Google Scholar]
- 12.Yoshikawa M, Iki T, Tsutsui Y, Miyashita K, Poethig RS, Habu Y, Ishikawa M. 2013. 3′ fragment of miR173-programmed RISC-cleaved RNA is protected from degradation in a complex with RISC and SGS3. Proc Natl Acad Sci U S A 110:4117–4122. doi: 10.1073/pnas.1217050110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang XB, Jovel J, Udomporn P, Wang Y, Wu Q, Li WX, Gasciolli V, Vaucheret H, Ding SW. 2011. The 21-nucleotide, but not 22-nucleotide, viral secondary small interfering RNAs direct potent antiviral defense by two cooperative argonautes in Arabidopsis thaliana. Plant Cell 23:1625–1638. doi: 10.1105/tpc.110.082305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qu F, Ye X, Hou G, Sato S, Clemente TE, Morris TJ. 2005. RDR6 has a broad-spectrum but temperature-dependent antiviral defense role in Nicotiana benthamiana. J Virol 79:15209–15217. doi: 10.1128/JVI.79.24.15209-15217.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qu F, Ye X, Morris TJ. 2008. Arabidopsis DRB4, AGO1, AGO7, and RDR6 participate in a DCL4-initiated antiviral RNA silencing pathway negatively regulated by DCL1. Proc Natl Acad Sci U S A 105:14732–14737. doi: 10.1073/pnas.0805760105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carbonell A, Fahlgren N, Garcia-Ruiz H, Gilbert KB, Montgomery TA, Nguyen T, Cuperus JT, Carrington JC. 2012. Functional analysis of three Arabidopsis ARGONAUTES using slicer-defective mutants. Plant Cell 24:3613–3629. doi: 10.1105/tpc.112.099945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blevins T, Rajeswaran R, Shivaprasad PV, Beknazariants D, Si-Ammour A, Park HS, Vazquez F, Robertson D, Meins F Jr, Hohn T, Pooggin MM. 2006. Four plant Dicers mediate viral small RNA biogenesis and DNA virus induced silencing. Nucleic Acids Res 34:6233–6246. doi: 10.1093/nar/gkl886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levy A, Dafny-Yelin M, Tzfira T. 2008. Attacking the defenders: plant viruses fight back. Trends Microbiol 16:194–197. doi: 10.1016/j.tim.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 19.Burgyán J, Havelda Z. 2011. Viral suppressors of RNA silencing. Trends Plant Sci 16:265–272. doi: 10.1016/j.tplants.2011.02.010. [DOI] [PubMed] [Google Scholar]
- 20.Wu Q, Wang X, Ding SW. 2010. Viral suppressors of RNA-based viral immunity: host targets. Cell Host Microbe 8:12–15. doi: 10.1016/j.chom.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Du Z, Xiao D, Wu J, Jia D, Yuan Z, Liu Y, Hu L, Han Z, Wei T, Lin Q, Wu Z, Xie L. 2011. p2 of Rice stripe virus (RSV) interacts with OsSGS3 and is a silencing suppressor. Mol Plant Pathol 12:808–814. doi: 10.1111/j.1364-3703.2011.00716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glick E, Zrachya A, Levy Y, Mett A, Gidoni D, Belausov E, Citovsky V, Gafni Y. 2008. Interaction with host SGS3 is required for suppression of RNA silencing by Tomato yellow leaf curl virus V2 protein. Proc Natl Acad Sci U S A 105:157–161. doi: 10.1073/pnas.0709036105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okano Y, Senshu H, Hashimoto M, Neriya Y, Netsu O, Minato N, Yoshida T, Maejima K, Oshima K, Komatsu K, Yamaji Y, Namba S. 2014. In planta recognition of a double-stranded RNA synthesis protein complex by a potexviral RNA silencing suppressor. Plant Cell 26:2168–2183. doi: 10.1105/tpc.113.120535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weinheimer I, Haikonen T, Ala-Poikela M, Moser M, Streng J, Rajamäki ML, Valkonen JPT. 2016. Viral RNase3 co-localizes and interacts with the antiviral defense protein SGS3 in plant cells. PLoS One 11:e0159080. doi: 10.1371/journal.pone.0159080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chung BY, Miller WA, Atkins JF, Firth AE. 2008. An overlapping essential gene in the Potyviridae. Proc Natl Acad Sci U S A 105:5897–5902. doi: 10.1073/pnas.0800468105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodamilans B, Valli A, Mingot A, San León D, Baulcombe D, López-Moya JJ, García JA. 2015. RNA polymerase slippage as a mechanism for the production of frameshift gene products in plant viruses of the Potyviridae family. J Virol 89:6965–6967. doi: 10.1128/JVI.00337-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olspert A, Chung BY, Atkins JF, Carr JP, Firth AE. 2015. Transcriptional slippage in the positive-sense RNA virus family Potyviridae. EMBO Rep 16:995–1004. doi: 10.15252/embr.201540509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Revers F, García JA. 2015. Molecular biology of potyviruses. Adv Virus Res 92:101–199. doi: 10.1016/bs.aivir.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 29.White KA. 2015. The polymerase slips and PIPO exists. EMBO Rep 16:885–886. doi: 10.15252/embr.201540871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mingota A, Vallib A, Rodamilansc B, Leónc DS, Baulcombeb DC, Garcíac JA, López-Moyaa JJ. 2016. The P1N-PISPO trans-frame gene of Sweet potato feathery mottle potyvirus is produced during virus infection and functions as an RNA silencing suppressor. J Virol 90:3543–3557. doi: 10.1128/JVI.02360-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Untiveros M, Olspert A, Artola K, Firth AE, Kreuze JF, Valkonen JPT. 2016. A novel sweet potato potyvirus open reading frame (ORF) is expressed via polymerase slippage and suppresses RNA silencing. Mol Plant Pathol 17:1111–1123. doi: 10.1111/mpp.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anandalakshmi R, Pruss GJ, Ge X, Marathe R, Mallory AC, Smith TH, Vance VB. 1998. A viral suppressor of gene silencing in plants. Proc Natl Acad Sci U S A 95:13079–13084. doi: 10.1073/pnas.95.22.13079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kasschau KD, Carrington JC. 1998. A counterdefensive strategy of plant viruses: suppression of posttranscriptional gene silencing. Cell 95:461–470. doi: 10.1016/S0092-8674(00)81614-1. [DOI] [PubMed] [Google Scholar]
- 34.Rajamäki M-L, Valkonen JPT. 2009. Control of nuclear and nucleolar localization of nuclear inclusion protein a of picorna-like Potato virus A in Nicotiana species. Plant Cell 21:2485–2502. doi: 10.1105/tpc.108.064147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ivanov KI, Eskelin K, Bašić M, De S, Lõhmus A, Varjosalo M, Mäkinen K. 2016. Molecular insights into the function of the viral RNA silencing suppressor HcPro. Plant J 85:30–45. doi: 10.1111/tpj.13088. [DOI] [PubMed] [Google Scholar]
- 36.Ballut L, Drucker M, Pugnière M, Cambon F, Blanc S, Roquet F, Candresse T, Schmid H-P, Nicolas P, Gall OL, Badaoui S. 2005. HcPro, a multifunctional protein encoded by a plant RNA virus, targets the 20S proteasome and affects its enzymic activities. J Gen Virol 86:2595–2603. doi: 10.1099/vir.0.81107-0. [DOI] [PubMed] [Google Scholar]
- 37.Kasschau KD, Xie Z, Allen E, Llave C, Chapman EJ, Krizan KA, Carrington JC. 2003. P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev Cell 4:205–217. doi: 10.1016/S1534-5807(03)00025-X. [DOI] [PubMed] [Google Scholar]
- 38.Shiboleth YM, Haronsky E, Leibman D, Arazi T, Wassenegger M, Whitham SA, Gaba V, Gal-On A. 2007. The conserved FRNK box in HC-Pro, a plant viral suppressor of gene silencing, is required for small RNA binding and mediates symptom development. J Virol 81:13135–13148. doi: 10.1128/JVI.01031-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Endres MW, Gregory BD, Gao Z, Foreman AW, Mlotshwa S, Ge X, Pruss GJ, Ecker JR, Bowman LH, Vance V. 2010. Two plant viral suppressors of silencing require the ethylene-inducible host transcription factor RAV2 to block RNA silencing. PLoS Pathog 6:e1000729. doi: 10.1371/journal.ppat.1000729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ala-Poikela M, Goytia E, Haikonen T, Rajamäki ML, Valkonen JP. 2011. Helper component proteinase of the genus Potyvirus is an interaction partner of translation initiation factors eIF(iso)4E and eIF4E and contains a 4E binding motif. J Virol 85:6784–6794. doi: 10.1128/JVI.00485-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anandalakshmi R, Marathe R, Ge X, Herr JM Jr, Mau C, Mallory A, Pruss G, Bowman L, Vance VB. 2000. A calmodulin-related protein that suppresses posttranscriptional gene silencing in plants. Science 290:142–144. doi: 10.1126/science.290.5489.142. [DOI] [PubMed] [Google Scholar]
- 42.Soitamo AJ, Jada B, Lehto K. 2011. HC-Pro silencing suppressor significantly alters the gene expression profile in tobacco leaves and flowers. BMC Plant Biol 11:68. doi: 10.1186/1471-2229-11-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schott G, Mari-Ordonez A, Himber C, Alioua A, Voinnet O, Dunoyer P. 2012. Differential effects of viral silencing suppressors on siRNA and miRNA loading support the existence of two distinct cellular pools of ARGONAUTE1. EMBO J 31:2553–2565. doi: 10.1038/emboj.2012.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chapman EJ, Prokhnevsky AI, Gopinath K, Dolja VV, Carrington JC. 2004. Viral RNA silencing suppressors inhibit the microRNA pathway at an intermediate step. Genes Dev 18:1179–1186. doi: 10.1101/gad.1201204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lakatos L, Csorba T, Pantaleo V, Chapman EJ, Carrington JC, Liu YP, Dolja VV, Calvino LF, López-Moya JJ, Burgyán J. 2006. Small RNA binding is a common strategy to suppress RNA silencing by several viral suppressors. EMBO J 25:2768–2780. doi: 10.1038/sj.emboj.7601164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mallory AC, Reinhart BJ, Bartel D, Vance VB, Bowman LH. 2002. A viral suppressor of RNA silencing differentially regulates the accumulation of short interfering RNAs and micro-RNAs in tobacco. Proc Natl Acad Sci U S A 99:15228–15233. doi: 10.1073/pnas.232434999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murphy JF, Klein PG, Hunt AG, Shaw JG. 1996. Replacement of the tyrosine residue that links a potyviral VPg to the viral RNA is lethal. Virology 220:535–538. doi: 10.1006/viro.1996.0344. [DOI] [PubMed] [Google Scholar]
- 48.Rantalainen KI, Eskelin K, Tompa P, Mäkinen K. 2011. Structural flexibility allows the functional diversity of potyvirus genome-linked protein VPg. J Virol 85:2449–2457. doi: 10.1128/JVI.02051-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rantalainen KI, Uversky VN, Permi P, Kalkkinen N, Dunker AK, Mäkinen K. 2008. Potato virus A genome-linked protein VPg is an intrinsically disordered molten globule-like protein with a hydrophobic core. Virology 377:280–288. doi: 10.1016/j.virol.2008.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grzela R, Szolajska E, Ebel C, Madern D, Favier A, Wojtal I, Zagorski W, Chroboczek J. 2008. Virulence factor of Potato virus Y, genome-attached terminal protein VPg, is a highly disordered protein. J Biol Chem 283:213–221. doi: 10.1074/jbc.M705666200. [DOI] [PubMed] [Google Scholar]
- 51.Hébrard E, Bessin Y, Michon T, Longhi S, Uversky VN, Delalande F, Van Dorsselaer A, Romero P, Walter J, Declerck N, Fargette D. 2009. Intrinsic disorder in viral proteins genome-linked: experimental and predictive analyses. Virol J 6:23. doi: 10.1186/1743-422X-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiang J, Laliberté JF. 2011. The genome-linked protein VPg of plant viruses—a protein with many partners. Curr Opin Virol 1:347–354. doi: 10.1016/j.coviro.2011.09.010. [DOI] [PubMed] [Google Scholar]
- 53.Elena SF, Rodrigo G. 2012. Towards an integrated molecular model of plant-virus interactions. Curr Opin Virol 2:719–724. doi: 10.1016/j.coviro.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 54.Lellis AD, Kasschau KD, Whitham SA, Carrington JC. 2002. Loss-of-susceptibility mutants of Arabidopsis thaliana reveal an essential role for eIF(iso)4E during potyvirus infection. Curr Biol 12:1046–1051. doi: 10.1016/S0960-9822(02)00898-9. [DOI] [PubMed] [Google Scholar]
- 55.Huang TS, Wei T, Laliberté JF, Wang A. 2010. A host RNA helicase-like protein, AtRH8, interacts with the potyviral genome-linked protein, VPg, associates with the virus accumulation complex, and is essential for infection. Plant Physiol 152:255–266. doi: 10.1104/pp.109.147983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Freire M. 2014. Potyviral VPg and HC-Pro proteins and the cellular translation initiation factor eIF(iso)4E interact with exoribonuclease Rrp6 and a small α-heat shock protein. Plant Mol Biol Rep 32:596–604. doi: 10.1007/s11105-013-0670-4. [DOI] [Google Scholar]
- 57.Dunoyer P, Thomas C, Harrison S, Revers F, Maule A. 2004. A cysteine-rich plant protein potentiates potyvirus movement through an interaction with the virus genome-linked protein VPg. J Virol 78:2301–2309. doi: 10.1128/JVI.78.5.2301-2309.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dufresne PJ, Ubalijoro E, Fortin MG, Laliberté JF. 2008. Arabidopsis thaliana class II poly(A)-binding proteins are required for efficient multiplication of turnip mosaic virus. J Gen Virol 89:2339–2348. doi: 10.1099/vir.0.2008/002139-0. [DOI] [PubMed] [Google Scholar]
- 59.Dufresne PJ, Thivierge K, Cotton S, Beauchemin C, Ide C, Ubalijoro E, Laliberté JF, Fortin MG. 2008. Heat shock 70 protein interaction with Turnip mosaic virus RNA-dependent RNA polymerase within virus-induced membrane vesicles. Virology 374:217–227. doi: 10.1016/j.virol.2007.12.014. [DOI] [PubMed] [Google Scholar]
- 60.Rajamäki ML, Streng J, Valkonen JP. 2014. Silencing suppressor protein VPg of a potyvirus interacts with the plant silencing-related protein SGS3. Mol Plant Microbe Interact 27:1199–1210. doi: 10.1094/MPMI-04-14-0109-R. [DOI] [PubMed] [Google Scholar]
- 61.Cheng X, Deng P, Cui H, Wang A. 2015. Visualizing double-stranded RNA distribution and dynamics in living cells by dsRNA binding-dependent fluorescence complementation. Virology 485:439–451. doi: 10.1016/j.virol.2015.08.023. [DOI] [PubMed] [Google Scholar]
- 62.Goldberg AL. 2012. Development of proteasome inhibitors as research tools and cancer drugs. J Cell Biol 199:583–588. doi: 10.1083/jcb.201210077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Seglen PO, Gordon PB. 1982. 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc Natl Acad Sci U S A 79:1889–1892. doi: 10.1073/pnas.79.6.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dalmay T, Hamilton A, Rudd S, Angell S, Baulcombe DC. 2000. An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 101:543–553. doi: 10.1016/S0092-8674(00)80864-8. [DOI] [PubMed] [Google Scholar]
- 65.Muangsan N, Beclin C, Vaucheret H, Robertson D. 2004. Geminivirus VIGS of endogenous genes requires SGS2/SDE1 and SGS3 and defines a new branch in the genetic pathway for silencing in plants. Plant J 38:1004–1014. doi: 10.1111/j.1365-313X.2004.02103.x. [DOI] [PubMed] [Google Scholar]
- 66.Fukunaga R, Doudna JA. 2009. dsRNA with 5′ overhangs contributes to endogenous and antiviral RNA silencing pathways in plants. EMBO J 28:545–555. doi: 10.1038/emboj.2009.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Beauchemin C, Boutet N, Laliberté JF. 2007. Visualization of the interaction between the precursors of VPg, the viral protein linked to the genome of turnip mosaic virus, and the translation eukaryotic initiation factor iso 4E in planta. J Virol 81:775–782. doi: 10.1128/JVI.01277-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Restrepo-Hartwig MA, Carrington JC. 1992. Regulation of nuclear transport of a plant potyvirus protein by autoproteolysis. J Virol 66:5662–5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bird SW, Kirkegaard K, Agbandje-McKenna M, Freed EO. 2014. The ins and outs of viral infection: keystone meeting review. Viruses 6:3652–3662. doi: 10.3390/v6093652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang D, Trudeau VL. 2008. The XS domain of a plant specific SGS3 protein adopts a unique RNA recognition motif (RRM) fold. Cell Cycle 7:2268–2270. doi: 10.4161/cc.7.14.6306. [DOI] [PubMed] [Google Scholar]
- 71.Xiong R, Wu J, Zhou Y, Zhou X. 2009. Characterization and subcellular localization of an RNA silencing suppressor encoded by Rice stripe tenuivirus. Virology 387:29–40. doi: 10.1016/j.virol.2009.01.045. [DOI] [PubMed] [Google Scholar]
- 72.Lu R, Folimonov A, Shintaku M, Li W-X, Falk BW, Dawson WO, Ding S-W. 2004. Three distinct suppressors of RNA silencing encoded by a 20-kb viral RNA genome. Proc Natl Acad Sci U S A 101:15742–15747. doi: 10.1073/pnas.0404940101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Garcia-Ruiz H, Takeda A, Chapman EJ, Sullivan CM, Fahlgren N, Brempelis KJ, Carrington JC. 2010. Arabidopsis RNA-dependent RNA polymerases and dicer-like proteins in antiviral defense and small interfering RNA biogenesis during turnip mosaic virus infection. Plant Cell 22:481–496. doi: 10.1105/tpc.109.073056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schwach F, Vaistij FE, Jones L, Baulcombe DC. 2005. An RNA-dependent RNA polymerase prevents meristem invasion by potato virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiol 138:1842–1852. doi: 10.1104/pp.105.063537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vaistij FE, Jones L. 2009. Compromised virus-induced gene silencing in RDR6-deficient plants. Plant Physiol 149:1399–1407. doi: 10.1104/pp.108.132688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang A, Krishnaswamy S. 2012. Eukaryotic translation initiation factor 4E-mediated recessive resistance to plant viruses and its utility in crop improvement. Mol Plant Pathol 13:795–803. doi: 10.1111/j.1364-3703.2012.00791.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hafrén A, Lõhmus A, Mäkinen K. 2015. Formation of Potato virus A-induced RNA granules and viral translation are interrelated processes required for optimal virus accumulation. PLoS Pathog 11:e1005314. doi: 10.1371/journal.ppat.1005314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wei T, Zhang C, Hou X, Sanfaçon H, Wang A. 2013. The SNARE protein Syp71 is essential for turnip mosaic virus infection by mediating fusion of virus-induced vesicles with chloroplasts. PLoS Pathog 9:e1003378. doi: 10.1371/journal.ppat.1003378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang L, Chen H, Brandizzi F, Verchot J, Wang A. 2015. The UPR branch IRE1-bZIP60 in plants plays an essential role in viral infection and is complementary to the only UPR pathway in yeast. PLoS Genet 11:e1005164. doi: 10.1371/journal.pgen.1005164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 81.Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS. 2006. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45:616–629. doi: 10.1111/j.1365-313X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- 82.Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T. 2007. Development of series of Gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104:34–41. doi: 10.1263/jbb.104.34. [DOI] [PubMed] [Google Scholar]
- 83.Zhu H, Hu F, Wang R, Zhou X, Sze S-H, Liou LW, Barefoot A, Dickman M, Zhang X. 2011. Arabidopsis argonaute10 specifically sequesters miR166/165 to regulate shoot apical meristem development. Cell 145:242–256. doi: 10.1016/j.cell.2011.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lu Q, Tang X, Tian G, Wang F, Liu K, Nguyen V, Kohalmi SE, Keller WA, Tsang EWT, Harada JJ, Rothstein SJ, Cui Y. 2010. Arabidopsis homolog of the yeast TREX-2 mRNA export complex: components and anchoring nucleoporin. Plant J 61:259–270. doi: 10.1111/j.1365-313X.2009.04048.x. [DOI] [PubMed] [Google Scholar]
- 85.Curtis MD, Grossniklaus U. 2003. A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133:462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wei T, Zhang C, Hong J, Xiong R, Kasschau KD, Zhou X, Carrington JC, Wang A. 2010. Formation of complexes at plasmodesmata for potyvirus intercellular movement is mediated by the viral protein P3N-PIPO. PLoS Pathog 6:e1000962. doi: 10.1371/journal.ppat.1000962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xiong R, Wang A. 2013. SCE1, the SUMO-conjugating enzyme in plants that interacts with NIb, the RNA-dependent RNA polymerase of Turnip mosaic virus, is required for viral infection. J Virol 87:4704–4715. doi: 10.1128/JVI.02828-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Leister RT, Dahlbeck D, Day B, Li Y, Chesnokova O, Staskawicz BJ. 2005. Molecular genetic evidence for the role of SGT1 in the intramolecular complementation of Bs2 protein activity in Nicotiana benthamiana. Plant Cell 17:1268–1278. doi: 10.1105/tpc.104.029637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cui H, Wang A. 2016. The plum pox virus 6K1 protein is required for viral replication and targets the viral replication complex at the early infection stage. J Virol 90:5119–5131. doi: 10.1128/JVI.00024-16. [DOI] [PMC free article] [PubMed] [Google Scholar]